Abstract
Single-cell transcriptome sequencing (scRNA-seq) is a high-throughput technique used to study gene expression at the single-cell level. Clustering analysis is a commonly used method in scRNA-seq data analysis, helping researchers identify cell types and uncover interactions between cells. However, the choice of a robust similarity metric in the clustering procedure is still an open challenge due to the complex underlying structures of the data and the inherent noise in data acquisition. Here, we propose a deep clustering method for scRNA-seq data called scRISE (scRNA-seq Iterative Smoothing and self-supervised discriminative Embedding model) to resolve this challenge. The model consists of two main modules: an iterative smoothing module based on graph autoencoders designed to denoise the data and refine the pairwise similarity in turn to gradually incorporate cell structural features and enrich the data information; and a self-supervised discriminative embedding module with adaptive similarity threshold for partitioning samples into correct clusters. Our approach has shown improved quality of data representation and clustering on seventeen scRNA-seq datasets against a number of state-of-the-art deep learning clustering methods. Furthermore, utilizing the scRISE method in biological analysis against the HNSCC dataset has unveiled 62 informative genes, highlighting their potential roles as therapeutic targets and biomarkers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
The advent of single-cell RNA sequencing (scRNA-seq) has revolutionized our understanding of cellular diversity and gene regulation [1]. This innovative technique enables the measurement of gene expression in individual cells, providing unprecedented insights into the complexity of biological systems [2]. By analyzing the expression of thousands of genes in tens of thousands of cells in a single experiment, scRNA-seq has emerged as a powerful tool for detecting cell-to-cell variability, identifying rare cell populations, and inferring cell lineage relationships [3, 4]. In multicellular organisms, a critical challenge in scRNA-seq data analysis is to accurately characterize different cell types and their lineage relationships [5, 6]. To this end, cell clustering has become an indispensable step in scRNA-seq data analysis. By grouping cells with similar gene expression patterns, cell clustering can help identify cell types and subpopulations, thus revealing the cellular heterogeneity and diversity present within a biological system [7, 8].
Despite the enormous potential of scRNA-seq, it poses multiple challenges in data processing, including high dimensionality, technical noise, missing events, and batch effects. The continuous increase in detection range and cell number leads to a significant rise in data dimensionality, thereby presenting considerable computational analysis challenges [9]. Additionally, sample preparation and sequencing processes can introduce biases and noise. Most gene expression values in the gene expression matrix are zero, which may result from biological or technical factors [10]. Due to relatively low mRNA expression, insufficient capture efficiency, or low sequencing depth, many genes exhibit low expression levels in scRNA-seq data. These low expression values do not necessarily reflect actual gene expression loss but may result from technical limitations, known as the “dropout” phenomenon [11]. Furthermore, it is necessary to consider the impact of cellular stress responses and batch effects on the cell state [12].
Traditionally, clustering methods such as K-means, and hierarchical clustering, have been used for scRNA-seq data analysis. However, these methods have limitations in handling high-dimensional and noisy scRNA-seq data. Several algorithms have been developed to address these challenges and are specifically designed for scRNA-seq data analysis. For example, pcaReduce [13] uses principal component analysis (PCA) for dimension reduction and k-means clustering for cell clustering. CIDR [14] is a fast and efficient method that considers zero-inflated expression data and uses implicit interpolation for single-cell clustering. SIMLR [15] combines multiple kernels to learn sample similarities and performs spectral clustering. SC3 [16] uses consensus clustering and PCA for dimension reduction to cluster single cells.
The powerful representation-learning ability of deep learning has provided more accurate and comprehensive results for the clustering analysis of single-cell transcriptomic data. In recent years, several methods utilizing deep embedding techniques for single-cell clustering have emerged. DCA [17] combines autoencoders and a negative binomial distribution model to model the count data and learn effective low-dimensional representations. This embedding representation can be used for subsequent clustering analysis. scDeepCluster [18] adds a clustering layer to the DCA model and performs cell cluster assignment after the initial denoising stage. scziDesk [19] employs a denoising autoencoder to characterize scRNA-seq data and then constructs a self-training K-means algorithm to cluster cell populations. This method aims to overcome the limitations of traditional K-means clustering, such as sensitivity to noise and initialization. scVI [20] is a deep embedding method that uses a VAE. scVI probabilistically models single-cell data and learns the distribution of its latent space.
Although these deep embedding clustering methods have made significant progress, they still have limitations in overlooking the structural relationships between data samples. To solve the challenge, scGNN [21] uses a GNN that iteratively constructs a cell graph using a multimodal autoencoder, dynamically prunes the cell graph during the iterative process, and finally clusters the feature data containing graph structure information using the K-means algorithm. Luo et al. [22] proposed a model based on graph autoencoders (scGAE), which constructs a cellular graph and uses graph autoencoders to preserve the features and topological structure information of scRNA-seq data. scTAG [23] optimizes a topologically adaptive graph convolutional autoencoder, which processes node features using polynomial convolution to generate latent embeddings for soft assignment clustering. scDSC [24] consists of a GNN module and a ZINB-based autoencoder and achieves end-to-end training using a multi-module mutual supervision strategy.
Currently, many graph neural network-based clustering techniques rely on constructing a cell graph from the input data, and the clustering performance heavily depends on the quality of the graph. Our work introduces a novel clustering strategy for scRNA-seq data called scRISE, which uses an autoencoder to iteratively denoise the data (with Laplacian smoothing) and construct the cell-graph reliably, while in the meantime seamlessly incorporating cell graph information with a self-supervised discriminative embedding technique that allows identifying correct clusters through adaptively determined similarity threshold.
