Abstract
Metastases are often the direct cause of death from pancreatic ductal adenocarcinoma (PDAC). The role of genomic alterations (GA) in mediating tropism and metastasis formation by PDAC cells is currently unknown. We aimed to identify GAs predisposing colonization of PDAC cells to the liver and decipher mechanisms enabling this process. In order to reveal specific genes, we studied the frequency of GA in 8,880 local and 7,983 metastatic PDAC samples. We observed differential pattern of GA in the local tumor and specific metastatic sites, with liver metastases characterized by deletion of CDKN2A/B (encoding p16/p15, respectively). The role of CDKN2A/B in promoting liver metastasis was evidenced by enhanced tumorigenic phenotype of p15/p16-deleted PDAC cells when exposed to hepatocytes conditioned media. The liver is characterized by high-ammonia low-glutamine environment and transcriptomic assays indicated unique adaptation of PDAC cells to these conditions, including regulation of genes leading to reduced glutaminolysis, like overexpression of GLUL and reduction in GLS2. Furthermore, metabolic assays indicated an increase in glutamate derived from [U-13C]-glucose in p15/p16-deleted cells. Importantly, these cells thrived under high ammonia condition. These data suggest a unique role for genomic alterations in mediating tropism of PDAC. Among these alterations, p15/16 deletion was identified as a promoter of liver metastases. Further studies indicated a unique role for p15/16 in regulating glutaminolysis. These findings reveal vulnerabilities in PDAC cells, which may pave the way for the development of novel therapeutic strategies aiming at the prevention of liver metastases formation.
Similar content being viewed by others
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive malignancies [1]. The majority of patients have metastases at diagnosis, and over 70% of those diagnosed at an early stage will also develop metastases [2]. Metastases are often the direct cause of death of these patients and the 5-year survival rate following development of metastases is less than 5% [3]. Hence, elucidating factors associated with metastases formation is of utmost importance. PDAC metastasizes primarily to the liver, followed by the peritoneum and lung [4].
The genomic landscape of local PDAC has been well-characterized, with KRAS, CDKN2A, TP53 and SMAD4, being the most commonly altered genes [5]. Yet, data regarding the role of genomic alterations (GA) in mediating PDAC metastases formation and predilection to specific organs (tropism) is scarce [6, 7]. It has been presumed, based on a small number of cases, that local and metastatic lesions of PDAC share a similar genomic landscape [7]. However, a recent study cast doubts on this assumption, demonstrating that cell cycle progression gene signature is markedly greater in PDAC metastases compared to local tumors [8]. In various malignancies, GA and molecular characteristics have been clearly associated with predilection to form metastases in specific organs. For example, in breast cancer, bone is the predominant site of metastasis for estrogen receptor-positive tumors, while brain and liver metastases are more common in HER2-overexpressing tumors [9]. Yet, interactions between specific GA and tropism of PDAC are currently unknown, possibly due to low number of specimens analyzed.
The liver micro-environment conditions are dramatically different than those encountered by PDAC cells in the pancreas [4]. Among the differences are the portal vein supply and the unique metabolic activities of the liver, which serves as a hub for glucose, glutamine and ammonia metabolism [10, 11]. Thus, liver colonization by pancreatic cancer cells requires complex adaptation processes providing survival advantage and allowing cells to thrive in this unique environment. These processes are likely to be mediated by GA leading to unique transcriptional reprogramming. Several factors have been implicated in the promotion of liver tropism. Ovarian cancer cells were shown to utilize glutamine and ammonia accumulated in the liver as building blocks for amino acid synthesis through increased activity of the enzyme glutamate dehydrogenase (GDH), thus supporting tumor growth and metastasis [10]. Another example is the crucial role played by CXCL8 and its receptor in the formation of liver metastasis by colorectal cancer [12].
