Abstract
Background
The site of prostate cancer metastasis is an important predictor of oncologic outcomes, however, the clinicogenomic characteristics associated with the site are not well-defined. Herein, we characterize the genomic alterations associated with the metastatic site of prostate cancer.
Methods
We analyzed clinical and genomic data from prostate cancer patients with metastatic disease and known metastatic sites from publicly available targeted sequencing data.
Results
Prostate cancer metastasis to the liver versus other sites of metastasis conferred a high hazard for death in patients with metastatic prostate cancer (HR: 3.96, 95% CI: 2.4–6.5, p < 0.0001). Genomic analysis of metastatic tissues of prostate cancer-specific genes demonstrated that liver metastases were more enriched with MYC amplification (29.5% vs. 9.8%, FDR = 0.001), PTEN deletion (42% vs. 20.8%, FDR = 0.005), and PIK3CB amplification (8.2% vs. 0.9, FDR = 0.005) compared to other sites. No point mutations were significantly associated with liver metastasis compared to other metastatic sites.
Conclusion
Liver metastases in prostate cancer are associated with poor survival and aggressive genomic features, including MYC-amplification, PTEN-deletion, and PIK3CB-amplification. These findings could have prognostic, treatment, and trial implications.
Similar content being viewed by others
Introduction
Metastatic prostate cancer is a heterogeneous disease with widely variable clinical outcomes. The site of prostate cancer metastasis is an important predictor of oncologic outcome and a likely source of heterogeneity [1]. Liver metastases may be a predictor of particularly adverse outcomes [1,2,3]. It also represents a subtype refractory to hormonal therapy and short-duration response to chemotherapy [2, 4], and is often associated with neuroendocrine characteristics [5]. Nevertheless, the genomic characteristics associated with prostatic liver metastasis are not well-defined. Here, we hypothesize that there are genomic alterations associated with prostatic liver metastasis that may explain their aggressiveness and lack of response to therapy.
Methods
We analyzed clinical (n = 280) and genomic (n = 608; 241 in common with clinical data) data from prostate cancer patients with metastatic disease across different sites (61 liver, 34 lungs, 231 LN, 125 bone, 157 “other sites”). The objective was to characterize the clinicogenomic features of prostatic liver metastasis [6, 7]. Genomic profiling and sequencing were conducted on the metastatic tissue using the MSK-IMPACT Next Generation Sequencing (NGS) assay (https://www.cbioportal.org/study/ clinicalData?id=prad_cdk12_mskcc_2020). More details on patients and sample processing of this cohort have been reported [7]. Genomic data for genes frequently altered in prostate cancer was obtained [8]. Cox proportional hazard analyses defined hazard ratios (HRs) and 95% confidence intervals (CIs) for overall mortality of liver metastases versus other sites of metastases, across seven different cancers from the MSK-IMPACT cohort. Survival data were calculated from the time of presenting with metastatic disease. X2 test was used for assessing associations between genomic alterations and metastatic sites. The Benjamini–Hochberg method controlled for false discovery rate (FDR).
Results
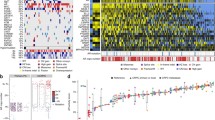
We characterized the clinical risk of liver metastasis across several cancers [6]. Metastasis to the liver versus other sites of metastasis conferred the highest hazard for death in patients with prostate cancer (HR: 3.96, 95% CI: 2.4–6.5, p < 0.0001), when compared across cancers (Fig. 1A). We further characterized the impact of site of metastasis on survival in a cohort of 280 patients with prostate adenocarcinoma, known metastasis site, and available data on time from metastasis diagnosis to death (Supplementary Table 1) [6].
A Hazard ratio for death for liver metastases in prostate [liver (33) vs. non-liver (247)], bladder [liver (13) vs. non-liver (83)], breast [liver (221) vs. non-liver (614)], colon [liver (292) vs. non-liver (197)], ovarian [liver (14) vs. non-liver (128)], non-small cell lung carcinoma [liver (83) vs. non-liver (589)], and melanoma [liver (40) vs. non-liver (233)]. B Overall survival (time from metastasis diagnosis to death) in patients with metastatic prostate cancer, by the site of disease. C Fraction of genome altered across the site of disease in metastatic prostate cancer. D Genomic alteration frequency across sites of metastasis. E Scatter plot of frequency, difference, and the ratio of genomic alterations in liver and non-liver metastatic sites. Abbreviations: L liver, NL non-liver, LN lymph node.