A distinctive feature of scRISE is its use of an iterative cycle-smoothing approach to achieve optimal clustering results during the data reconstruction phase. Through the application of a self-supervised discriminative embedding learning technique, scRISE guides the clustering of the reconstructed data, ensuring a more precise and insightful representation of the underlying cellular structures. Importantly, scRISE is versatile in correcting various types of noise and non-signal fluctuations, as it does not assume any specific form of data distribution. We conducted a comprehensive performance study, comparing scRISE with five state-of-the-art deep clustering techniques to assess its effectiveness in identifying meaningful clusters in the data. Our findings unequivocally demonstrate the superior benefits of scRISE for scRNA-seq data processing. Additionally, we showcase the powerful denoising capability of scRISE through visually compelling visualizations and dimensionality reduction studies, emphasizing its effectiveness in extracting biologically significant insights from noisy and complex single-cell transcriptome datasets.
Results
The framework of scRISE
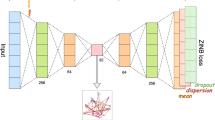
The scRISE method consists of two main modules, as shown in Fig. 1. Firstly, we use an iterative smoothing module based on graph autoencoder, which combines the autoencoder with Laplacian smoothing filters to smooth and reconstruct potentially noisy, incomplete, or rough data, while also incorporating intercellular structural information. The autoencoder accurately reconstructs the main signals in the data, and the Laplacian smoothing filters further improve the data quality by smoothing, reducing the impact of noise. This iterative process continuously updates the reconstructed data, gradually improving the accuracy and stability of single-cell data. Then we use a self-supervised discriminative embedding module, which utilizes the similarity between cells to select positive and negative sample pairs to determine the inherent clusters in the data. In this module, the threshold for positive and negative sample pairs are chosen adaptively, so that samples belonging to the same cluster are naturally pushed together, while those from different clusters will be expelled from each other in the embedding space. This module aims to enhance clustering performance by learning the intrinsic similarity structures embedded in the data distribution. By combining these two modules, scRISE effectively removes incompatible and noisy signals in the data and achieves self-supervised clustering without having to resort to extensive human interventions.
The framework of scRISE includes the iterative smoothing module based on graph autoencoder and the self-supervised discriminative embedding learning module. The iterative smoothing module consists of an autoencoder and a Laplacian filter connected. In each iteration, a cell graph is constructed from the input data, and the reconstructed data from the autoencoder is smoothed using the Laplacian filter. The smoothed data are fed back to the autoencoder for further processing. The output of the autoencoder is then processed through the self-supervised discriminative embedding module, which adopts an adaptive threshold to identify positive and negative sample pairs to compute the final clustering.
Evaluation of the iterative smoothing module
Autoencoders reassemble data in an unrefined manner that could include noise and missing information. As a result, Laplacian smoothing filters are required for scRNA-seq data processing to update and smooth the data. To progressively increase the precision and stability of single-cell data, this procedure must be repeated several times. The smoothed data are passed back to the autoencoder for reconstruction in each iteration, and more precise and trustworthy data are produced by doing this repeatedly. We used scRISE on five simulated scRNA-seq datasets to assess its performance in order to look into how the number of smoothing iterations affected the clustering performance. We determined the ideal number of smoothing iterations through rigorous testing that produced integrated information that improved clustering performance.
Figure 2a–d shows the clustering performance (ACC, ARI, NMI and Silhouette Coefficient) across five simulated single-cell datasets for different numbers of smoothing iterations. We noticed that setting the smoothing iteration to 1 resulted in less favorable clustering performance for scRISE across most datasets. As the number of iterations increases to 2, there is an obvious improvement in clustering performance. Different datasets show varying sensitivity to the smoothing iterations. For the sim_1000 dataset, the ACC metric gradually decreases, but remains relatively stable within 3 iterations. The NMI and ARI metrics reach their maximum values at 3 iterations, while the Silhouette Coefficient reaches its peak. For the simulated single-cell datasets with cell numbers ranging from 2500 to 7500, the ACC, NMI, and ARI metrics reach their maximum values at two iterations, with ACC values above 0.95 and NMI and ARI values above 0.89. These metrics show little change with increasing iteration numbers, while the Silhouette Coefficient gradually increases. Overall, scRISE performs well on datasets with a larger number of cells. Additionally, we analyzed the impact of different iteration numbers on runtime, as shown in Supplementary Fig. S1. The iteration number and runtime show a linear growth trend. Considering both clustering metrics and runtime, selecting a smoothing iteration of three for clustering simulated single-cell data achieves good results while shortening the runtime and improving analysis efficiency without compromising the results.
Comparison of scRISE with prior methods on scRNA-seq datasets
We conducted clustering comparisons between scRISE predictions and five recently proposed deep learning clustering methods on the seventeen scRNA-Seq datasets. The results show that scRISE improves the clustering performance on the aforementioned seventeen scRNA-Seq datasets. In the comparative analysis, we utilized three metrics (NMI, ARI, silhouette coefficient) to evaluate the clustering performance of each method.
In these seventeen real datasets, the ARI and NMI values for various methods are presented in Tables 1 and 2, respectively. Consistent with the NMI results, scRISE demonstrates top-notch clustering performance across all datasets, ranking first in eight datasets and second in five datasets, with all ARI and NMI values surpassing 0.5. While scTAG and scziDesk exhibit relatively good performance, their applicability is limited due to their excessive reliance on assumptions about data distribution. scDeepcluster shows good performance in only a few datasets, with subpar clustering performance in most. scGMAI and scGAE show relatively poor performance in most datasets, with both algorithms having low ARI and NMI values. scGMAI is a combination of multiple algorithms [25], which may affect the clustering effect. scGAE is constrained by the construction of cell graphs and does not correctly analyze the structural similarities between cells [22], resulting in poor clustering accuracy.
Figure 3a, b illustrates the overall performance of the six methods across the seventeen datasets. The proposed scRISE exhibits the highest average values in terms of ARI and NMI compared to other methods. The scTAG and scziDesk methods demonstrate competitive performance too, but they may perform poorly on some datasets. The comparison results for NMI align closely with those of ARI. Additionally, the bar chart in Supplementary Fig. S2a illustrates the Silhouette Coefficient values for the six methods. In measuring the compactness and separation of cell clusters, scDeepCluster demonstrates strong competitiveness, slightly surpassing the scRISE method on most datasets. In Supplementary Fig. S2b, we compared the runtime of existing deep learning clustering methods. ScRISE outperforms existing graph embedding clustering models (scTAG and scGAE). Overall, scRISE’s efficiency is at an upper-middle level, with relatively stable performance across different datasets.