Alterations in CDKN2A occur in ~40% of PDAC [13]. CDKN2A and CDKN2B (encoding p16 and p15 proteins, respectively) are adjacent on chromosome 9p, share high homology [14] and are commonly co-deleted [15]. The proteins encoded by these genes, p15 and p16, belong to the class of G1 cell cycle inhibitors, interfering with the catalytic activity of cyclin dependent kinases CDK4 and CDK6 [16]. In the context of PDAC, most attention was focused on p16, with hardly any study focusing on p15. However, the predominance of homozygous deletions of both genes suggests functional advantage of simultaneous inactivation in mediating pro-tumorigenic processes [15]. We hypothesized that promotion of organ-specific tropism by PDAC cells requires acquisition of specific GA enabling adaptation to the unique micro-environment of each hosting organ and supporting pro-metastatic niche formation. To identify such alterations, we studied a large dataset of PDAC clinical samples and identified, for the first time, site-specific patterns of GAs, including higher rate of CDKN2A and CDKN2B deletion in liver metastases.
Studies in PDAC cells indicated association between p15/p16 not only with cell proliferation, but also with aggressive phenotype and predilection to grow under liver conditions. Furthermore, manipulation of p15/p16 expression revealed unique transcriptional and metabolic rewiring, supporting growth in the liver micro-environment. Thus, our data suggests a role for CDKN2A/CDKN2B deletion in mediating liver tropism of PDAC, partly independent to their well-known role as cell cycle inhibitors. These observations may pave the way for the design of rational therapies aimed at the prevention of metastases formation, thus leading to improved survival of patients with PDAC.
Results
Increased prevalence p15/p16 co-deletion in PDAC liver metastasis
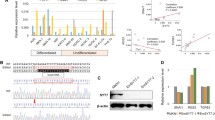
Data on the prevalence of GA in 16,863 PDAC clinical samples derived from local (n = 8,880) and metastatic (n = 7,983) sites was obtained from a large real-world database (Foundation Medicine) through a collaborative agreement. Distribution of metastatic sites is presented in Fig. 1A. Most of the metastatic samples were obtained from the liver (n = 5,883; 73.6%), followed by lungs (n = 705; 8.8%), lymph nodes (n = 465; 5.8%), omentum (n = 365; 4.5%), duodenum (n = 288, 3.6%) and stomach (n = 100, 1.2%) with rare representation of bone and brain metastases (n = 52, n = 17, respectively). Patients with liver metastatic biopsies were significantly younger compared to lung biopsies with a median age of 65 vs. 68, respectively (p = 2.05e−13). Median tumor mutation burden (TMB) was modestly higher in liver metastasis samples compared to other metastasis (1.7 vs 1.2, respectively p = 0.00017) (Table 1). In total 27,136 GA were detected (including short varaints, copy number and rearrangement/fusion mutations), and their prevalence in metastatic samples was compared to local tumor samples. The most significant differences were observed in MYC, CDKN2B and CDKN2A (Fig. 1B), suggesting they confer a more aggressive disease. As our aim was to reveal GA assoaicated with predilection to specific metastatic sites, particularely to the liver, we further analyzed GA prevalence according to local and different metastatic sites. The results showed that most GA are evenly distributed among the local and specific metastatic sites (Fig. 1C). Yet, differences were observed in specific genes. In accordance with Fig. 1B, KRAS alterations (SV, CN, RE) are more prevalent in the local tumor than the metastatic sites, especially when compared to lymph nodes. However, p53 alterations (SV, CN, RE) are less prevalent in lung and omentum metastases compared to other tumor sites.
A Distribution of 16,863 PDAC clinical samples, analyzed for genomic alterations, according to biopsy site. B Analysis of the most significant gene alterations in PDAC local vs metastatic sites. C Analysis of alteration distribution between local and most common metastatic sites. D A comparison of CDKN2A/p16 and CDKN2B/p15 alteration prevalence between the different metastatic sites.
