Abstract
Cellular communication between gastric cancer (GC) cells with different metastatic potentials and microenvironments and resultant cancer progression is not fully understood. Circular RNAs (circRNAs) and exosomal circRNAs are known to play extremely important regulatory roles in GC occurrence and progression. Here, we revealed significant differences in coronin-like actin-binding protein 1C (CORO1C) derived circRNA hsa_circ_0000437 between GC and para-cancer tissues. Hsa_circ_0000437 regulated GC cell proliferation, invasion, migration and apoptosis by targeting Ser/Arg-rich splicing factor 3 (SRSF3) and inhibiting programmed cell death 4 (PDCD4). The ectopic expression of hsa_circ_0000437 dramatically promoted tumor growth in nude mice in vivo. Furthermore, both gain-of-function and loss-of-function experiments demonstrated that hsa_circ_0000437 promoted human lymphatic endothelial cells (HLECs) invasion, migration, and tube formation in vitro and also promoted lymphangiogenesis and lymph node metastasis (LNM) in popliteal LNM model in vivo, when it was enriched in GC-secreted exosomes and transferred into HLECs. Mechanistically, exosomal hsa_circ_0000437 induced LNM via HSPA2-ERK signaling pathway independent of VEGF-C. Clinical data showed that exosomal hsa_circ_0000437 was enriched in the serum of GC patients, which was associated with LNM. In summary, these findings highlight the potential role of hsa_circ_0000437 as an outcome biomarker in GC patients with LNM, which may provide a novel target for GC therapy.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a common malignant tumor of the gastrointestinal tract, the third most common cause of cancer-related deaths. Although the combination of surgical resection and chemotherapy has improved the clinical outcome of patients with early-stage GC, the prevention and treatment of GC recurrence and metastasis remain a clinical challenge, knowing that the 5-year survival rate of these patients is still less than 30% [1, 2]. It is therefore critical to identify factors contributing to GC recurrence and metastasis and explore potential biomarkers and underlying molecular mechanisms.
Circular RNAs (circRNAs) are a class of single-stranded RNAs generated from precursor mRNA back-splicing with a covalently closed loop structure without 5′-3′ polarity or a 3′ poly A tail [3], featured by a stable structure, high abundance, and tissue-specific expression in eukaryotic cells [4]. CircRNAs participate in gene expression and regulation in several ways by acting as a sponge to competitively inhibit miRNA, interacting with nuclear ribonuclein or RNA polymerase II, and directly binding with transcription factors to regulate the splicing of classical RNA [5,6,7]. Our previous study [8] demonstrated that enhanced circHAS2 promoted the proliferation and invasion of GC cells and the role of PPM1E through competitive binding with miR-944. Liang et al. [9] reported that circDCUN1D4 participated in metastasis and glycolysis of lung adenocarcinoma by acting as a scaffold to promote the interaction between HuR protein and thioredoxin-interacting protein (TXNIP) mRNA, thus enhancing TXNIP mRNA stability. Wang et al. [10] found that EIF4A3, an RNA-binding protein, inhibited the formation of circRNA_100290 that promoted GC cell proliferation, invasion, and epithelial-mesenchymal transition. Despite the extensive research on the molecular mechanism of dysregulated circRNA-related pathways, there is little knowledge about circRNA-activated oncogenic pathways that participate in GC progression.
As a member of the WD repeat protein family that regulates actin-dependent processes by assembling F-actin, coronin-like actin-binding protein 1C (CORO1C) was reported to promote metastasis in colorectal cancer, breast cancer, and GC [11,12,13]. However, the biological function of CORO1C derived circRNAs has not been thoroughly studied. Knowing that CORO1C-derived circRNA (hsa_circ_0008714) could promote the phenotype of laryngeal squamous cell carcinoma proliferation, migration and invasion via let-7c-5p-PBX3 axis [14]. Interestingly, we found CORO1C-derived circRNAs were also differential expression in our circRNA-seq analysis results. In this study, we explored the molecular mechanism underlying the role of hsa_circ_0000437 in GC, hoping that the results could provide useful clues for its clinical application in the diagnosis and treatment of this devastating malignancy.
Results
Hsa_circ_0000437 is upregulated in GC
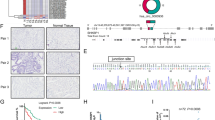
The circRNAs expression profiles obtained from circRNA-seq analysis on the GC tissue and corresponding non-tumor tissue in our previous study [15]. CORO1C-derived circRNAs were displayed in Table S1. CircRNAs with high read count scores and included in Circbase database were selected. Four circRNAs (hsa_circ_0028081, hsa_circ_0007830, hsa_circ_0000437 and hsa_circ_0008081) in GC tissues were identified by qRT-PCR assay, of which hsa_circ_0000437 was significantly upregulated (Fig. S1). Hsa_circ_0000437 was formed by head-to-tail splicing of CORO1C exon 4 and exon 5, and the spliced mature sequence length was 251 bp according to the Circbase database (http://www.circbase.org/) (Fig. 1A). A qRT-PCR assay was performed with divergent and convergent primers and agarose gel electrophoresis to confirm its circularity (Fig. 1B). Head-to-tail splicing of qRT-PCR products was found via Sanger sequencing (Fig. 1C). Then, 48 pairs of tissues from the GC patients were selected to identify the expression of hsa_circ_0000437 (Fig. 1D). It was also found that hsa_circ_0000437 was highly expressed in BGC-823 cells, HGC-27, MKN-1, and MKN-45 cells relative to GES-1 cells (Fig. 1E).