The Baron_Human dataset consists of fourteen different cell types. Among them, there are seven cell types with larger quantities: ‘acinar’, ‘alpha’, ‘beta’, ‘ductal’, ‘delta’, ‘gamma’, and ‘endothelial’, and seven cell types with smaller quantities, including ‘quiescent stellate’, ‘mast’, ‘T cell’, ‘activate stellate’, ‘schwann’, ‘epsilon’, and ‘macrophage’. On this dataset, the scRISE method demonstrates high accuracy (ARI 0.8155, NMI 0.8327) and effectively separates each cell type. To visually compare the accuracy of different clustering methods, we use a Sankey diagram to illustrate the differences between the clustering results and the ground truth labels (Fig. 3c–h). In the Sankey diagram, each box represents a cluster, and the width and height of the boxes indicate the variation in cell quantities within the clusters, while the colors represent the similarity and dissimilarity between different clusters. The observations reveal that the scGMAI (Fig. 3c) and scGAE (Fig. 3f) methods tend to divide cell types with larger quantities into multiple clusters, especially the ‘alpha’ and ‘beta’ types being divided into multiple clusters. The scDeepCluster (Fig. 3d) method divides ‘beta’ and ‘ductal’ into multiple clusters, while clustering ‘quiescent stellate’, ‘activate stellate’, and ‘schwann’ into the same category. The scziDesk (Fig. 3e) method clusters the four cell types with larger quantities, ‘beta’, ‘alpha’, ‘delta’, and ‘gamma’, into a single category, resulting in significant errors. Although the scTAG (Fig. 3g) method achieves relatively high accuracy (ARI 0.6054, NMI 0.6907), it suffers from the same problem as other methods, i.e., dividing cell types with large number of samples into multiple categories, and some cell types with small number of samples are easily mixed with others. In contrast, scRISE (Fig. 3h) clearly reveals distinct clusters for the aforementioned cell types. We also generated a Sankey plot for the prediction results of the Baron_Mouse dataset (Supplementary Fig. S3). We note that for some clustering methods, a significant number of cells are incorrectly clustered, and some certain cell population could be divided into multiple categories. When clustering the Baron_Mouse dataset with scRISE, the resulting number of clusters is lower than the expected number, indicating the merging of several cell clusters. This merging enhances the identification of rare cell populations. Essentially, it merges some cells that are likely to be small populations of rare cells rather than larger, common cell types, suggesting that scRISE could help identify and study these rare cell types.
To demonstrate the clustering performance and validate the effectiveness of the proposed model, as well as to extract low-dimensional representations of high-dimensional data, we used the t-SNE algorithm to project the features from the adaptive encoder onto a two-dimensional space and visualize the final data embedding results. This allows for a more intuitive observation of the clustering patterns and the performance of the model. Figure 4 presents the t-SNE visualizations of three datasets: Klein, HNSCC, and Baron_Human. In Fig. 4a, we observe that scRISE clearly separates different cell types, while the clusters identified by other methods are scattered, and the boundaries between clusters are mixed. The boundaries between the ‘2d’ and ‘4d’ cell clusters are not distinct. As shown in Fig. 4b, we can see that scRISE achieves better inter-cluster compactness. Compared to scRISE, although scDeepCluster also shows clear cluster boundaries, there are multiple cell types mixed together, such as the ‘Fibroblast’, ‘Endothelial’, and ‘tumor’ clusters represented by pink, orange-red, and cyan dots in HNSCC. In Fig. 4c, for the Baron_Human dataset, methods other than scRISE fail to accurately identify the cell types. For example, in scGAE, the alpha and delta intermediate neuronal cells represented by brown and light blue are mixed together and cannot be well distinguished. In scGMAI, scziDesk, and scTAG, the clustering results are unclear. The beta cells represented by yellow are distributed throughout the entire plot and mixed with other cell types. Compared to other clustering methods, the proposed method scRISE identifies clear clusters with distinct boundaries between them.
In scRISE, we enhance the clustering process by incorporating supervised training. This involves dynamically selecting positive and negative samples based on K-means soft clustering. By doing so, we aim to refine the node embeddings, making them more representative and ultimately enhancing the performance of clustering. After a comprehensive comparison and analysis of seventeen datasets, it’s evident that scRISE stands out prominently in single-cell RNA sequencing data clustering. Across various metrics, including ARI and NMI, scRISE consistently outperforms other commonly used methods, showcasing its exceptional effectiveness. Notably, on the Baron_Human dataset, scRISE achieves remarkable ARI and NMI scores of 0.8155 and 0.8327 respectively, demonstrating its ability to accurately segregate distinct cell types with high precision. In Fig. 3, the Sankey plots demonstrate scRISE’s capacity to establish well-defined cluster boundaries, leading to enhanced clarity in distinguishing between different cell types. The distinct paths in the Sankey plots illustrate the robustness of scRISE in segregating cells into discrete groups with minimal overlap, a feat that is crucial for accurate downstream analysis. Furthermore, in Fig. 4, the t-SNE visualization provides a comprehensive view of how scRISE excels in achieving compact clusters with minimal dispersion between clusters. The tight clustering of data points in the t-SNE plot reflects scRISE’s effectiveness in capturing the underlying structure of the single-cell RNA sequencing data, thereby facilitating precise cell type identification. Comparatively, when juxtaposed with other existing clustering methods, scRISE’s performance shines through as it consistently achieves superior accuracy in delineating cell subtypes. While alternative methods may exhibit confusion or errors in this task, scRISE stands out for its ability to provide researchers with reliable and interpretable clustering results. Overall, the combination of Sankey plots and t-SNE visualizations serves to underscore scRISE’s proficiency in single-cell RNA sequencing data clustering, emphasizing its role as a powerful tool for unraveling the complexities of cellular heterogeneity and advancing our understanding of biological systems at the single-cell level.