We focused on alterations of CDKN2A and CDKN2B, as these showed the most pronounced differences between the liver metastasis and local tumors as well as other metastatic sites (Fig. 1C). Consistent with their role as tumor suppressors [15], the GA observed in these genes were primarily deletions but included also rarer loss of function mutations (nonsense, frameshift, splice, truncating rearrangements). Thus, deletion of CDKN2A or CDKN2B was noted in 49% and 22% of local tumors, and in 63% and 38% in liver metastases, respectively (Fig. 1D). The prevalence of CDKN2A or CDKN2B deletion was significantly lower in other metastatic sites (CDKN2B or example, lungs 23%, lymph nodes and omentum 18%, and duodenum 15% Fig. 1D), suggesting a unique role for p15/p16 in liver tropism. Moreover, liver metastasis were not enriched with alterations in the PDAC associated genes KRAS, p53 or SMAD4 (Fig. 1B, C), emphasizing the unique role of p15/p16 in liver predilection.
In order to evaluate whether CDKN2A/B deletions are a general characteristic of liver metastasis, we analyzed the prevalence of these deletions in metastases originated from other malignancies, and discovered enrichment only in liver metastases obtained from other pancreatic malignancies and gastric adenocarcinoma. In contrast, lower prevalence of CDKN2A/B deletions were detected in liver metastases of melanoma, lung or ovarian cancers compared to other metastatic sites. For example only 23% of melanoma liver metastasis harbored CDKN2A/B deletions, compared to 42% in other metastatic sites (Table 2). Taken together, these data describe an association between CDKN2A/B deletion and PDAC liver metastases.
CDKN2B/p15 and CDKN2A/p16 co-deletion enhances aggressive phenotype of PDAC cells
CDKN2A and CDKN2B are often co-deleted, and while the role of CDKN2A in tumorigenic phenotype was widely explored, the contribution of CDKN2B silencing to PDAC development was less studied [15]. We therefore examined the role of p15 and p16 either alone or in combination in PDAC cell lines phenotype. We first analyzed p15 and p16 mRNA and protein expression in CAPAN-2, MIA PaCa-2 and PANC-1 cells, originating from high grade PDAC local tumor [17] and COLO-357 cells, originating from low grade PDAC lymph node metastasis [18]. As previously reported [19] no mRNA (Fig. 2A) or protein (Fig. 2B) expression of either p15 or p16 was detected in CAPAN-2, MIA PaCa-2 or PANC-1 cells, whereas high mRNA and protein levels of both genes were detected in COLO-357 cells. We first generated PDAC isogenic cell lines, with or without p15 and/or p16 expression. We overexpressed either p15, p16 or their combination in the p15/p16-deleted cell lines PANC-1 and MIA PaCa-2 (Fig. 2D). The opposite model was generated, where we employed the COLO-357 cells, and stably silenced either p15 (COLO-357sh-p15), p16 (COLO-357sh-p16), or their combination (COLO-357sh-p15/p16) and selected single clones for further studies (Fig. 2C, and detailed in Figs. S1, S2). Silencing of p15 or p16 increased colony formation while their co-silencing further increased colony number (Fig. S3A). Silencing of p15 alone yielded a less pronounced effect, possibly due to compensatory increase in p15 level following p16 silencing (Fig. 2C). While the role of p15 and p16 in cell cycle regulation and hence in proliferation is established, the role it plays in migration, invasion and metastasis formation is less characterized [15, 20]. We first studied the effect of p15/p16 co-deletion on cell migration using a wound healing assay. In order to neutralize the effects of p15 and p16 on cell proliferation, the assay was performed in the presence of the cell-cycle inhibitor mitomycin C. The results show that following mitomycin C treatment, p16 silencing increased migration, whereas p15 silencing did not affect it. Yet, co-silencing increased migration beyond that of p16 silencing (Fig. 2E). Trans-well migration and invasion assays exhibited a similar trend, thus whereas p15 silencing only slightly increased these properties, p16 exhibited a stronger effect and co-silencing markedly increased cell migration and invasion (Fig. S3B,C). We next studied the ability of the manipulated COLO-357 cells to form 3D spheres and noted that co-silencing of both p16/p15 enabled cells to form 3D spheres. Analysis of viability of sphere cells indicated a twofold higher cell number of p15/p16 co-silenced cells compared to sh-NS and also compared to p16 single silencing (Fig. 2F). Therefore, only co-silencing allowed sphere formation, which tests cells’ aggressive phenotype.