A Schematic diagram of the formation of hsa_circ_0000437. B hsa_circ_0000437 and GAPDH were amplified with divergent primers and convergent primers using cDNA and gDNA of BGC-823 cells as templates. C Sanger sequencing of the RT-PCR products of hsa_circ_0000437. The black arrow indicates the splicing site of hsa_circ_0000437. D Hsa_circ_0000437 expression in GC tissues and corresponding nontumor tissues was analyzed by qRT-PCR (n = 48, ***P < 0.001). E qRT-PCR analysis of hsa_circ_0000437 expression in GC cell lines and a normal gastric mucosa epithelium cell line (*P < 0.05, **P < 0.01, ***P < 0.001).
Hsa_circ_0000437 plays an oncogene role in vitro and in vivo
Hsa_circ_0000437 (Lv-circ)/control (Lv-NC) was transfected into BGC-823 cells, and sh-hsa_circ_0000437 (Lv-sh)/control (Lv-sh-NC) was transfected into MKN-45 cells to determine the role of hsa_circ_0000437 in GC. It was found that Lv-sh significantly reduced the expression of hsa_circ_0000437, while the level of hsa_circ_0000437 was significantly increased in Lv-circ group (Fig. S2A). Colony formation and CCK-8 assay showed that over-expression of hsa_circ_0000437 (Lv-circ) promoted cell proliferation. In contrast, hsa_circ_0000437 (Lv-sh) knockdown inhibited cell proliferation (Fig. S2B, C). The invasion and migration rates in Lv-circ group were significantly higher than those in the control group (Fig. 2A), and silencing hsa_circ_0000437 showed the opposite result. Flow cytometry showed that the apoptotic percentage in Lv-sh group was significantly higher than that in Lv-circ group (Fig. 2B). Western blotting analysis showed the same result for apoptotic proteins Bcl-2 and Bax (Fig. S2D). To determine whether hsa_circ_0000437 affected tumourigenesis, BGC-823 cells were transfected with Lv-NC or Lv-circ and then injected into the nude mice. It was found that the tumors in Lv-circ group were larger in size (Fig. 2C, D) and weight (Fig. 2E), and grew faster (Fig. 2F). Ki-67 and caspase-3 IHC staining showed that hsa_circ_0000437 overexpression increased the tumor proliferation and reduced apoptosis (Fig. S2E). All these results demonstrated that hsa_circ_0000437 functioned as an oncogene in GC.
Hsa_circ_0000437 (Lv-circ)/control (Lv-NC) was transfected into BGC-823 cells, and sh-hsa_circ_0000437 (Lv-sh)/control (Lv-sh-NC) was transfected into MKN-45 cells. A Invasion and migration assay in transfected cells. Scale bar: 100 μm. B Apoptosis was detected by flow cytometry in transfected cells. C–F The effect of overexpression of hsa_circ_0000437 on GC tumor growth was assayed in the nude xenograft tumor model. The data are presented as the mean ± SD, *P < 0.05, **P < 0.01.
Hsa_circ_0000437 directly targets SRSF3
It is known that the function of circRNAs depends on their varying localizations, and circRNAs in the cytoplasm usually serve as the “molecular sponges” of miRNA and interact with specific proteins in the nucleus. Using fluorescence in situ hybridization assay, we ascertained the subcellular localization of hsa_circ_000437 in GC cells (Fig. 3A). Nuclear and cytoplasmic abundance of hsa_circ_0000437 in BGC-823 and MKN-45 cells were also estimated using qRT-PCR. We found hsa_circ_000437 was mainly distributed in the nucleus of GC cells (Fig. 3B). Therefore, we performed chromatin isolation by RNA purification and massspectrometry analysis (ChIRP-MS) to identify the binding protein of hsa_circ_000437 in GC. The cellular proteins relatively enriched by hsa_circ_000437 probe and control probe are summarized in Fig. 3C. Through ChIRP-MS, it was found that 29 specific proteins were enriched by the hsa_circ_000437 probe, including JUP, CPEB4, HNRNPA2B1, DSG1, YBX1, and SRSF3, among others. The above-enriched proteins were further confirmed by absolute quantitative parallel reaction monitoring (PRM), and it was only found that hsa_circ_000437 could enrich SRSF3 (Fig. 3D). In addition, SRSF3 was increased by hsa_circ_000437 upregulation and reduced by hsa_circ_000437 downregulation (Fig. 3E).
The localization of hsa_circ_0000437 in GC cells was determined by fluorescence in situ hybridization. Scale bar: 50 μm (A) and nucleoplasmic separation assay (B). C ChIRP-MS was performed to identify the binding protein of hsa_circ_000437 in BGC-823 cells. D The binding protein was further verified by parallel reaction monitoring analysis. E SRSF3 was detected by Western blotting analysis. All data are means ± SD, *P < 0.05.