Ablation study and scalability
We conducted ablation studies using seventeen real datasets to further understand the impact of the data smoothing task and clustering module in scRISE on clustering performance and the resulting improvements. Figure 5a, b shows the scatter plot of corresponding NMI and ARI values for scRISE with and without the data smoothing task. The results demonstrate that the data smoothing task significantly improves clustering accuracy and brings significant improvements in clustering precision across all tested datasets (Fig. 5c, d). The results of clustering studies with and without the clustering module indicate that the clustering module improves clustering accuracy in most datasets but does not enhance clustering performance in some datasets. This is because, in some datasets, the model has already achieved significant improvements in clustering accuracy through the data smoothing task, and the clustering module does not provide significant performance gains for these datasets. These results are consistent with our expectations. The data smoothing task effectively filters out high-frequency noise present in the data, while the clustering module considers the similarity between cells and captures discriminative expression patterns. Based on the ablation studies conducted on these real datasets, we can conclude that both the data smoothing task and the clustering module make significant contributions to the enhanced clustering performance of scRISE.
The comparison of ARI (a) and NMI (b) values with and without the iterative smoothing module. The comparison of ARI (c) and NMI (d) values with and without the self-supervised discriminative embedding module for clustering. Red points indicate that the addition of the module leads to better clustering results, while blue points indicate the opposite. e The runtime of scRISE on different-scale real datasets, including the time for the iterative smoothing module, the self-supervised discriminative embedding learning module, and the total runtime.
To evaluate the scalability of scRISE, we tested the running time on seventeen real scRNA-seq datasets with cell numbers ranging from 268 to 23184. We compared the total running time of the scRISE model, the running time of the autoencoder-based cycle-smoothing module, and the running time of the adaptive encoder clustering module (Fig. 5e). The results showed that scRISE has good scalability, and its running time is similar to a binomial relationship with the size of the dataset. In datasets with a large number of cells, the clustering module becomes the main step controlling the speed, as this module requires time-consuming calculations of the similarity between nodes. Therefore, scRISE can efficiently handle large-scale scRNA-seq datasets. Overall, these results indicate that scRISE is a scalable and efficient clustering method suitable for processing large-scale single-cell datasets.
Biological analysis
Information genes are a set of genes that show significant expression differences among different cell types and can be used to distinguish between them. In scRNA-seq analysis, information genes can be identified by analyzing gene expression data and used to determine different cell types. Non-negative matrix factorization (NMF) is a data analysis method used for dimensionality reduction and feature extraction, which can factorize a non-negative matrix into two non-negative matrices [26]. In scRNA-seq data analysis, NMF can be used to factorize the original cell-gene expression matrix into two matrices, one representing cell features and the other representing gene expression features. Here, we replace the cell feature matrix obtained from NMF factorization with the cell cluster feature matrix obtained from the scRISE model, and reconstruct the original scRNA-seq data to obtain a new cell-gene expression matrix that includes the cluster features extracted by the scRISE model. We then use Lasso [27] to further analyze this new cell-gene expression matrix to identify information genes that can best distinguish between different cell subtypes.
Using Lasso and the scRISE method for gene selection can help us identify potential therapeutic targets and biomarkers. In the HNSCC dataset, information genes were extracted using Lasso regression with a regularization strength (λ) set to 0.001, resulting in 62 genes (Supplementary Table S2). The top 10 positively and negatively correlated genes were selected for plotting (Fig. 6a). Subsequently, we performed COX regression analysis on these 62 informative genes (Supplementary Fig. S4). COX regression analysis is a commonly used survival analysis method used to evaluate the impact of gene expression or other factors on patient survival time or survival status. Through this analysis, we identified these informative genes to be associated with survival in patients with head and neck squamous cell carcinoma (HNSCC), and these differences were statistically significant, which provides the basis for further investigation of the potential role of these genes in HNSCC treatment and prognosis. Among these 20 genes, ZNF331 is a potential anti-tumor therapeutic target as it is involved in the development and progression of various cancers [28]. CD52 is a glycoprotein widely expressed in lymphocytes and monocytes and has been used as a therapeutic target and marker in lymphoma treatment [29]. PTPRC, also known as CD45, is considered an important T-cell antigen in the immune system and plays a crucial role in immune regulation [30]. PRKCQ plays a role in immune modulation and can regulate inflammatory responses, making it a potential target for the treatment of inflammatory diseases [31]. TACSTD2 (also known as TROP2) is an epithelial cell adhesion molecule that is highly expressed in various tumors, making it a research target for cancer treatment [32]. In the HNSCC dataset, these genes may play important roles in tumor cell proliferation, metastasis, and immune evasion. These findings suggest that the scRISE method can provide us with effective biomarkers for guiding tumor treatment and autoimmune disease therapy.
Next, further gene ontology (GO) and KEGG enrichment analysis can be performed on the obtained 62 information genes to explore their functional profiles, search for enriched biological processes, and uncover potential biological pathways. Figure 6b displays the gene distribution under GO enrichment, with the top 10 terms sorted by p-values in the categories of biological processes, cellular components, and molecular functions. For the biological processes category, the most common and enriched GO term is ‘myeloid leukocyte activation’ (GO:0002274). Myeloid leukocyte activation is an immune response process that involves the activation and differentiation of myeloid lineage white blood cells, including monocytes, macrophages, dendritic cells, etc., and their enhanced recognition and attack capabilities against pathogens and tumor cells [33]. In the cellular components category, the most enriched and concentrated GO terms are ‘membrane raft’ (GO:0045121) and ‘membrane microdomain’ (GO:0098857), which are both special regions of the plasma membrane enriched with cholesterol and sphingolipids. They are involved in various cellular processes, including signal transduction, transport, and membrane organization. They are also related to the pathogenesis of various cancers, including breast cancer, lung cancer, colorectal cancer, and melanoma [34]. For the molecular functions category, the most enriched and concentrated GO term is ‘scaffold protein binding’ (GO:0097110). Scaffold proteins are proteins that provide structural stability and serve as a support and framework in the cell. Scaffold proteins play important roles within the cell by forming complex networks through interactions with other proteins [35]. ‘Modified amino acid binding’ (GO:0072341) indicates the binding of modified amino acids with other molecules. These results provide an overview of the functional characteristics of the 62 information genes and shed light on their interrelationships, revealing potential biological processes.