The mRNA (A) and protein (B) level of CDKN2A/p16 and CDKN2B/p15 was studied in PDAC cells. β-actin was used as loading control. C COLO-357 cells were infected with shRNA againt p15, p16 their combination (sh-p15, sh-p16, sh-p15/p16) or control anti sense (sh-NS) and Western blot analysis was conducted. Quantitation relative to sh-NS and β-actin is depicted. D PANC-1 and MiaPaCa-2 cells were transfected with p15, p16 or their combination and their expression was validated using Western blot analysis. E A wound healing assay (scratch) was conducted with COLO-357sh-NS compared to silenced cells. Cells were photographed at 0 and 24 h and migratory lengths were quantified. F Cell spheres of COLO-357sh-NS and silenced cells or (G) MIAPaCa-2 cells transfected with control vector or p15/p16, were generated as described in “Materials and Methods”, and their viability was evaluated after seven days. The figures show representative results of three independent experiments. Each bar represents ± SD, statistical analysis was performed using one way Anova or two way Anova (***, p < 0.001).
The opposite was observed in PANC-1 and MIA PaCa-2 cells with p15 and p16 overexpression. Expression of both or either protein significantly decreased colony formation of both cell lines (Fig. S3D, E), interestingly, p15 expression exerted the strongest inhibitory effect. Co-overexpression of p15/p16 significantly decreased 3D sphere cell number of MIA PaCa-2 cells (Fig. 2G). Moreover, co-overexpression decreased PANC-1 cell migration by 50% (Fig. S3F) and inhibited MIA PaCa-2 sphere formation by 51% (Fig. S3G). Taken together, these data indicate a role for p15/p16 in regulating aggressive phenotype of PDAC cells, beyond their effect on cell cycle.
p15/p16 co-deletion enhanced tumorigenicity of PDAC cells in hepatocyte-conditioned media
We next aimed assess the effect of p15 and p16 deletion on PDAC cell tumorigenic phenotypes within the liver microenvironment. For this aim, we studied the effects of conditioned media (CM) derived from human hepatocytes (hCM) on the above described manipulated cells. hCM reduced viability of COLO-357sh-NS cells by fivefold, but did not affect viability of COLO-357sh-p15/p16 cells (Fig. 3A). Reciprocal experiments using PANC-1 cells revealed that while hCM elevated proliferation of cells transfected with a control vehicle by 1.7-fold compared to control media (Fig. 3B), it reduced proliferation of cells co-expressing p15/p16 by twofold. We assessed sphere formation of COLO-357sh-p15, COLO-357sh-p16, COLO-357sh-p15/p16 compared to control cells, and used mouse CM (mCM) for these experiments, as the cells failed to form colonies in human CM. Under these conditions, only COLO-357sh-p16 and COLO-357sh-p15/p16 were able to form spheres in mCM (Fig. 3C) and cell viability in COLO-357sh-p15/p16 spheres was threefold higher than in control or p15-silenced cells (Fig. 3D). While p16 silencing led to sphere formation and increased viability compared to p15-silencing or control cells, its maximal effect was observed at 24 hours, and cell viability declined afterwards (Fig. 3D). Thus, in hepatocyte conditions, co-deletion is required for sphere formation. As expected, the opposite was observed using MIA PaCa-2 cells overexpressing p15/p16. Thus, mCM elevated viability of pCDNA3 transfected MIA PaCa-2 sphere cells 1.5-fold more than p15/p16 expressing cells (Fig. 3E). Taken together, these data indicate that p15/p16 co-deletion may provide an advantage for PDAC cells to thrive in the liver microenvironment.