Hsa_circ_0000437 regulates GC cell function by targeting SRSF3/PDCD4 complex
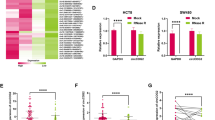
To further investigate the regulatory relationship between hsa_circ_0000437 and SRSF3, we conducted functional recovery experiments. hsa_circ_0000437 overexpression was used to respond to the functional changes induced by the interference of SRSF3 in GC cells. It was found that the proliferation ability of GC cells in sh-SRSF3 transfection group was significantly lower than that in sh-NC group, and overexpression of hsa_circ_0000437 (Lv-circ) restored the proliferation inhibition of sh-SRSF3 GC cells (Fig. 4A, S3A). Overexpression of hsa_circ_0000437 (Lv-circ) reduced cell apoptosis and pro-apoptotic associated proteins induced by sh-SRSF3 (Fig. 4B, S3B). The invasion and migration rates of sh-SRSF3 and Lv-circ co-transfection group were significantly higher than those in sh-SRSF3 group (Fig. 4C). Kim et al. found that SRSF3 interacted directly with PDCD4 mRNA by binding to the 5′-UTR and mediated translational repression [16]. The RIP assay also revealed that the anti-SRSF3 antibody pulled down abundant SRSF3 compared to IgG, (Fig. 4D). qRT-PCR results showed that PDCD4 could be detected in the immunoprecipitation complex bound by anti-SRSF3 antibody, but hardly in the IgG control group, confirming the direct interaction between SRSF3 and PDCD4 mRNA (Fig. 4E). Western blotting showed that the expression of PDCD4 in GC cells transfected with sh-SRSF3 was higher than that in sh-NC group (Fig. 4F). To determine whether SRSF3 affected GC cell function by regulating PDCD4, SRSF3 was overexpressed (OE-SRSF3) in PDCD4-overexpressing (OE-PDCD4) GC cells. Colony formation and CCK-8 assay showed that SRSF3 enhancement partly reversed the inhibitory effect of PDCD4 in cell proliferation (Fig. S3C, D). Concordantly, the same effect was found for cell migration and invasion (Fig. S3E). However, no significant difference was observed for apoptosis (Fig. S3F). Overexpression of hsa_circ_0000437 (Lv-circ) attenuated increased PDCD4 expression in sh-SRSF3 GC cells (Fig. 4F). SRSF3 and PDCD4 IHC staining in nude mice also suggested that overexpressing hsa_circ_0000437 increased SRSF3 expression and reduced PDCD4 expression (Fig. 4G). All these findings suggest that hsa_circ_0000437 promoted GC growth by regulating the SRSF3/PDCD4 axis.
BGC-823 cells transfected with SRSF3 shRNA or control shRNA or co-transfected with hsa_circ_0000437 (Lv-circ) or the control (Lv-NC), cell colony formation assay (A), cell apoptosis by flow cytometry (B) and migration and invasion assays (C) were used. D Anti-SRSF3 antibody pulled down abundant SRSF3 by RIP assay. E Levels of PDCD4 mRNA detected by qRT-PCR after RIP for SRSF3 in BGC823 cells. F The expression of SRSF3 and PDCD4 were detected using Western blotting analysis. G Nude mouse xenograft tumors were detected by H & E staining and IHC analysis of SRSF3 and PDCD4. All data shown are means ± SD, *P < 0.05, **P < 0.01.
GC-secreted exosomal hsa_circ_0000437 can be transferred to HLECs
Upregulation of hsa_circ_0000437 in tissues positively correlated with LNM in GC (Fig. S4). Previous studies reported that circRNA transfer between cells was through exosomes by mediating intercellular communication [17]. Therefore, we wanted to see whether exosomes participated in hsa_circ_0000437 delivery from GC cells to HLECs, using exosomes purified from the supernatant of MKN-45 and BGC-823 cells. TEM showed that exosomes were present as a cup-shaped morphology (Fig. 5A). The size was measured using a Nan tracer (Fig. 5B). The expression of exosomes surface marker proteins CD9 and CD63 was determined by Western blotting analysis (Fig. 5C). The purified exosomes were labeled with green fluorescent film tracer PKH67 and then incubated with HLECs for 48 h. Many exosomes from GC cells entered HLECs (Fig. 5D). To confirm whether GC secreted hsa_circ_0000437 could be transferred to HLECs via exosomes, exosomes from BGC-823 cells transfected with Lv-circ/control Lv-NC, and MKN45 cells transfected with Lv-sh/control Lv-sh-NC (Fig. 5E), the corresponding expression of exosomal hsa_circ_0000437 in different groups was changed. Then, the level of hsa_circ_0000437 in HLECs pre-treated with these exosomes (823EXOLv-circ, 823EXOLv-NC, 45EXOLv-sh and 45EXOLv-sh-NC) from the above transfected cells was measured. The expression level of hsa_circ_0000437 in HLECs was significantly increased in 823EXOLv-circ group, and significantly decreased in 45EXOLv-sh group (Fig. 5F). All these results demonstrate that hsa_circ_0000437 transferred from GC cells to HLECs via exosomes.
A Electron microscopy micrographs of BGC-823 and MKN-45 cell conditioning medium secreted exosomes. Scale bar: 200 nm. B Exosome size distribution and number were analyzed by NanoSight particle tracker. C The expression of CD63 and CD9 in cells and exosomes was detected by Western blotting analysis. D HLECs were pre-treated with PKH67-labeled exosomes secreted by BGC-823 and MKN-45 for 48 h. Scale bar: 50 µm. E Exosomal hsa_circ_0000437 levels in BGC-823 cells transfected with Lv-circ/Lv-NC and MKN45 cells transfected with Lv-sh/Lv-sh-NC were measured using qRT-PCR. F hsa_circ_0000437 levels in HLECs pre-treated with indicated exosomes for 48 h were measured by qRT-PCR. All data are the means ± SD, *P < 0.05, **P < 0.01.