KEGG enrichment analysis can help identify biological processes and pathways influenced by the input gene set, providing insights into potential biological mechanisms related to specific diseases or biological processes. Figure 6c displays the relevant pathways enriched by KEGG, sorted by adjusted p-values, showing the top 15 pathways. ‘T-cell receptor signaling pathway’ (hsa04660) is a pathway that involves a series of proteins and molecules related to T-cell receptor (TCR) activation and downstream signaling. Genes involved in the TCR signaling pathway can provide insights into potential molecular mechanisms of T-cell activation and differentiation, making them potential therapeutic targets for T-cell dysfunction-related diseases such as autoimmune diseases and cancer [36]. ‘Th17 cell differentiation’ (hsa04659) includes a series of cellular factors, transcription factors, and signaling pathways involved in the differentiation and activation of Th17 cells. It plays an important role in the immune system, particularly in combating bacterial and fungal infections and tumor immune responses [37]. ‘PD-L1 expression and PD-1 checkpoint pathway in cancer’ (hsa05235) is an important pathway related to tumor immune evasion. High expression of PD-L1 inhibits the activity of immune cells, thereby promoting immune evasion by tumor cells. PD-1 is one of the checkpoint molecules highly expressed in the tumor microenvironment. When PD-1 binds to its ligand PD-L1, it inhibits the activity of T cells, suppressing their attack on tumor cells [38]. In cancer treatment, enhancing T-cell immune activity by inhibiting the PD-L1 and PD-1 pathway has become an important therapeutic strategy. In Supplementary Fig. S5, PRKCQ, LAT, and MAPK13 are genes associated with the PD-L1 and PD-1 pathway in the informative gene set. These findings highlight relevant pathways identified through KEGG enrichment analysis, providing insights into potential therapeutic targets and biological mechanisms associated with specific diseases and biological processes. These results demonstrate that scRISE can capture key representations and patterns of scRNA-seq data. The results of GSEA and GSVA enrichment analysis of 62 informative genes are shown in Supplementary Fig. S6. In the GSEA analysis, KEGG background gene set and immune-related set were used as preset gene sets to explore the impact of these genes on metabolic pathways and immune-related pathways. The results show that these information genes are closely related to multiple cancer pathways and associated regulation. Among them, the enrichment scores of cancer pathways such as breast cancer and pancreatic cancer are significant, suggesting that these genes may play an important role in the occurrence and development of cancers. Further analysis showed that the T-cell receptor (TCR) signaling pathway plays a key role in the biological processes regulated by these information genes. The activation of the TCR signaling pathway regulates the differentiation and activation of T cells through a variety of protein kinases and signaling molecules (such as LCK, ZAP70, PI3K-AKT, MAPK, etc.) and transcription factors (such as NF-κB, AP-1, etc.). The findings further support the importance of these informative genes in immune regulation and tumor immunology. Analysis of immune-related sets also showed similar results, indicating that these information genes are important for the regulation of immune cells. In immune cells, these genes participate in multiple regulatory effects, affecting the development, function, and immune response of immune cells [36, 37]. These findings were further supported by GSVA analysis, which showed that these informative genes are closely associated with small-cell lung cancer and other cancer-related pathways. This indicates that these genes may play an important role in the occurrence and development of cancer, providing important clues for further revealing their role in tumorigenesis mechanisms and immune regulation. In summary, scRISE is highly practical in interpreting biological processes and can serve as an effective analytical tool in biological research.
Discussion
In this study, we propose a deep learning clustering method called scRISE for scRNA-seq data. It utilizes Laplacian data smoothing and adaptive learning. scRISE exhibits novelty in several aspects. Firstly, we use autoencoders to learn the relationships between the data, allowing the reconstruction of single-cell data without assuming data distribution. Secondly, we apply Laplacian smoothing filters in scRNA-seq clustering analysis. This step reduces high-frequency noise in the data, improving data quality, while maintaining data dimensionality. Thirdly, scRISE gradually improves the accuracy and stability of single-cell data through iterative cycles of autoencoder and Laplacian smoothing filters. This iterative approach helps enhance the accuracy of clustering results. Additionally, the adaptive encoder constructs a similarity matrix and adaptively selects positive and negative samples to extract low-dimensional embeddings that represent the intrinsic features of the data. This enhances clustering effectiveness and accuracy. The clustering results demonstrate that scRISE outperforms other deep learning algorithms in various biological scenarios. To provide better biological interpretations of the results, we conducted biological analyses, including inference of informative genes, gene ontology, and KEGG pathway enrichment analysis.
Our current scRISE method has some limitations. The selection of positive and negative samples in the adaptive clustering module relies entirely on the similarity calculation method, which can be computationally time-consuming. Therefore, we will explore more accurate and comprehensive similarity calculation methods to improve clustering performance. In the future, we plan to apply our proposed clustering framework to the field of multi-omics research, integrating different omics data sources such as Bulk RNA-seq, spatial transcriptomics, etc. This integration will help us gain a deeper understanding of biological systems from multiple perspectives. It can uncover correlations and interactions between different omics layers, providing a more comprehensive view.
Methods
Datasets and preprocessing
To determine the optimal number of iterations in the graph autoencoder cycle-smoothing module, we conducted experiments using the R package Splatter [39] to generate five simulated datasets. Each dataset was configured with 8 clusters, with varying numbers of cells ranging from 1000 to 7500, as outlined in Supplementary Table S1. Each cell in the datasets contained 2500 genes. The proportions of cells in each category were set as follows: 0.1, 0.15, 0.1, 0.1, 0.1, 0.1, 0.2, 0.1. The dropout rate used in the experiments was approximately 65%, with the specific splatter parameter set as dropout.mid = 2.5.
