Abstract
Genetic variations in CYP3A4, CYP3A5, and m-TOR could contribute to interpatient variability regarding m-TOR inhibitors pharmacokinetics or cellular effects. The purpose of this study was to evaluate the influence of selected candidate variations in these genes on everolimus pharmacokinetics, efficacy, and toxicity in cancer patients. Thirty-four patients receiving everolimus for breast (n = 22) or renal (n = 10) cancers, or neuroendocrine tumors of pancreatic origin (n = 2) were included in the study. Six variants in genes related to everolimus pharmacokinetics (CYP3A4*22 and CYP3A5*3) or pharmacodynamics (m-TOR rs2295079, rs2295080, rs2024627 and rs1057079) were genotyped. Associations with trough concentrations (C0), dose reductions, or treatment interruptions due to toxicity and progression-free survival were investigated using generalized estimating equations and Cox models. CYP3A5 nonexpressers had significantly higher C0 as compared with expressers (βGG vs AG = + 6.32 ± 2.22 ng/mL, p = 0.004). m-TOR rs2024627 was significantly associated with an increased risk of cancer progression studied alone or as part of an haplotype (T vs C: HR = 2.60, 95% CI [1.16–5.80], p = 0.020; CTCG vs other haplotypes HR = 2.29, 95% CI [1.06–4.95], p = 0.035, respectively). This study showed that CYP3A5 expression impacts everolimus pharmacokinetics in cancer patients and identified a genetic variation in m-TOR associated with the risk of cancer progression.
Similar content being viewed by others
Introduction
The m-TOR pathway is deregulated in many types of human cancers, which explains why everolimus (EVR) is used in oncology [1]. The efficacy of EVR has been demonstrated in the treatment of hormone receptor positive and HER2 negative advanced breast cancer [2], neuroendocrine tumors of pancreatic origin [3, 4] and metastatic renal cell cancer [5].
Variability in blood exposure or clinical response to EVR involves several factors, including genetic factors and drug–drug interactions. EVR is metabolized by CYP3A4 and CYP3A5 in the gut and liver with a stronger contribution of CYP3A4 as compared with CYP3A5 found in vitro [6]. The CYP3A4*22 (rs35599367) is a deficient allele associated to a decreased metabolism [7], while the CYP3A5*3 (rs7767465) allele causes a splicing defect that leads to a truncated protein with no enzymatic activity [8]. However, the majority of studies carried out in organ transplantation (kidney, heart, or lung) do not support an effect of CYP3A4*22 [9,10,11] or CYP3A5*3 [6, 9, 11,12,13,14] variants on the pharmacokinetics (PK) of EVR. Only one study in heart transplant recipients showed that carriers of a CYP3A5*1 allele required a higher dose of EVR to reach the targeted C0 (p = 0.041) [15] compared with patients with the CYP3A5*3/*3 genotype. In oncology, a study showed that CYP3A4*22 carriers had higher blood EVR concentrations than wild-type patients (p = 0.019) [16], while no influence of CYP3A5 genotype was shown. CYP3A4*22 and CYP3A5*3 variations have also been studied together as a metabolic status based on the functionality of the combination [17].
On the other hand, polymorphisms in genes of the m-TOR pathway may be responsible for variations in EVR efficacy or associated with the occurrence of adverse events [16, 18]. Three single-nucleotide variations (SNV) in FRAP1 (m-TOR gene) exhibited an adequate level of recommendation for research according to a classification proposed in organ transplantation (rs2295080, rs2024627, and rs1057079) [19] and they are in strong linkage disequilibrium with another m-TOR variant: rs2295079.
The aim of this study was to investigate the effect of CYP3A4*22 and CYP3A5*3 variants on EVR blood concentrations as well as the influence of the four abovementioned SNV candidates in m-TOR on EVR toxicity and efficacy in cancer patients.