GC-secreted exosomal hsa_circ_0000437 promotes LNM in vitro and in vivo
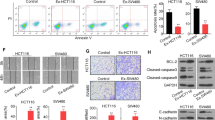
To investigate the role of exosomal hsa_circ_0000437 in lymphangiogenesis, HLECs were incubated with 823EXOLv-circ, 823EXOLv-NC, 45EXOLv-sh and 45EXOLv-sh-NC. The proliferation, migration, invasion, and tube formation of HLECs were analyzed. The results showed that there was no significant difference in HLEC proliferation between exosomes with high or low hsa_circ_0000437 expression level (Fig. S5A). Exosomes with high level of hsa_circ_0000437 dramatically promoted HLECs migration, invasion, and tube formation as compared with low hsa_circ_0000437 level group (Fig. 6A, B), suggesting that exosomal hsa_circ_0000437 promoted lymphangiogenesis in vitro. The effects of exosomal hsa_circ_0000437 on GC lymphangiogenesis and lymph node metastasis (LNM) were determined in the popliteal LNM model in vivo. BGC-823 cells were cultured, harvested, and injected intratumorally into the footpads of BALB/c nude mice. The mice were then equally randomized into three groups: injection with a PBS group, an 823EXOLv-NC group, and an 823EXOLv-circ group. When the primary tumors reached 150 mm3, the animals were sacrificed to obtain both the tumors and footpad-draining (popliteal and inguinal) LNs. Compared with the control or 823EXOLv-NC group, 823EXOLv-circ markedly enhanced the ability of GC cells to transfer to LNs (Fig. 6C–F). IHC showed that LNM and the expression level of LYVE-1 were increased in 823EXOLv-circ group (Fig. S5B). These findings suggest that GC cell-secreted exosomal hsa_circ_0000437 could promote lymphangiogenesis and LNM.
A Migration and invasion assays in HLECs pre-treated with indicated exosomes. The mean number of migrated/invaded cells per field was calculated. B Tube formation assay was performed in HLECs pre-treated with indicated exosomes. The mean number of branches per field was calculated. C The representative image of the popliteal lymph node metastasis model. D The representative image of inoculated popliteal lymph nodes. E Lymph node volume of all groups (n = 12). F The percentage of the lymph node metastasis in all groups (n = 12). The data are presented as the mean ± SD, *P < 0.05.
GC-secreted exosomal hsa_circ_0000437 induces LNM via HSPA2-ERK signaling pathway independent of VEGF-C
A previous study [18] showed that AKT and ERK pathways played important roles in tumor lymphangiogenesis and lymphatic metastasis. Therefore, we determined whether the ERK and AKT pathways in HLECs were activated by exosomal hsa_circ_0000437. HLECs were incubated with 823EXOLv-NC, 823EXOLv-circ, 45EXOLv-sh-NC and 45EXOLv-sh. The results indicated that phosphorylation of ERK1/2 was higher in exosomes with high hsa_circ_0000437 expression than that in exosomes with low hsa_circ_0000437 expression (Fig. 7A). Tumor-derived VEGF-C, a classic lymphangiogenic growth factor, induced lymphangiogenesis by activating the ERK pathway [19, 20]. It was found in our study that hsa_circ_0000437 overexpression in BGC823 or hsa_circ_0000437 knockdown in MKN-45 did not lead to VEGF-C mRNA or protein changes (Fig. S6A, B), suggesting that exosomal hsa_circ_0000437 acted through a VEGF-C-independent mechanism.
A The expression of ERK and p-ERK was detected by Western blotting analysis in HLECs incubated with PBS alone, 823EXOLv-circ, 823EXOLv-NC, 45EXOLv-sh, or 45EXOLv-sh-NC. B Silver staining of hsa_circ_0000437 pulldown. Arrows show different bands between the sense and antisense lanes. C Western blotting analysis of hsa_circ_0000437-enriched proteins after RNA pull-down. D The expression of HSPA2 were detected using Western blotting analysis. E The expression of ERK and p-ERK was detected by Western blotting analysis in HLECs. All data are the means ± SD, *P < 0.05, **P < 0.01, **P < 0.001.
Next, we investigated the molecular mechanism and interacting partners of hsa_circ_0000437 in HLECs cells incubated with 823EXOLv-circ. In vitro RNA pull-down assays with biotinylated hsa_circ_0000437 and antisense control showed obvious different bands (Fig. 7B). Protein mass spectrometry analysis was used to identify differentially expressed proteins. In the ranking list of recognized proteins, HSPA2 ranked first (Table S2). Western blotting analysis of hsa_circ_0000437-enriched proteins after RNA pull-down indicated that hsa_circ_0000437 bound specifically to HSPA2 (Fig. 7C). Moreover, overexpression of hsa_circ_0000437 slightly increased the total amount of HSPA2, and hsa_circ_0000437 knockdown inhibited its expression (Fig. 7D). Collectively, these results validated the interaction between hsa_circ_0000437 and HSPA2.
It has been proposed that HSPA2 plays an important role in the origin and development of lung adenocarcinoma via ERK1/2 pathway [21]. We observed the phosphorylation of ERK1/2 was lower in sh-HSPA2 transfection group than that in sh-NC group, and overexpression of hsa_circ_0000437 (Lv-circ) restored the expression inhibition of phosphorylation of ERK1/2 in sh-HSPA2 transfection group (Fig. 7E). Moreover, we also assessed whether HSPA2 regulates the LNM. Compared with the control group, knockdown of HSPA2 significantly suppressed the expression of LNM-related protein MMP-2, LYVE-1 and ICAM-1, and hsa_circ_0000437 (Lv-circ) also reversed the inhibitory effect of sh-HSPA2 on the expression of these LNM-related protein to a certain extent in HLECs (Fig. S6C). Taken together, these results indicate that GC cells induce LNM by transmitting exosomal hsa_circ_0000437 to HLECs via HSPA2-ERK signaling pathway independent of VEGF-C.