As shown in Table 3, We compared the performance of our model with other benchmark methods on seventeen real scRNA-seq datasets from several representative sequencing platforms. This includes fifteen medium-scale datasets and two large-scale datasets. The medium-scale datasets consist of nine mouse scRNA-seq datasets: Deng [40], mESC [41], Tabula_Heart_and_Aorta [42], Tabula_Liver, Baron_Mouse [43], Klein [44], Romanov [45], Zeisel [46], and Tabula_Spleen; and six human datasets: Li [47], Chu [48], Petropoulos [49], HNSCC [50], Tirosh [51], and Baron_Human. The two large-scale datasets are the turtle dataset Tosches [52] and the mouse dataset Bach [53]. The annotation of cell types from the original publications is utilized as the ground truth for cell type identification.
Before performing clustering, the data underwent quality control and normalization procedures. Firstly, gene filtering was applied to retain genes that were expressed in at least one or more cells. After quality control, the read counts were divided by the library size, multiplied by 100,000, and transformed into a logarithmic value with a base of 10, with the addition of pseudo-count 1. The data from HNSCC and Triosh have already undergone expression normalization and logarithmic transformation and do not require further processing at this step. From this, an expression matrix consisting of the top 2,000 highly variable genes was selected as the input for the network. Subsequently, the filtered scRNA-seq data underwent normalization, scaling the values to be within the range of [0, 1]. All of these data preprocessing steps were performed using the Python package Scanpy [54].
Autoencoder module
Autoencoder is an unsupervised deep learning algorithm for learning a compact representation of the data while attempting to maximize the preservation of the input data information. In cases where the original scRNA-seq data contains a significant amount of redundant information and dropout events, the autoencoder is trained to reconstruct the expression matrix of each cell population while learning representative embeddings of the expressions.
The autoencoder (AE) consists of an encoder and a decoder. The encoder compresses the input data into a low-dimensional encoding, while the decoder maps this encoding back to the original data space. Take \(X={E}^{m\times n}\) as a raw gene expression matrix where m is the number of cells, n is the number of genes. The encoder contains a hidden layer and an output layer that constructs low-dimensional embeddings H from the input gene expression X. The decoder accepts these embeddings H as input and passes them to a hidden layer and an output layer that produces a reconstruction \(\widetilde{X}\) of the original sample. Assuming the encoder has L layers, each layer l learns a data representation denoted as \({H}^{\left(l\right)}\), the weights are denoted as \({W}^{\left(l\right)}\), and the bias vector is denoted as \({b}^{\left(l\right)}\). The learning process of each layer in the autoencoder can be described as follows:
Where \(s(\cdot )\) is the activation function applied element-wise to the weighted sum of the inputs and biases in the l-th layer. The encoder stage of the autoencoder transforms the input data X into a latent representation H, which can be expressed as:
Where Wenc represents the encoder weights, benc represents the encoder biases.
The decoder stage maps H to the reconstructed input \(\widetilde{X}\) as:
Where Wdec represents the decoder weights, bdec represents the decoder biases. The autoencoder is trained by minimizing the reconstruction error between X and\(\widetilde{X}\), typically measured by mean squared error (MSE) loss:
Construction of the KNN graph
K-nearest neighbor graph (KNN) is an undirected graph based on the nearest neighbor distances, used to transform scRNA-seq datasets into a graphical structure that describes the relationships between cells in the dataset. We first reduce the dimensionality of the scRNA-seq data using PCA. Each node in the graph represents a cell, and if cell xi is one of the k-nearest neighbors of cell xj, we assign an edge between them. Here, we set the value of k to 15. In previous studies, the Pearson correlation coefficient has been found to better calculate the similarity between cells for constructing the KNN graph [24]. Therefore, we use the Pearson correlation coefficient to compute the similarity between cells and construct the KNN graph. Pearson correlation coefficient is defined as:
Laplacian smoothing filter
Laplacian smoothing is a graph-based signal processing method used to smooth the feature information of nodes in graph data [55]. It iteratively computes a weighted average of a node’s feature with its neighboring nodes’ features, leveraging the adjacency relationship of the graph and the connectivity between nodes to enhance feature consistency and stability.
Given an attributed graph G with an adjacency matrix A and an identity matrix I, by employing the renormalization trick, we define the modified adjacency matrix as \({A}^{{\prime} }=A+I\). \({D}^{{\prime} }\) is the degree matrix corresponding to\({A}^{{\prime} }\). Consequently, the formula for computing the normalized graph Laplacian matrix, Lnorm is as follows:
The definition of the Laplacian smoothing filter is as follows:
Following the adaptive graph encoder (AGE) algorithm [56], with a setting of α = 2/3 and applying the Laplacian smoothing filter iteratively for t times, the filtered representation of the reconstruction matrix \(\hat{X}\) can be denoted as:
The Self-supervised discriminative embedding
To enhance the effectiveness of node embedding learning and improve clustering performance, we employ a self-supervised discriminative embedding learning method. In the encoder, we adaptively select highly similar node pairs as positive training samples and choose low-similarity node pairs as negative samples, enabling supervised training. Through this approach, the adaptive encoder can better learn representations of nodes, thereby improving the quality of node embeddings and enhancing clustering performance. Given filtered reconstruction matrix \(\hat{X}\), the node embeddings are encoded by a non-linear encoder \(g(\cdot )\) and a linear encoding layer \(h(\cdot )\), resulting in the feature matrix Z.
To measure the pairwise similarity matrix sij between nodes, the Pearson correlation coefficient is used as the similarity metric. After computing the similarity matrix, we sort the pairwise similarity sequences in descending order. Here, rij represents the ranking position of cell pair (vi, vj). We set the maximum ranking position of positive samples as rpos and the minimum ranking position for negative samples as rneg. Therefore, the label generated for (vi, vj) is:
The training set consists of \({r}_{{\rm{pos}}}\) positive samples and \({n}^{2}-{r}_{{\rm{neg}}}\) negative samples. At the beginning of the training, selecting a larger number of samples provides more information and diversity. As the training process progresses, the value of rpos decreases, while rneg increases.