A COLO-357sh-NS and COLO-357sh-p15/p16 cells were grown with human heptocytes conditioned media (hCM) or the respective control media and viability was assessed by methylene blue assay. B PANC-1 cells were co-transfected with p15 and p16 or pCDNA3 and medium was changed to hCM or the respective control media. Viability was assessed as described in (A). C, D Sphere of COLO-357sh-NS, COLO-357sh-p15, COLO-35sh-p16 or COLO-357sh-p15/p16 cells were generated, and medium was changed to hepatocyte mouse CM (mCM) or control media. D Viability was assayed by Realtime-Glow MT at 0 h, 24 h, 48 h and 72 h (mCM, marked as “+”, control media marked as “−”) and (C) spheres were photographed at the end of the experiment (72 h). E Spheres of MIA PaCa-2 cells co-transfected with p15 + p16 or control pCDNA3 were processed as in (C), and viability determined similarly at 72 h. Three independent experiments were performed and a representative experiment is shown. Each bar represents ± SD, statistical analysis was performed using two-way Anova (**, p < 0.01, ***, p < 0.001).
p15/p16 co-deletion in PDAC cells enhanced expression of metastasis-associated genes
In order to gain insight into the mechanism underlying the aggressive phenotype of p15/p16-deleted PDAC cells and the advantage this co-deletion confers to growth in the liver, we performed global transcriptomic analysis, using RNAseq, on COLO-357sh-NS, COLO-357sh-p15, COLO-357sh-p16 and COLO-357sh-p15/p16 cells. Cluster analysis and correlation map indicated separation of these four cell genotypes into mainly three distinct groups: (i) COLO-357sh-NS (ii) COLO-357sh-p15 and (iii) COLO-357sh-p16/COLO-357sh-p15/p16 (Fig. 4A, B). This clustering has a similar pattern observed in the phenotypic assay (Fig. 3D), of reduced effect of p15 silencing compared to p16 or p15/p16 co-silencing. A comparison between COLO-357sh-p15/p16 and COLO-357sh-NS cells revealed differential expression of 271 genes (Fig. 4C, Supplementary Table 6). Of these genes, 65% (177 genes) were shared with p16-deleted cells, 37% (100 genes) shared with p15-deleted cells and 24% (66 genes) were shared by all three comparison groups. In accordance with the above described aggressive phenotype, pathway enrichment analysis revealed enrichment not only of cell proliferation associated genes, but also of adhesion, migration, communication, angiogenesis and stress response pathways (Fig. 4D, Fig. S4). Verification of the RNAseq data by qPCR analysis revealed a significant increase in the expression of several genes related to aggressiveness and metastasis formation (Fig. 4E). Moreover, these data indicate a role for cell cycle-independent activity of p15/p16 co-deletion in PDAC. Global transcriptomic analysis was also performed with mCM treatment and gene expression differences between COLO-357sh-p15/p16 and COLO-357sh-NS cells were similar in mCM compared to control media (data not shown).
A transcriptomic analysis of COLO-357sh-NS, COLO-357sh-p15, COLO-357sh-p16 and COLO-357sh-p15/p16 cells was performed using RNAseq. A A heatmap of all 1257 differentially expressed genes was generated (B) and a correlation analysis, of Similarity Matrix using Pearson’s correlation was performed. High correlation (1) is marked in red and low correlation (0) is marked in green. C Venn diagram representing comparisons between differentially expressed genes (DEGs) of the manipulated cells (pFDR < 0.05 and FC difference ≥ 2). Genes are listed in supplementary Table 6. D Pathway enrichment analysis was conducted using Gorilla gene ontology for 271 genes regarding biological processes of COLO-357sh-p15/p16 cells vs. COLO-357sh-NS cells, pFDR < 0.05 and FC ≥ 2. E Verification of selected upregulated genes related to metastasis development in COLO-357sh-p15/p16 and COLO-357sh-NS cells.