Materials/subjects and methods
Patients
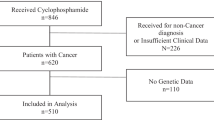
This monocentric retrospective observational study was performed in 34 patients receiving EVR for the treatment of breast cancer (n = 22), renal cancer (n = 10), or neuroendocrine tumors of pancreatic origin (n = 2). Patients had been receiving EVR for at least 15 days and were thus at the PK steady-state. This study was part of a previously published investigation on the relationship between exposure and effect for EVR in oncology. The present study is the pharmacogenetics investigation of the patients included in the previous study with available DNA, as no pharmacogenetics study had been conducted before on this cohort [20]. The main result of the previous study was to report EVR thresholds, for toxicity (C0 > 26.3 ng/ml was associated with an increased risk of toxicity: HR = 4.12, 95% CI [1.48–11.5], p = 0.007) and for efficacy (C0 < 11.9 ng/ml was associated with an increased risk of progression: HR = 3.2, 95% CI [1.33–7.81], p = 0.001) [20]).
This study was declared to the National Commission on Computer Technology and Freedom (CNIL) under the number 1948009v0 and approved by the Hospital Ethics Committee under the number 198-2016-12. All 34 patients enrolled signed an informed consent before treatment with EVR. Blood samples were collected in a biological collection declared to the competent authorities (DC 2010-1074).
Data collected from patient medical records
Clinical, biological, and radiologic data were collected from patients’ medical records. Adverse events were graded using the National Cancer Institute Common Terminology Criteria for Adverse Events, version 4.3. The efficacy was defined as progression-free survival (PFS) corresponding to the time elapsed between EVR initiation and tumor progression or death from any cause and toxicity was defined as the termination, temporary interruption, and/or dose reduction of EVR.
The clinical data included in the analysis were collected between the date of cancer diagnosis and the end of the study (25-04-2016): demographic characteristics (age, sex, type of cancer, date of cancer diagnosis based on a biopsy); number of metastases and localization; cancer treatment (adjuvant hormonotherapy (HT) or chemotherapy (CT), number of HT lines in metastatic situation, and number of CT lines or target therapies lines in metastatic situation prior to EVR treatment, use of a concomitant treatment with biphosphonates; EVR treatment (date of introduction of EVR, initial dose (mg), EVR dose at the time of C0 measurement, C0 threshold determined in the previous study [20], date and reason for EVR discontinuation, temporary interruption, and/or dose reduction).
The biological data collected were: hemoglobin levels, platelet, white cell, and neutrophil counts; albumin, glycaemia, triglycerides, and cholesterol levels; glomerular filtration rate calculated according to the Modification of Diet in Renal Disease equation, transaminases, alkaline phosphatase levels.
Pharmacogenetic analyses
Patients’ DNA was extracted and purified from EDTA-treated blood using the mini Qiagen blood kit (Qiagen, Courtaboeuf, France) and quantified by spectrophotometry on a nanodrop 2000c® (Thermo Scientific™). Genotyping was performed with 10 ng of DNA using Life Technologies validated Taqman® allelic discrimination (CYP3A4*22 (rs35599367): C_59013445_10, CYP3A5*3 (rs7767465): C_26201809_30), rs2295079: C_16189145_10, rs2295080: C_16189146_10, rs1057079: C_8862305_1 and rs2024627: C_11647371_10). TaqMan discrimination assays were performed using a Rotor Gene Q® (Qiagen) for m-TOR SNV or an ABI PRISM 7500 Sequence Detection System (Life Technologies) for CYP3A SNV. CYP3A4 and CYP3A5 were gathered depending on their functionality in a metabolic status covariate with three categories: poor metabolizers (PM) corresponding to CYP3A4*22 and CYP3A5*3, intermediate metabolizers (IM) corresponding to CYP3A4*22 and CYP3A5*1 or CYP3A4 *1 and CYP3A5*3, and extensive metabolizers (EM) corresponding to CYP3A4*1 and CYP3A5*1 [11] (of note, we called the noncarrier of the variants, “*1”, in order to simplify the reading but no sequencing was performed and theoretically, “non CYP3A4*22” or “non CYP3A5*3 carrier” would have been more correct).