During the training process of the encoder, we compare the sample labels with the similarity of the nodes generated by the encoder to measure the difference between the learned node representations by the encoder and the true similarity. Accordingly, our cross-entropy loss is given by
Self-optimizing clustering
After training the adaptive encoder, the latent representation Z can capture the relationship between cells and gene expressions. By performing k-means clustering on Z, a simple clustering result can be obtained. However, this result may not be optimal due to the lack of interaction between the clustering module and the feature learning module. To address this issue, we applied a self-optimizing embedding algorithm where the latent embedding is fed into a self-optimizing clustering module. The objective function of this module is represented using Kullback-Leibler (KL) divergence. Since the target distribution P is defined based on Q, the embedding learning of Q is supervised in a self-optimizing manner, aiming to make it approach the target distribution P, as shown in the following expression:
where qij is the soft label of the embedding node zi. This label measures the similarity between zi and the cluster central embedding uj by a Student’s t-distribution, which can be described as follows:
Additionally, pij is an auxiliary target distribution that emphasizes assigning high-confidence similar data points based on qij, as shown below:
Throughout the entire training process, similarity and clustering learning are jointly optimized. We minimize the following overall objective function:
Where Losssi is the similarity loss, Lossclu is the clustering loss, α and \(\beta\) are hyperparameters that balance the two losses. The loss function integrates latent representation learning and clustering into a unified framework, thereby promoting the final clustering result.
Baseline
To validate the clustering performance of the scRISE algorithm, we compared it with five deep learning clustering methods. These methods can be categorized into deep embedding clustering methods and deep graph-based clustering methods.scGMAI [25] utilizes an autoencoder network to reconstruct gene expression values from scRNA-Seq data. It employs FastICA to reduce the dimensionality of the reconstructed data and subsequently applies a Gaussian Mixture Clustering (GMC) method for clustering. scDeepCluster incorporates the ZINB model to simulate the distribution of scRNA-seq data within the denoising autoencoder. By explicitly modeling scRNA-seq data, it learns feature representations and performs clustering tasks. scziDesk utilizes a denoising autoencoder to represent scRNA-seq data, and then constructs a self-training k-means algorithm to perform cell clustering. scGAE employs a multi-task graph autoencoder to simultaneously capture the topological structure information and feature information in scRNA-Seq data. scTAG is a method that integrates the ZINB model into a topologically adaptive graph convolutional autoencoder to learn low-dimensional latent representations, and employs the KL divergence for clustering tasks.
Statistics and reproducibility
scRISE was implemented in Python 3 (version 3.8) using PyTorch (version 2.0). The size of the encoding layers in the autoencoder was set as (256, 64, 32), and the decoding layers had the opposite structure. We initially set the learning rate as lr=0.001, epoch=100, and batch size = 256, and then used Adam optimizer to adjust the learning rate. The size of the adaptive encoder was set as 32. The learning rate for the adaptive encoder was lr=0.0005, and an initial threshold was set \({r}_{{\rm{pos}}}^{\rm{st}}=0.0015\) and \({r}_{\rm{neg}}^{\rm{st}}=0.3\), while the final threshold is set to \({r}_{\rm{pos}}^{\rm{ed}}=0.001\) and \({r}_{\rm{neg}}^{\rm{ed}}=0.7\), the number of update iterations (T) to 40, and the batch size for sample pairs to 10,000. We trained the model for 400 epochs using the Adam optimizer. The hyperparameters α and β were both set to 10. We ran the experiments 10 times on all datasets and reported the median results to ensure the accuracy of the data. All experiments were conducted on an NVIDIA Tesla-V100-PCLe-32GB.
Data availability
The authors declare that all data supporting the findings of this study are available within the article or from the corresponding author upon reasonable request.
Code availability
The scRISE software package and source code are available in Github (https://github.com/LiLab-ssruan/scRISE).
References
Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:1–14.
Wen L, Li G, Huang T, Geng W, Pei H, Yang J, et al. Single-cell technologies: from research to application. Innovation. 2022;3:100342.
Eraslan G, Drokhlyansky E, Anand S, Fiskin E, Subramanian A, Slyper M, et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science. 2022;376:eabl4290.
Iacono G, Mereu E, Guillaumet-Adkins A, Corominas R, Cuscó I, Rodríguez-Esteban G, et al. bigSCale: an analytical framework for big-scale single-cell data. Genome Res. 2018;28:878–90.
Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front Genet. 2019;10:317.
Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat Commun. 2017;8:1627.
Wen J, Ling R, Chen R, Zhang S, Dai Y, Zhang T, et al. Diversity of arterial cell and phenotypic heterogeneity induced by high-fat and high-cholesterol diet. Front Cell Dev Biol. 2023;11:971091.
Yang L, Liu J, Lu Q, Riggs AD, Wu X. SAIC: an iterative clustering approach for analysis of single cell RNA-seq data. BMC Genomics. 2017;18:689.
Pal S, Mondal S, Das G, Khatua S, Ghosh Z. Big data in biology: The hope and present-day challenges in it. Gene Rep. 2020;21:100869.
Lingxue Z, Jing L, Bernie D, Kathryn R. A unified statistical framework for single cell and bulk RNA sequencing data. Ann Appl Stat. 2018;12:609–32.
Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–2.
Tung P-Y, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK, et al. Batch effects and the effective design of single-cell gene expression studies. Sci Rep. 2017;7:39921.
žurauskienė J, Yau C. pcaReduce: hierarchical clustering of single cell transcriptional profiles. BMC Bioinform. 2016;17:140.
Lin P, Troup M, Ho JWK. CIDR: Ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol. 2017;18:59.
Wang B, Ramazzotti D, De Sano L, Zhu J, Pierson E, Batzoglou S. SIMLR: a tool for large-scale genomic analyses by multi-kernel learning. Proteomics. 2018;18:1700232.
Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–6.
Eraslan G, Simon LM, Mircea M, Mueller NS, Theis FJ. Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun. 2019;10:390.
Tian T, Wan J, Song Q, Wei Z. Clustering single-cell RNA-seq data with a model-based deep learning approach. Nat Mach Intell. 2019;1:191–8.
Chen L, Wang W, Zhai Y, Deng M. Deep soft K-means clustering with self-training for single-cell RNA sequence data. NAR Genomics Bioinform. 2020;2:lqaa039.
Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15:1053–8.
Wang J, Ma A, Chang Y, Gong J, Jiang Y, Qi R, et al. scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nat Commun. 2021;12:1882.
Luo Z, Xu C, Zhang Z, Jin W. A topology-preserving dimensionality reduction method for single-cell RNA-seq data using graph autoencoder. Sci Rep. 2021;11:20028.
Yu Z, Lu Y, Wang Y, Tang F, Wong K-C, Li X. ZINB-based graph embedding autoencoder for single-cell RNA-seq interpretations. Proc AAAI Confer Artif Intell. 2022;36:4671–9.
Gan Y, Huang X, Zou G, Zhou S, Guan J. Deep structural clustering for single-cell RNA-seq data jointly through autoencoder and graph neural network. Brief Bioinform. 2022;23:bbac018.
Yu B, Chen C, Qi R, Zheng RQ, Skillman-Lawrence PJ, Wang XL, et al. scGMAI: a Gaussian mixture model for clustering single-cell RNA-Seq data based on deep autoencoder. Brief Bioinform. 2021;22:bbaa316.
Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–91.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc: Ser B (Methodol). 1996;58:267–88.
Yu J, Liang QY, Wang J, Cheng Y, Wang S, Poon TCW, et al. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2013;32:307–17.
Wang J, Zhang G, Sui Y, Yang Z, Chu Y, Tang H, et al. CD52 is a prognostic biomarker and associated with tumor microenvironment in breast cancer. Front Genet. 2020;11:578002.
Ma Y-F, Chen Y, Fang D, Huang Q, Luo Z, Qin Q, et al. The immune-related gene CD52 is a favorable Biomark breast cancer prognosis. Gland Surg. 2021;10:780–98.
Byerly JH, Port ER, Irie HY. PRKCQ inhibition enhances chemosensitivity of triple-negative breast cancer by regulating Bim. Breast Cancer Res. 2020;22:72.
Katzendorn O, Peters I, Dubrowinskaja N, Tezval H, Tabrizi PF, von Klot CA, et al. DNA methylation of tumor associated calcium signal transducer 2 (TACSTD2) loci shows association with clinically aggressive renal cell cancers. BMC Cancer. 2021;21:444.
Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–S23.
Greenlee JD, Subramanian T, Liu K, King MR. Rafting down the metastatic cascade: the role of lipid rafts in cancer metastasis, cell death, and clinical outcomes. Cancer Res. 2021;81:5–17.
DiRusso CJ, Dashtiahangar M, Gilmore TD. Scaffold proteins as dynamic integrators of biological processes. J Biol Chem. 2022;298:102628.
Shah K, Al-Haidari A, Sun J, Kazi JU. T cell receptor (TCR) signaling in health and disease. Signal Transduct Target Ther. 2021;6:412.
Bhaumik S, Basu R. Cellular and molecular dynamics of Th17 differentiation and its developmental plasticity in the intestinal immune response. Front Immunol. 2017;8:254.
Kim MJ, Ha S-J. Differential role of PD-1 expressed by various immune and tumor cells in the tumor immune microenvironment: expression, function, therapeutic efficacy, and resistance to cancer immunotherapy. Front Cell Dev Biol. 2021;9:767466.
Zappia L, Phipson B, Oshlack A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 2017;18:174.
Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-Seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–6.
Hayashi T, Ozaki H, Sasagawa Y, Umeda M, Danno H, Nikaido I. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun. 2018;9:619.
Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–72.
Baron M, Veres A, Wolock Samuel L, Faust Aubrey L, Gaujoux R, Vetere A, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e344.
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–201.
Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R, et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci. 2017;20:176–88.
Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–42.
Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49:708–18.
Chu L-F, Leng N, Zhang J, Hou Z, Mamott D, Vereide DT, et al. Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol. 2016;17:173.
Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SaritaP, Codeluppi S, et al. Single-Cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–26.
Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e1624.
Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96.
Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science. 2018;360:881–8.
Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat Commun. 2017;8:2128.
Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15.
Taubin G. A signal processing approach to fair surface design. In: Proceedings of the 22nd Annual Conference on Computer Graphics and Interactive Techniques. Association for Computing Machinery (ACM); 1995. p. 351–8.
Cui G, Zhou J, Yang C, Liu Z. Adaptive graph encoder for attributed graph embedding. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. Association for Computing Machinery: Virtual Event, CA, USA, 2020, pp 976-85.
Funding
This work was supported in part by the National Natural Science Foundation of China (grants 82425104 and 82150208 to HL, 82173690 to SLL); the National Key R&D Program of China (2022YFC3400501, 2022YFC3400504); SLL is also sponsored by the Shanghai Rising-Star Program (23QA1402800).
Author information
Authors and Affiliations
Contributions
Jinxin Xie: Conceptualization, methodology, software, writing - original draft. Shanshan Ruan: Data curation, formal analysis, visualization, writing - review & editing. Mingyan Tu: Validation, investigation, software. Zhen Yuan, Jianguo Hu: Investigation. Honglin Li, Shiliang Li: Supervision, project administration, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41388_2024_3074_MOESM1_ESM.docx
Supporting Information for Clustering single-cell RNA sequencing data via iterative smoothing and self-supervised discriminative embedding
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, J., Ruan, S., Tu, M. et al. Clustering single-cell RNA sequencing data via iterative smoothing and self-supervised discriminative embedding. Oncogene 43, 2279–2292 (2024). https://doi.org/10.1038/s41388-024-03074-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-03074-5
- Springer Nature Limited