p15/p16 silencing differentially regulates glutamine metabolism and abrogates ammonia cytotoxicity
In accordance with the low glutamine—high ammonia conditions of the liver micro-environment [21, 22], and the unique requirements of cells thriving under these conditions, we analyzed the transcriptomic for genes associated with ammonia metabolism. A heatmap of 19 ammonia metabolism associated genes revealed 9 down-regulated and 10 up-regulated genes in COLO-357sh-p15/p16 cells compared to the control cells (FDR, q = 0.02) (Fig. 5A). Interestingly, the up-regulated genes are associated with ammonia consumption, and include genes like GLUL, BCAT and ASNS, while genes associated with de-ammination like GLS2 and GLUD were down-regulated. We focused on the two key genes in glutamine and ammonia metabolism GLUL, encoding for glutamine synthetase (GS) which adds an amine group to glutamate to generate glutamine thus may reduce ammonia levels, and GLS2, which catalyzes the reverse reaction, i.e., de-aminates glutamine to generate glutamate thus may generate ammonia access (Fig. 5B). RNAseq data were verified, and qRT-PCR and western blot showed upregulation of GLUL and down-regulation of GLS2 in mRNA (Fig. 5C) and protein (Fig. 5D) levels in COLO-357sh-p15/p16 cells. The net effect of these changes is expected to be reduction of ammonia levels.
A A heatmap, based on transcriptomic data from Fig. 4, of differentially expressed genes related to ammonia metabolism pathway. B Schematic overview of GLUL and GLS2 enzymatic activities is depicted. C mRNA expression of GLUL and GLS2 in COLO357 compared to silenced cells. D Protein expression of GLUL and GLS2 in these cells. E COLO-357 sh-NS and COLO-357sh-p15/p16 cells were plated and cultured in regular DMEM with 10% FBS, 5 mM labeled [U − 13C] glucose and 1 mM glutamine. Metabolites were extracted and collected for subsequent LC-MS analysis. Labeling of indicated metabolites from [U − 13C] glucose is presented.
Our next aim was to reveal whether the transcriptome data were indeed translated into decreased glutamynolysis and increased metabolism of α-KG to glutamate and further to glutamine. For this aim, we fed cells with [U-13C]-glucose, and probed metabolite flux of TCA cycle intermediates and of glutamate (Fig. 5E). We detected a significant 24% decrease in secreted m + 3 lactate in COLO-357sh-p15/p16 cells (Fig. 5a, p = 0.012), indicating reduced glycolysis. On the other hand, we observed an increase in the fractional labeling of m + 2 (23%) and m + 4 (55%) citrate and of m + 2 (16%) and m + 4 (48%) glutamic acid (Fig. 5F, p < 0.05 for all comparisons). While we did not detect a change in α-KG, a decrease of 16% in fractional labeling of m + 2 succinate was observed (p = 0.018). The increase in relative labeling of citrate and of glutamic acid in parallel with the decrease in succinate labeling from [U-13C]-glucose, supports a drop in the relative contribution of glutaminolysis to TCA cycle anaploeriss flux, in accordance with the drop in GLS-2 expression. As the media contained unlabeled glutamine, labeled glutamine could not be measured. These results supported the transcriptome data, and indicate that p15/p16 silencing can lead to better utilization of ammonia.
Based on these observations, we next hypothesized that co-silenced PDAC cells will survive better under glutamine starvation and exposure to high ammonia levels. Indeed, proliferation assays indicated that COLO-357sh-p15/p16 cells are less sensitive to glutamine depletion compared to their WT counterparts (Fig. 6A). Moreover, while glutamine starvation inhibited colony formation of COLO-357sh-NS cells by 40%, it did not affect COLO-357sh-p15/p16 cells (Fig. 6B, p < 0.001).