Exosomal hsa_circ_0000437 is associated with LNM in GC patients
Serum exosomal hsa_circ_0000437 expression in GC patients was higher than that in healthy controls. In addition, serum exosomal hsa_circ_0000437 expression GC patients with LNM was also significantly higher than that in GC patients without LNM (Fig. 8A, B). The correlation between the serum exosomal hsa_circ_0000437 level and the clinicopathological features was determined by dividing patients into a high-expression group and a low-expression group according to the median hsa_circ_0000437 expression level. There was a significant correlation between high serum exosomal hsa_circ_0000437 expression and T stage, N stage, and differentiation (Table 1). Receiver operating characteristic (ROC) analysis indicated that serum exosomal hsa_circ_0000437 was able to discriminate between GC patients and healthy controls. AUC of serum exosomal hsa_circ_0000437 was 0.808 with 95% CI 0.734-0.882, indicating a high diagnostic accuracy (Fig. 8C). Comparison between GC patients with and without LNM showed that AUC of serum exosomal hsa_circ_0000437 was 0.716 with 95% CI 0.596-0.837 (Fig. 8D). Kaplan-Meier survival curve results showed that high serum exosomal hsa_circ_0000437 level was associated with poor survival outcomes in GC (Fig. 8E, F). All these results suggest that exosomal hsa_circ_0000437 may prove to be a potential diagnostic biomarker and therapeutic target for LNM GC (Fig. 8G).
Serum exosomal hsa_circ_0000437 expression in GC patients and healthy controls (A) and GC patients with or without lymphatic metastasis (B) was detected by qRT-PCR. ROC analysis of serum exosomal hsa_circ_0000437 for GC (C) and LN metastasis (D). Overall survival curves of GC patients according to the relative serum exosomal hsa_circ_0000437 expression (E) and lymphatic metastasis states (F). G The schematic model of hsa_circ_0000437 enhances tumorigenesis and promotes lymphatic metastasis in GC. hsa_circ_0000437 acted as a tumor oncogene in GC by forming a regulatory loop consisting of hsa_circ_0000437/SRSF3/PDCD4. GC-secreted exosomal hsa_circ_0000437 promoted lymphangiogenesis and lymphatic metastasis via the ERK pathway by transfer into HLECs. The data are presented as the mean ± SD, **P < 0.01, ***P < 0.001.
Discussion
Recent studies [22] have demonstrated that circRNAs are differentially expressed in multiple human malignant tumors, playing indispensable roles in cancer occurrence and progression. A previous study [23] conducted large-scale RNA sequencing of GC and established the circRNA expression profile, which found that CORO1C derived circRNAs were significantly different between GC patients and healthy controls. However, their functions have not yet been reported in GC. Combined with our previous results of circRNA-seq analysis [15], it was found in this study that CORO1C-derived circRNA hsa_circ_0000437 expression was significantly increased in GC tissues and cells, originating from the back-splicing of CORO1C exons 4 and 5. The results of biological function assay in vitro showed that hsa_circ_0000437 promoted GC cell proliferation, invasion, and migration, which was later verified by the in vivo nude mouse experiments.
Many circRNAs exert their biological functions by acting as microRNA or protein inhibitors (‘sponges’) by regulating protein functions or via self-translation [24,25,26]. Rong et al. discovered that circPSMC3 promoted GC proliferation by sponging miRNA-296-5p with phosphatase and tensin homolog [27]. Another circRNA, circDONSON, induced cisplatin resistance of GC cells by regulating the miR-802/BMI1 axis [28]. Yang et al. reported that circZKSCAN1 competitively bound FMRP against its complex with CCAR1 to negatively regulate hepatocellular carcinoma (HCC) stem cells [29]. In the present study, our fluorescence in situ hybridization showed that the content of hsa_circ_0000437 was almost distributed in the nucleus. The results of ChIRP/MS and PRM showed that hsa_circ_0000437 bound SRSF3, the smallest member of the Ser/Arg-rich protein family that worked as a constitutive and alternative splicing regulator to regulate several aspects of the gene expression program, and that the aberrant expression of SRSF3 was associated with breast cancer [30], head & neck SCC [31], and glioblastoma [32], colorectal cancer [33]. Park et al. found that SRSF3 participated in PDCD4 mRNA alternative splicing, export and translation, resulting in the downregulation of the PDCD4 protein in GC [34]. Our results suggest that hsa_circ_0000437 and SRSF3 act to control GC cell growth similarly, such that the decreased growth induced by SRSF3 knockout could be ameliorated by hsa_circ_0000437 over-expression. Hsa_circ_0000437 also affected PDCD4 protein expression by regulating the expression of SRSF3. Wu et al. [35] found that the expression of PDCD4 in GC metastatic peritoneal tissues was significantly lower than that in normal tissues, which promoted the apoptosis of peritoneal mesothelial cells. We demonstrated that PDCD4 acted as a tumor suppressor gene and could be partially reversed by SRSF3 in GC. This finding suggests that hsa_circ_0000437 may suppress PDCD4 expression through recruiting SRSF3 in cells. Hsa_circ_0000437 may form a triple complex with SRSF3 and PDCD4 and inhibits the translation of the tumor suppressor gene PDCD4, thereby promoting tumor progression.