With a median survival of 13 months, patients with metastatic prostate cancer to the liver had worse 2-year overall survival (22%) compared with bone alone (57%), lung alone (54%), and lymph node (LN) metastases (63%) (p < 0.0001, Fig. 1B). We found no association between race (White vs. African American) and the site of prostate cancer metastases (p = 0.52). Prostate tumors metastasized to the liver were more enriched with small cell/neuroendocrine carcinoma (SC/NE) (15% for liver, 1% for bone, 0 for lung, and 3% for LN).
On sensitivity analysis, after adjusting for age, liver metastasis remained significantly associated with a higher hazard for death when compared with other metastatic sites (HR: 3.2, 95% CI: 1.74–5.9, p = 0.0001). Furthermore, when excluding patients with SC/NE, patients with metastatic prostate cancer to the liver continued to have a higher hazard for death compared with other sites of metastatic disease (HR: 3.85, 95% CI: 2.2–6.7, p < 0.0001). Information on treatment and other clinical characteristics were not available for further adjustment.
To define the genomic characteristics of metastatic prostate cancer to the liver, we analyzed targeted DNA sequencing data from a larger set of metastatic tissues of 608 unique patients with metastatic prostate adenocarcinoma and known metastasis site (61 liver, 34 lungs, 231 LN, 125 bone, 157 “other sites” [abdomen, adrenal gland, brain, bladder, colon, epidural, pelvis, soft tissue, and retroperitoneum]) [7]. Of note, there were 241 patients in common with the 280 patients used for survival analysis; the remaining samples had no survival data available. None of the samples used for genomic analyses were neuroendocrine or SC carcinoma.
Genomic alterations analysis demonstrated that prostatic liver metastases tissues were associated with a higher fraction of genomic alterations compared to bone (p < 0.0001), LN (p < 0.0001), lung (p = 0.0008), and other site metastases (p = 0.009 using Wilcoxon test) (Fig. 1C). In comparing the genomic alteration frequency of prostate cancer genes across sites of metastatic disease (Supplementary Table 2), we did not find any mutation (including mutations in TP53, FOXA1, SPOP, and AR) to be enriched in liver metastases (FDR > 0.2 for all, Supplementary Table 3).
When characterizing copy number alterations, we found prostate tumors metastasized to the liver to be enriched with AR-amplification (39% for liver vs 31% for bone, 31% for LN, 8.8% for lung and 28% for other sites group), MYC-amplication (29% for liver vs. 7.2% for bone, 10.4% for LN, 0 for lung and 13.3% for other sites group), PTEN-deletion (42.6% for liver vs. 16% for bone, 21% for LN, 26 for lung and 22% for other sites group, and PIK3CB-amplification (8.2% for liver vs. 1% for bone, 1.3% for LN, 0 for lung and 0.6% for other sites group) (Fig. 1D). When comparing liver metastases to all other metastatic sites, only MYC-amplification (29.5% vs. 9.8%, X2 FDR = 0.001), PTEN-deletion (42% vs. 20.8%, X2 FDR = 0.005), and PIK3CB-amplification (8.2% vs. 0.9%, X2 FDR = 0.005) were significantly associated with liver metastases after FDR correction (Fig. 1E and Supplementary Table 3). Notably, AR-amplification was not significantly associated with liver metastasis compared to other sites (39% vs. 30, p = 0.13). Furthermore, 62% of liver metastases had at least one of these alterations compared with 28% of metastases from all other sites (X2 p < 0.001).
Discussion and conclusion
To the best of our knowledge, the present study is the first to report these genomic associations with prostate cancer liver metastases and builds on existing literature that has suggested liver metastases are a poor prognostic feature but lack genomic profiling of tumor tissues. Our genomic analyses of the actual metastatic site revealed a unique signature of liver metastasis. Disease state (CRPC vs. HSPC) could be a confounding variable of our analyses. But, given that AR-amplification, a surrogate feature of CRPC, is equally enriched across metastatic sites, the observed genomic differences are likely driven by its tumor–host site. These findings could inform genomic risk prognostication, treatment decisions, and the design of future clinical trials of combined therapies for metastatic prostate cancer that may be molecularly distinct.
For example, patients with prostatic liver metastasis could benefit from selective inhibitors targeting MYC and PI3K pathways that are more activated in liver metastatic tissues. MYC is upregulating EZH2 [9], which is associated with prostate cancer progression [10]. Given this recent revelation, MYC-dependent PC may be particularly sensitive to treatment with selective EZH2 inhibitors, some of which are in clinical trials to treat other malignancies [11]. Similarly, amplification of PIK3CB and loss of PTEN in liver metastasis lead to activation of PI3K/AKT pathways. Thus, nominating this aggressive subgroup for PI3K inhibitors that are already used in other solid tumors [12].