Statistical analyses
The statistical analyses were performed using the R software (version 3.4.3). Conformity to the Hardy–Weinberg equilibrium was checked. M-TOR haplotypes were inferred using the “haplo.stat” R package. Risk factors for toxicity were investigated using Cox models allowing taking into account repeated events. Progression-free survival was studied using the Cox model censured at the first event.
The influence of CYP3A4*22 and CYP3A5*3, studied independently and gathered as metabolic status, on EVR exposure was explored using a generalized estimation equation (“geepack” R package). Boxplots were drawn to represent the difference in C0 as function of the genotype or phenotype status (to draw the boxplot, several C0 values measured in a same patient were used, leading to a higher number of values than patients). Univariate analysis was performed first and variables characterized by a p value < 0.1 were included in an intermediate model. The final model was selected by a backward stepwise approach based on the Bayesian information criterion (BIC). The robustness of the results was assessed by 1000 bootstraps followed by 1000 backward stepwise. For Cox models, the proportionality of risks was assessed on the final model using the Schoenfeld residues and for the significant polymorphisms, time-to-event data were estimated using Kaplan–Meier analysis as function of genotype and groups were compared using the log-rank test for trend. An additive genetic model was used to investigate the influence of SNV and haplotype on outcomes.
Results
Patients and clinical data description
Characteristics of the 34 patients studied are presented in Table 1.
SNV allele frequencies and linkage disequilibrium
The frequencies of the m-TOR SNVs, CYP3A4*22, and CYP3A5*3 alleles were similar to those described for Caucasians in the 1000 Genomes project and were in conformity with the Hardy–Weinberg equilibrium (Table 2).
A significant linkage disequilibrium was found between the four m-TOR SNVs (D′ > 0.9, r2 > 0.8 two by two; supplemental Table 1). The D′ and r2 obtained in the present study were similar to those described in the 1000 Genomes project for Caucasians. Haplotype frequencies and their descriptions are presented in Supplemental Table 2.
Influence of CYP3A variants on EVR trough level
CYP3A4*22
Among the 34 patients enrolled in the study, 30 were noncarriers of the CYP3A4*22 allele (CC genotype; 162 C0 values) and four were heterozygous (CT genotype; 24 C0 values). No significant association between CYP3A4 genotype and EVR C0 was observed (intercept [genotype CC] = 15.97 ± 1.71 ng/mL, β genotype CT = 1.21 ± 2.51 ng/mL, p = 0.63) (Fig. 1a).
Boxplots representing EVR trough concentration as function of a CYP3A4*22 status, b CYP3A5*3 status (CYP3A5 expressors are patients who are carriers of at least one CYP3A5*1 allele, patient nonexpressors carry two CYP3A5*3 alleles), or c metabolic status for combined CYP3A4*22 and CYP3A5*3 status (poor metabolizers (PM) corresponding to CYP3A4*22 and CYP3A5*3, intermediate metabolizers (IM) corresponding to CYP3A4*22 and CYP3A5*1 or CYP3A4 *1 and CYP3A5*3, and extensive metabolizers (EM) corresponding to CYP3A4*1 and CYP3A5*1).
CYP3A5*3
Thirty patients were identified as CYP3A5 nonexpressers (CYP3A5*3/*3 GG genotype; 161 C0 values) and four patients as CYP3A5 expressers (CYP3A5*1/*3 AG genotype 25 C0 values). Patients with the CYP3A5*1/*3 genotype had significantly lower EVR C0 as compared with nonexpressers (intercept [genotype AG] = 10.72 ± 1.45 ng/mL, β[genotype GG] = 6.32 ± 2.22 ng/mL, p = 0.004) (Fig. 1b).