Regional LNM occurs in early stages of GC [36]. Cancer-related mortality and communication with the tumor microenvironment are critical factors in tumor metastasis [37]. Exosomes play a critical role in remodeling of the premetastatic microenvironment [38]. Some recent studies have identified circulating exosomal circRNAs as promising cancer biomarkers in that they can reflect dysregulated expression patterns in a variety of human cancer types. In addition, they can be obtained through a non-invasive way and remain highly stable in circulation [38,39,40]. We analyzed the expression levels of hsa_circ_0000437 with clinical data and found that the level of hsa_circ_0000437 was associated with LNM in GC. It was found that hsa_circ_0000437 was overexpressed in serum exosomes from GC patients and was associated with LNM, suggesting that hsa_circ_0000437 may be a potential molecular target for clinical intervention in GC patients with LNM. We also found that HLECs internalized GC-secreted exosomal hsa_circ_0000437 to promote migration, invasion, and tube formation but not proliferation, thereby facilitating lymphangiogenesis and lymphatic metastasis. The popliteal LNM animal model confirmed this result in vivo.
VEGF-C is an important lymphangiogenic growth cytokine [41]. Some studies have demonstrated that VEGF-C signaling induces lymphangiogenesis by activating ERK and AKT pathways [42]. In the present study, HLECs treated with exosomes with high hsa_circ_0000437 increased the phosphorylation of ERK1/2 proteins but not AKT proteins (date not listed), suggesting a potential mechanism for hsa_circ_0000437-induced lymphangiogenesis in GC. Interestingly, overexpression and knockdown of hsa_circ_0000437 did not cause the changes of VEGF-C mRNA or protein in GC cells, suggesting that hsa_circ_0000437 activated the ERK signaling pathway and might act in HLECs in a VEGF-C independent manner. RNA pull-down indicated that hsa_circ_0000437 bound specifically to HSPA2, which regulated the expression of LNM-related protein in HLECs by ERK1/2 pathway. Antagonistic hsa_circ_0000437 combined with anti-VEGF-C therapy may represent a new anti-lymphangiogenetic target, which appears to be a promising direction for the treatment of early-stage GC patients with LNM.
Conclusion
The presents study demonstrated that hsa_circ_0000437 acted as a tumor oncogene in GC by forming a regulatory loop that consisted of hsa_circ_0000437/SRSF3/PDCD4. HLECs promoted lymphangiogenesis and LNM by internalizing GC-secreted exosomal hsa_circ_0000437 via the ERK pathway in a VEGF-C-independent manner. These findings suggest that hsa_circ_0000437 may prove to be a potential biomarker for survival of GC patients with LNM, and a target for GC therapy as well.
Materials and methods
The main methods used in this study are listed in Supplementary Materials and Methods, including cell line RNA isolation, quantitative real-time PCR (qRT-PCR), lentivirus infection, cell lines, cells transfection and proliferation assays, transwell migration, invasion and apoptosis assays, immunohistochemistry (IHC), Western blotting analysis, enzyme-linked immunosorbent assay (ELISA), isolation and characterization of exosomes, and exosome labeling and uptake.
Clinical specimens
48 pairs of GC tissues and corresponding adjacent noncancerous tissues were collected from the Affiliated Hospital of Nantong University (Nantong, China) from 2018 to 2020 following the Helsinki Declaration. During the same period, a 5-ml peripheral blood sample was collected from each of 76 GC patients who were not enrolled in this study and had not received any chemo- or radiotherapy before surgery or had any other malignancies. All specimens were stably stored at −80 °C until use. Informed consent was obtained from all the subjects before specimen collection, and the research protocol was approved by the Ethics Committee of the said hospital.
Animal experiments
The animal experimental procedures were approved by the animal management committee of Nantong University and followed the ethics guidelines for animal experiments. A xenograft tumor model was established in 12 4-week-old female nude mice. They were equally randomized into three groups: a hsa_circ_0000437 overexpression group (Lv-circ), a control (Lv-NC) group, and an untransfected (NC) group. Approximately 1 × 107 BGC-823 cells transfected with Lv-NC, Lv-circ, or NC were subcutaneously injected into the axilla. Tumor volumes were measured every 4 days as follows: volume = (width)2 × length/2. After 28 days, all injected nude mice were sacrificed. The subcutaneous tumors were excised, weighed, and stained with hematoxylin and eosin (H&E), and IHC. BGC-823 cells (5 × 106) were injected into the footpad of the 12 female nude mice for the popliteal lymph node metastasis (LNM) model. These mice were divided into three (NC, Lv-NC and Lv-circ) groups randomly. When the tumor volume of NC group reached 50 mm3, animals in the three groups were respectively injected with PBS, exosomes from BGC-823 cells transfected with Lv-NC (823EXOLv-NC), or Lv-circ (823EXOLv-circ) into the tumor twice a week. After five injections, when the tumors reached a comparable size of 150 mm3 in PBS group, the mice were sacrificed. Popliteal lymph nodes (LNs) were paraffin-embedded and analyzed by H&E staining. The proportion of metastasis-positive to total LNs including inguinal LNs and popliteal LNs was calculated. IHC staining for LYVE-1 was analyzed.