A Cell growth following glutamine depletion was assessed in COLO357 cells with p15/p16 silencing. Cells were grown with or without glutamine (4 mM) and growth was assessed after 48 h using methylene blue assay. B For colony assay, cells were seeded at low density with or without glutamine and 15 days later cells were fixed and colonies stained with crystal violet and quantified. C COLO-357sh-NS and COLO-357sh-p15/p16 cells were grown in medium containing ammonia (NH4) at indicated concentrations. After 24 h, viability was assessed using Realtime-Glow MT assay. D Colony assay was conducted as in (B), cells were treated with or without ammonia for 15 days. E MIA PaCa-2 cells were co-transfected with p15 + p16 or pCDNA3. Transfected cells were treated as in (C), and viability determined similarly. F COLO-357sh-NS and COLO-357sh-p15/p16 cells were infected with two shRNA against GLUL (sh-GLUL-1 and sh-GLUL-2) or nonspecific control shRNA (sh-control). Silencing efficiency was evaluated by Western blot. G Cell growth and (H) viability assays of COLO-357sh-NS and COLO-357sh-p15/p16 cells harboring GLUL silencing was performed after 24 h. Each bar represents ± SD, statistical analysis was performed using two-way Anova (**, p < 0.01, ***, p < 0.001). I Correlation analysis between GLUL and CDKN2A in TCGA PDAC, colorectal, breast and prostate cancer samples was performed with XENA browser using Spearman’s correlation A negative correlation was detected in all types of cancer analyzed.
Next, we assessed the effect of ammonia on these cells. While the abundance in ammonia in the liver [23] may support cancer cells by providing essential building blocks [24], it may also be toxic [25]. We found that while ammonia treatment decreased viability of COLO-357sh-NS cells in a dose dependent manner, it did not affect viability of COLO-357sh-p15/p16 cells, nor affected growth of these cells (Fig. 6C and Fig. S5B). Moreover, while ammonia (0.5 nM) decreased colony formation of COLO-357sh-NS cells, it did not affect COLO-357sh-p15/p16 cells (Fig. 6D). Interestingly, viability and colony assays showed that COLO-357sh-p15 and COLO-357sh-p16 cells are not resistant to ammonia as the co-deleted cells (Fig. S5), suggesting co-deletion is required to modulate glutamine metabolism. As expected, the opposite was observed upon p15/p16 overexpression in MIA PaCa-2 cells, where p15/p16 expression decreased cells’ ability to grow with ammonia (Fig. 6E). In conclusion, these results suggest that p15 and p16 co-deletion provides growth advantage in glutamine low, ammonia enriched microenvironment.
As the results suggest that glutamine and ammonia metabolism are markedly altered upon p15/p16 silencing, and it was recently reported that GLUL plays an important role in PDAC aggressive behavior [26], we focused on this gene and silenced it in COLO-357sh-p15/p16 and COLO-357sh-NS cells (Fig. 6F). We found that GLUL silencing inhibited growth and viability of COLO-357sh-p15/p16 significantly more than in COLO-357sh-NS, 60% vs 20%, respectively (p < 0.001, Fig. 6G, H). These results suggest that GLUL plays a role in mediating enhanced aggressive phenotype in p15/p16 deleted COLO-357 cells.
Last, we aimed to demonstrate the association between expression of CDKN2A and GLUL in clinical samples. To this aim, we correlated expression of CDKN2A and GLUL using TCGA datasets of pancreatic cancer samples. The results show a negative correlation of −0.1482 between CDKN2A and GLUL expression (Fig. 6I, p < 0.05). We further aimed to determine this negative correlation in other cancer types, and indeed observed the same trend in colorectal (r = −0.2014, p = 0.00002367), breast (−0.2129, p = 5.975e–14) and prostate (rho = −0.2493, p = 3.006e–9) cancer datasets.
Discussion
Despite extensive research of local and metastatic PDAC tumors, little is known regarding mechanisms underlying the predilection of tumor cells to specific metastatic sites. An important step in the understanding of site-specific metastasis formation of PDAC is the identification of GAs which may facilitate adaptation to the new environment.