Metabolic status
Five patients were EM (25 C0 values), 25 IM (137 C0 values), and 4 PM (24 C0 values). Patients with the IM or PM phenotype had a significantly higher EVR C0 as compared with EM (intercept [EM phenotype] = 10.72 ± 1.45 ng/mL, β[IM phenotype] = 6.30 ± 2.43 ng/mL, p = 0.0095, β[PM phenotype] = 6.46 ± 2.35 ng/mL, p = 0.0059) (Fig. 1c).
Influence of m-TOR SNVs on EVR toxicity
No significant association was found between SNVs and the toxicity of EVR (Table 3).
Influence of m-TOR SNPs on the EVR efficacy (progression-free survival)
SNV study
rs2024627 was significantly associated with an increased risk of progression (T vs C: HR = 2.60, 95% CI [1.16–5.80], p = 0.020; Fig. 2). This result was confirmed by bootstrap analysis as rs2024627 was selected in 64.6% of the 1000 bootstrap models. Details of the results are presented in Table 4.
Haplotype study
The CTCG haplotype was significantly associated with an increased risk of progression in univariate analysis (CTCG vs other: HR = 2.30, 95% CI [1.09–4.89], p = 0.030) and in multivariate analysis (CTCG vs other: HR = 2.29, 95% CI [1.06–4.95], p = 0.035) after adjustment on the significant covariates (metastatic initially, EVR concentration threshold).
Discussion
We found that cancer patients with a CYP3A5 expresser status had decreased EVR C0 in comparison with nonexpressers and that a variant in the m-TOR gene (rs2024627) was associated with a decreased PFS.
Our study is one of the first investigating the influence of CYP3A genotypes on EVR PK. In metastatic breast cancer patients, Pascual et al. found that carriers of the deficient CYP3A4*22 allele had EVR dose adjusted C0 2.7 times higher than noncarrier patients (median 69.1 vs 25.7 ng/mL, respectively), while no significant association between CYP3A5 status and EVR concentration was found [16]. We could not confirm this association in the present study but we found that the CYP3A5*3 allele (i.e., the absence of functional CYP3A5 enzyme) is associated with increased EVR C0. This result was unexpected since an in vitro study using liver microsomes performed by our group showed that CYP3A5 might not contribute in a large extent to EVR metabolism in comparison with CYP3A4. Also, no significant association between CYP3A5*3 and the PK of EVR was found in renal transplant recipients [6]. In oncology, EVR is prescribed at doses 5–10 times higher than in transplantation. This might modify the relative contribution of CYP3A4 and 3A5 in these two different clinical settings. A saturation of EVR CYP3A4-medicated metabolism could occur at the doses used in oncology, resulting in a secondary involvement of CYP3A5. Nonetheless, the difference between our results and those reported by the Pascual et al. are difficult to explain. While the number of subjects investigated in our study and in Pascual et al. was similar, and the clinical outcomes studied comparable (blood exposure, toxicities and PFS), it has to be noted that the authors did not perform statistical corrections. As discussed by the authors themselves, part of the significant findings may be due to multiple testing. On the contrary, we chose to reduce the number of investigations (indeed, instead of studying every organ toxicity separately we only studied toxicity as one outcome) to prevent the risk of false findings. We also performed bootstrap analyses to assess the robustness of our results. However, even with bootstrap analyses and due to the quite small number of patients in both trials, we could not exclude that one of these finding was obtained by chance. Further studies are required to confirm our results and they should be carried out in a larger number of patients. Interestingly, we observed a phenotype effect when studying the metabolic status with an increase of the C0 in both IM and PM vs. EM. However, the values of the C0 regression coefficients observed were very similar to the ones in the CYP3A5 analysis, meaning that the effect of the metabolic status is probably mainly due to the CYP3A5.