ChIRP-MS
It was performed as described previously [43]. Briefly, about 5 × 109 GC cells were ground into powder and resuspended with pre-cooled PBS. The formaldehyde solution was cross-linked at room temperature for 30 min. Glycine was added to terminate the reaction, and precipitation was obtained by centrifugation. After combining the biotin-labeled probe with the magnetic beads for 30 min, the probe was mixed with the sample and hybridized overnight at 37 °C. Subsequently, the beads were washed by elution buffer containing 20 U benzonase at 37 °C for 1 h. The crosslinking reaction was carried out at 95 °C for 30 min. Finally, the protein was precipitated using 0.1% sodium deoxycholate and 10% trichloroacetic acid. The polypeptides were separated using a nano-UltraPerformance LC liquid phase system (EASY-NLC1200) and detected with a mass spectrometer (Q-Exactive).
PRM
PRM experiments were carried out as previously described [44]. A 5-μL sample of tryptic polypeptide was dissolved in a 0.1% formic acid acetonitrile aqueous solution. The sample was injected and separated using a 100 μm ID × 15-cm reverse-phase chromatographic column (Reprosil-Pur 120 C18). After nano-UltraPerformance LC separation, PRM data were collected by MS and imported into skyline software for further analysis. The experiment was assisted by Kangchen Biotech (Shanghai, China). The identification criteria of differentially expressed peptide were set at P < 0.05 and fold-change >2. Gene ontology enrichment analysis was performed by Blast2GO software and KEGG pathway analysis was performed by KEGG software.
Tube formation assay
75 μl precoated growth factor-reduced Matrigel (BD Biosciences, CA, USA) was placed in a 48-well plate and solidified at 37 °C for 2 h, on which 2 × 105 HLECs were seeded and incubated with either PBS or exosomes for 12 h. The number of lymphatic tubes was determined using an inverted microscope (Olympus, Tokyo, Japan).
RNA fluorescence in situ hybridization
The U6 probe, 18S probe and hsa_circ_0000437 probe labeled by Cy3 were designed and synthesized by GenePharma Biotechnology Co., Ltd. (Shanghai, China). GC cells were seeded onto round coverslips overnight. After fixation with 4% paraformaldehyde and permeabilization with 0.5% Triton-100, hybridization was carried out with the probes overnight at 37 °C. After hybridization, the slice was stained with DAPI at room temperature for 10 min, sealed and observed under the confocal microscope (Leica Microsystems, Germany).
Statistical analysis
SPSS 20.0 and GraphPad Prism v7.0 software were used for statistical analysis. Data are shown as means ± standard deviation (SD). Comparisons between different groups were analyzed using paired or unpaired t-tests. The χ 2-test was applied for categorical variables. Survival curves were drawn according to the Kaplan-Meier method. Sensitivity, specificity, and are under the curve (AUC) were computed using the constructed ROC curves and optimal cut-off values were calculated according to Youden index. Differences were statistically significant when P < 0.05.
References
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet 2020;396:635–48.
Feng W, Ding Y, Zong W, Ju S. Non-coding RNAs in regulating gastric cancer metastasis. Clin Chim Acta. 2019;496:125–33.
Cao YZ, Sun JY, Chen YX, Wen CC, Wei L. The roles of circRNAs in cancers: Perspectives from molecular functions. Gene 2021;767:145182.
Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19:14.
Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90.
Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30.
Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics 2020;10:3503–17.
Ma S, Gu X, Shen L, Chen Y, Qian C, Shen X, et al. CircHAS2 promotes the proliferation, migration, and invasion of gastric cancer cells by regulating PPM1E mediated by hsa-miR-944. Cell Death Dis. 2021;12:863.
Liang Y, Wang H, Chen B, Mao Q, Xia W, Zhang T, et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Ther Nucleic Acids. 2020;23:355–68.
Wang G, Sun D, Li W, Xin Y. CircRNA_100290 promotes GC cell proliferation and invasion via the miR-29b-3p/ITGA11 axis and is regulated by EIF4A3. Cancer Cell Int. 2021;21:324.
Cheng X, Wang X, Wu Z, Tan S, Zhu T, Ding K. CORO1C expression is associated with poor survival rates in gastric cancer and promotes metastasis in vitro. FEBS Open Biol. 2019;9:1097–108.
Lim JP, Shyamasundar S, Gunaratne J, Scully OJ, Matsumoto K, Bay BH. YBX1gene silencing inhibits migratory and invasive potential via CORO1C in breast cancer in vitro. BMC Cancer. 2017;17:201.
Wang Z, Jia L, Sun Y, Li C, Zhang L, Wang X, et al. CORO1C is associated with poor prognosis and promotes metastasis through PI3K/AKT pathway in colorectal cancer. Front Mol Biosci. 2021;8:682594.
Wu Y, Zhang Y, Zheng X, Dai F, Lu Y, Dai L, et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol Cancer. 2020;19:99.
Ma S, Kong S, Gu X, Xu Y, Tao M, Shen L, et al. As a biomarker for gastric cancer, circPTPN22 regulates the progression of gastric cancer through the EMT pathway. Cancer Cell Int. 2021;21:44.
Kim J, Park RY, Chen JK, Kim J, Jeong S, Ohn T. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5’-UTR region. Cell Death Differ. 2014;21:481–90.
Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and exosomes: A mysterious frontier for human cancer. Mol Ther Nucleic Acids. 2020;19:384–92.
Maeng YS, Aguilar B, Choi SI, Kim EK. Inhibition of TGFBIp expression reduces lymphangiogenesis and tumor metastasis. Oncogene 2016;35:196–205.