Limitations to this study include not having access to certain clinical factors (Gleason score, disease volume, etc.) or prior treatment history of the patients included in the analyses, nevertheless, our survival analyses largely validate existing literature. Furthermore, liver metastasis is more likely accompanied by other sites of disease that may not have been profiled. Nevertheless, this study focused on comparing clinicogenomic characteristics of tissues from liver metastases to tissues from other metastases and defined liver metastases as an adverse feature, and therefore other sites of disease would be expected and would likely have little impact on the conclusions of our findings. Future work will be needed to determine the impact of prior treatment history, other clinical factors, and multiple sites of disease on the clinicogenomics of liver metastases.
References
Halabi S, Kelly WK, Ma H, Zhou H, Solomon NC, Fizazi K, et al. Meta-analysis evaluating the impact of site of metastasis on overall survival in men with castration-resistant prostate cancer. J Clin Oncol. 2016;34:1652–9.
Pond GR, Sonpavde G, De Wit R, Eisenberger MA, Tannock IF, Armstrong AJ. The prognostic importance of metastatic site in men with metastatic castration-resistant prostate cancer. Eur Urol. 2014;65:3–6.
Zhao F, Wang J, Chen M, Chen D, Ye S, Li X, et al. Sites of synchronous distant metastases and prognosis in prostate cancer patients with bone metastases at initial diagnosis: a population-based study of 16,643 patients. Clin Transl Med. 2019;8:30.
Singh A, Cheedella NKS, Shakil SA, Gulmi F, Kim DS, Wang JC. Liver metastases in prostate carcinoma represent a relatively aggressive subtype refractory to hormonal therapy and short-duration response to docetaxel monotherapy. Can J Addict Med. 2015;6:265–9.
Pouessel D, Gallet B, Bibeau F, Avancès C, Iborra F, Sénesse P, et al. Liver metastases in prostate carcinoma: Clinical characteristics and outcome. BJU Int. 2007;99:807–11.
Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Nguyen B, Mota JM, Nandakumar S, Stopsack KH, Weg E, Rathkopf D, et al. Pan-cancer Analysis of CDK12 alterations identifies a subset of prostate cancers with distinct genomic and clinical characteristics. Eur Urol. 2020. https://doi.org/10.1016/j.eururo.2020.03.024.
Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–57.
Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–83.
Labbé DP, Sweeney CJ, Brown M, Galbo P, Rosario S, Wadosky KM, et al. TOP2A and EZH2 provide early detection of an aggressive prostate cancer subgroup. Clin Cancer Res. 2017;23:7072–83.
Kim KH, Roberts CWM. Targeting EZH2 in cancer. Nat Med. 2016;22:128–34.
LoRusso PM. Inhibition of the PI3K/AKT/mTOR pathway in solid tumors. J Clin Oncol. 2016;34:3803–15.
Funding
Dr. Mahal is funded by the Prostate Cancer Foundation, American Society for Radiation Oncology, Department of Defense, and the Sylvester Comprehensive Cancer Center.
Author information
Authors and Affiliations
Contributions
MA, CS, IF, and BM conducted the analysis, drafted the paper, and contributed to revisions. MA and AP contributed to data acquisition and critical revision for important content. RV, RC, SP, SK, RD, AK, DS, JS, and ADP contributed to the conception of the study, data acquisition, and critical revision for important content. MA, CS, and BM have responsibility for the final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
Dr. Mahal is funded by the Prostate Cancer Foundation-American Society for Radiation Oncology Award to End Prostate Cancer (grant support). Dr. Franco is funded by an NIH NCI Center to Reduce Cancer Health Disparities Diversity Supplement, and ACRO Luther Brady Educational Award (grant support). Dr. Spratt has received personal fees: Janssen, Blue Earth, Boston Scientific, AstraZeneca, and funding from Janssen. The remaining authors declare that they have no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Alshalalfa, M., Seldon, C., Franco, I. et al. Clinicogenomic characterization of prostate cancer liver metastases. Prostate Cancer Prostatic Dis 25, 366–369 (2022). https://doi.org/10.1038/s41391-021-00486-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-021-00486-2
- Springer Nature Limited
This article is cited by
-
From biology to the clinic — exploring liver metastasis in prostate cancer
Nature Reviews Urology (2024)
-
Autophagy, a critical element in the aging male reproductive disorders and prostate cancer: a therapeutic point of view
Reproductive Biology and Endocrinology (2023)
-
Incidence and survival of castration-resistant prostate cancer patients with visceral metastases: results from the Dutch CAPRI-registry
Prostate Cancer and Prostatic Diseases (2023)