We hypothesized that SNVs in the gene encoding the m-TOR protein, which is directly involved in the mechanism of action of EVR, could explain the difference in outcome in EVR-treated patients. Among the four SNVs we studied, a significant association was observed between rs2024627 (C>T) and PFS with a 2.5 increase in cancer progression in patient carriers of the variant allele. It can be hypothesized that this variant decreases m-TOR inhibition by EVR, as a study showed that homozygous carriers of rs2024627 variant had decreased m-TOR expression (p = 0.011) [21]. In addition, studies in oncology conducted on the rs2295080 (T>G) (which is in strong linkage disequilibrium with the rs2024627), showed that the presence of a T nucleotide was significantly associated with an increase in m-TOR mRNA expression in tumor tissues in a Chinese population [22, 23]. The authors suggested that rs2295080 would be a functional variant regulating the transcriptional activity and expression levels of m-TOR [22, 23]. In the present study, we observed an effect of the rs2024627 studied alone or included in a haplotype together with rs2295080 suggesting that rs2024627 could influence the transcriptional activity and expression of m-TOR. However, as the rs2295080 has been shown to exhibit the functional effect, we were waiting for an effect of this SNP more than the other ones constituting the haplotype. The only hypothesis would be that the effect observed came from the entire haplotype rather than one of the SNP (i.e., as it has been previously shown for ABCB1) [24].
We chose to investigate the effect of m-TOR genetic variants on indirect indicators of EVR toxicity (i.e., termination, temporary interruption and/or dose reduction of EVR) in order to avoid multiple analyses for each type of toxicities. No significant association was observed. Our study has some limitations. The cohort of 34 patients is relatively small but in order to decrease the risk of false finding, bootstrap analyses were performed on significant results as the effect of one covariate might be largely influenced by one single observation or patient. In conclusion, cancer patients with a CYP3A5 expresser status were found to have a decreased trough concentration of EVR in comparison with nonexpressers but the clinical relevance of this association is still questionable. An m-TOR SNV (rs2024627) was found to associate with decreased PFS. These results need to be confirmed in further studies.
Code availability
R code used in the present article is available under request.
References
Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol J Am Soc Clin Oncol. 2009;27:2278–87.
Baselga J, Campone M, Piccart M, Burris HA, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor–positive advanced breast cancer. N Engl J Med. 2012;366:520–9.
Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, Van Cutsem E, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:514–23.
Pavel ME, Hainsworth JD, Baudin E, Peeters M, Hörsch D, Winkler RE, et al. Everolimus plus octreotide long-acting repeatable for the treatment of advanced neuroendocrine tumours associated with carcinoid syndrome (RADIANT-2): a randomised, placebo-controlled, phase 3 study. Lancet Lond Engl. 2011;378:2005–12.
Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet Lond Engl. 2008;372:449–56.
Picard N, Rouguieg-Malki K, Kamar N, Rostaing L, Marquet P. CYP3A5 genotype does not influence everolimus in vitro metabolism and clinical pharmacokinetics in renal transplant recipients. Transplantation. 2011;91:652–6.
Wang D, Guo Y, Wrighton S, Cooke G, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11:274–86.
Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91.
Moes DJAR Swen JJ, den Hartigh J, van der Straaten T, van der Heide JJH, Sanders JS, et al. Effect of CYP3A4*22, CYP3A5*3, and CYP3A combined genotypes on cyclosporine, everolimus, and tacrolimus pharmacokinetics in renal transplantation. CPT Pharmacometrics Syst Pharm. 2014;3:e100.
Shipkova M, Hesselink DA, Holt DW, Billaud EM, van Gelder T, Kunicki PK, et al. Therapeutic drug monitoring of everolimus: a consensus report. Ther Drug Monit. 2016;38:143–69.
Woillard J-B, Chouchana L, Picard N, Loriot M-A. French network of pharmacogenetics (RNPGX). Pharmacogenetics of immunosuppressants: state of the art and clinical implementation—recommendations from the French National Network of Pharmacogenetics (RNPGx). Therapie. 2017;72:285–99.