Ishii M, Takahashi M, Murakami J, Yanagisawa T, Nishimura M. Vascular endothelial growth factor-C promotes human mesenchymal stem cell migration via an ERK-and FAK-dependent mechanism. Mol Cell Biochem. 2019;455:185–93.
Liu L, Lin C, Liang W, Wu S, Liu A, Wu J, et al. TBL1XR1 promotes lymphangiogenesis and lymphatic metastasis in esophageal squamous cell carcinoma. Gut 2015;64:26–36.
Cao L, Yuan X, Bao F, Lv W, He Z, Tang J, et al. Downregulation of HSPA2 inhibits proliferation via ERK1/2 pathway and endoplasmic reticular stress in lung adenocarcinoma. Ann Transl Med. 2019;7:540.
Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16:292–301.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71.
Zhang J, Hou L, Liang R, Chen X, Zhang R, Chen W, et al. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR-502-5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 2019;18:80.
Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. 23
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18:25.
Liu Y, Xu J, Jiang M, Ni L, Ling Y. CircRNA DONSON contributes to cisplatin resistance in gastric cancer cells by regulating miR-802/BMI1 axis. Cancer Cell Int. 2020;20:261.
Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics 2019;9:3526–40.
Ke H, Zhao L, Zhang H, Feng X, Xu H, Hao J, et al. Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc Natl Acad Sci USA. 2018;115:E3426–E3435.
Xu L, Shen J, Jia J, Jia R. Inclusion of hnRNPL alternative exon 7 is associated with good prognosis and inhibited by oncogene SRSF3 in head and neck squamous cell carcinoma. Biomed Res Int. 2019;2019:9612425.
Song X, Wan X, Huang T, Zeng C, Sastry N, Wu B, et al. SRSF3-regulated RNA alternative splicing promotes glioblastoma tumorigenicity by affecting multiple cellular processes. Cancer Res. 2019;79:5288–301.
Kuranaga Y, Sugito N, Shinohara H, Tsujino T, Taniguchi K, Komura K, et al. SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer-specific energy metabolism in colon cancer cells. Int J Mol Sci. 2018;19:3012.
Park SK, Jeong S. SRSF3 represses the expression of PDCD4 protein by coordinated regulation of alternative splicing, export and translation. Biochem Biophys Res Commun. 2016;470:431–8.
Wu P, Wang J, Mao X, Xu H, Zhu Z. PDCD4 regulates apoptosis in human peritoneal mesothelial cells and promotes gastric cancer peritoneal metastasis. Histol Histopathol. 2021;36:447–57.
Mu GC, Huang Y, Liu ZM, Wu XH, Qin XG, Chen ZB. Application value of nomogram and prognostic factors of gastric cancer patients who underwent D2 radical lymphadenectomy. BMC Gastroenterol. 2019;19:188.
Jung HY, Fattet L, Yang J. Molecular pathways: linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin Cancer Res. 2015;21:962–8.
Kok VC, Yu CC. Cancer-derived exosomes: Their role in cancer biology and biomarker development. Int J Nanomed. 2020;15:8019–36.
Guo X, Tan W, Wang C. The emerging roles of exosomal circRNAs in diseases. Clin Transl Oncol. 2021;23:1020–33.
Wang M, Yu F, Li P, Wang K. Emerging function and clinical significance of exosomal circRNAs in eancer. Mol Ther Nucleic Acids. 2020;21:367–83.
Schwager S, Detmar M. Inflammation and lymphatic function. Front Immunol. 2019;10:308.
Deng Y, Zhang X, Simons M. Molecular controls of lymphatic VEGFR3 signaling. Arterioscler Thromb Vasc Biol. 2015;35:421–9.
Chai P, Jia R, Jia R, Pan H, Wang S, Ni H, et al. Dynamic chromosomal tuning of a novel GAU1 lncing driver at chr12p13.32 accelerates tumorigenesis. Nucleic Acids Res. 2018;46:6041–56.
Zhu Y, Li J, Bo H, He D, Xiao M, Xiang L, et al. LINC00467 is up-regulated by TDG-mediated acetylation in non-small cell lung cancer and promotes tumor progression. Oncogene 2020;39:6071–84.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81871720, 82072363), Nantong Science and Technology Project (MS12021002). The project of Nantong Commission of Health (MB2021061).
Author information
Authors and Affiliations
Contributions
XS and SK designed the project. XS and SM performed the experiments. LS collected the GC samples and clinical information. JQ and QW processed the data. XS and SJ wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shen, X., Kong, S., Ma, S. et al. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis. Oncogene 41, 4724–4735 (2022). https://doi.org/10.1038/s41388-022-02449-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02449-w
- Springer Nature Limited
This article is cited by
-
Cancer-derived exosomes as novel biomarkers in metastatic gastrointestinal cancer
Molecular Cancer (2024)
-
N6-methyladenosine modified circPAK2 promotes lymph node metastasis via targeting IGF2BPs/VEGFA signaling in gastric cancer
Oncogene (2024)
-
Circular RNAs in EMT-driven metastasis regulation: modulation of cancer cell plasticity, tumorigenesis and therapy resistance
Cellular and Molecular Life Sciences (2024)
-
Helicobacter pylori upregulates circPGD and promotes development of gastric cancer
Journal of Cancer Research and Clinical Oncology (2024)
-
Exosomal circRNA: emerging insights into cancer progression and clinical application potential
Journal of Hematology & Oncology (2023)