Moes DJAR, Press RR, den Hartigh J, van der Straaten T, de Fijter JW, Guchelaar H-J. Population pharmacokinetics and pharmacogenetics of everolimus in renal transplant patients. Clin Pharmacokinet. 2012;51:467–80.
Kniepeiss D, Renner W, Trummer O, Wagner D, Wasler A, Khoschsorur GA, et al. The role of CYP3A5 genotypes in dose requirements of tacrolimus and everolimus after heart transplantation. Clin Transpl. 2011;25:146–50.
Schoeppler KE, Aquilante CL, Kiser TH, Fish DN, Zamora MR. The impact of genetic polymorphisms, diltiazem, and demographic variables on everolimus trough concentrations in lung transplant recipients. Clin Transpl. 2014;28:590–7.
Lesche D, Sigurdardottir V, Setoud R, Englberger L, Fiedler GM, Largiadèr CR, et al. Influence of CYP3A5 genetic variation on everolimus maintenance dosing after cardiac transplantation. Clin Transpl. 2015;29:1213–20.
Pascual T, Apellániz-Ruiz M, Pernaut C, Cueto-Felgueroso C, Villalba P, Álvarez C, et al. Polymorphisms associated with everolimus pharmacokinetics, toxicity and survival in metastatic breast cancer. PLoS ONE. 2017;12. https://doi.org/10.1371/journal.pone.0180192.
Elens L, Haufroid V. Genotype-based tacrolimus dosing guidelines: with or without CYP3A4*22? Pharmacogenomics. 2017;18:1473–80.
Woillard J-B, Kamar N, Rousseau A, Rostaing L, Marquet P, Picard N. Association of sirolimus adverse effects with m-TOR, p70S6K or Raptor polymorphisms in kidney transplant recipients. Pharmacogenet Genomics. 2012;22:725–32.
Pouché L, Stojanova J, Marquet P, Picard N. New challenges and promises in solid organ transplantation pharmacogenetics: the genetic variability of proteins involved in the pharmacodynamics of immunosuppressive drugs. Pharmacogenomics. 2016;17:277–96.
Deppenweiler M, Falkowski S, Saint-Marcoux F, Monchaud C, Picard N, Laroche M-L, et al. Towards therapeutic drug monitoring of everolimus in cancer? Results of an exploratory study of exposure-effect relationship. Pharm Res. 2017;121:138–44.
Slattery ML, Lundgreen A, Mullany LE, Penney RB, Wolff RK. Influence of CHIEF pathway genes on gene expression: a pathway approach to functionality. Int J Mol Epidemiol Genet. 2014;5:100–11.
Cao Q, Ju X, Li P, Meng X, Shao P, Cai H, et al. A functional variant in the MTOR promoter modulates its expression and is associated with renal cell cancer risk. PLoS ONE. 2012;7. https://doi.org/10.1371/journal.pone.0050302.
Zhao Y, Diao Y, Wang X, Lin S, Wang M, Kang H, et al. Impacts of the mTOR gene polymorphisms rs2536 and rs2295080 on breast cancer risk in the Chinese population. Oncotarget. 2016;7:58174–80.
Speidel JT, Xu M, Abdel-Rahman SZ. Differential effect of ABCB1 haplotypes on promoter activity. Pharmacogenet Genomics. 2018;28:69–77.
Acknowledgements
We are grateful to K. Poole for manuscript editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
SF receives funding from Novartis, Pfizer, Sanofi, BMS, Jansen, and Amgen; the other authors have no conflict of interest to declare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bonnet, S., Falkowski, S., Deppenweiler, M. et al. Effect of genetic polymorphisms in CYP3A4, CYP3A5, and m-TOR on everolimus blood exposure and clinical outcomes in cancer patients. Pharmacogenomics J 20, 647–654 (2020). https://doi.org/10.1038/s41397-020-0152-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-020-0152-7
- Springer Nature Limited