Analysis of 26 metastasis samples derived from four patients, noted that the GAs fraction in classical PDAC mutated genes did not differ between pancreatic liver, pelvic lung and peritoneum metastasis [7]. A recent study analyzed GAs in 76 metastases from 20 untreated patients with eight different cancer types, concluded that all metastasis within a patient shared the same functional drivers [27]. We noted highly statistically significant differences in the prevalence of GA between different metastatic sites. As the differences ranged from 49% to 63% (for CDKN2A for example), these differences could only be detected using a very large dataset. Importantly, although small, these differences may have biological implications.
As our dataset did not include matched local and metastatic samples, we could not differentiate between three possible conditions: (i) specific GAs evolve during tumorigenesis and promote metastases formation; (ii) tumors harboring specific genomic alterations are more prone to metastasize to specific organs; (iii) the liver microenvironment drives the GAs which evolve after the metastatic deposit has developed and the tumor adapts to its new microenvironment. However, all the above possibilities may take place in different patients. Data obtained from a previous study using matched samples support the second possibility in some of the cases [8]. This recently published analysis of clinical, genomic and transcriptomic data of 319, mostly unmatched, PDAC samples strongly supports our findings [8]. It revealed numerically higher (though not statistically significant) differences between CDKN2A alterations in local compared to metastatic sites (CDKN2B alterations were not reported) and revealed higher cell cycle progression score in metastases.
This study shows that the landscape of GAs was dependent upon metastatic site. Thus, while alterations in CDKN2A/B were more common in liver metastases, alterations in GNAS were significantly more common in omental and thoracic metastases (Fig. 1). This finding suggests a unique role for these alterations in mediating tissue tropism, rather than simply mediating a more aggressive disease phenotype. Indeed, previous study showed that SMAD4 loss in PDAC is implicated in metastasis formation [6]. Accordingly, while p15/16 are traditionally associated with regulation of cell cycle, our study, as well as the clinical data, clearly indicated their role in promoting aggressiveness, as evident by colony formation, migration, invasion and 3D sphere formation and even more importantly, adaptation to the liver microenvironment (Figs. 2, 3). Indeed, whole transcriptome analysis following p15/p16 manipulations indicated differential expression not only of cell cycle-related genes, but also of genes regulating adhesion, migration, cell communication, angiogenesis and stress response, including CXCL8, FOS and JUN, S100 family and SerpinE1 (Fig. 4). A recent study has reported that PDAC liver metastasis recurrence was associated with aggressive phenotype and poor prognosis compared to PDAC lung metastasis [28]. Hence, the observation that p15/p16 deletion enhanced aggressive phenotype of PDAC cells may provide an explanation to the increased occurrence of this GA in liver metastasis samples.
In order to reveal a mechanism through which p15/p16 co-deletion mediates liver predilection, we conducted an unbiased transcriptomic screen, using RNAseq, of COLO-357sh-NS compared to COLO-357sh-p15/p16, in regular medium and hepatocytes CM. We noted activation of pathways not directly related to cell cycle, among them the glutamine and ammonia metabolic pathway (Fig. 5A). The liver is a hub for glutamine and ammonia metabolism [10, 11] and due to extensive use of glutamine by hepatocytes liver microenvironment is glutamine poor and ammonia rich [29, 30]. Our data indicate adaptation of PDAC cells harboring p15/p16-co-deletion to these conditions (Figs. 5, 6). Thus, p15/p16-co-deletion increased colony formation and viability in glutamine-depleted media and promoted resistance to ammonia, while opposite effects were observed in PDAC cells over-expressing p15/p16 (Fig. 6). Previous studies in various cancer types demonstrated that GAs enhanced glutamine metabolism reprogramming resulting in aggressive phenotype. For instance, we discovered that glutamine, by itself, can mediate aggressive phenotype acquired by breast cancer cells harboring activating mutations in the estrogen receptor [31]. A correlation between glutamine dependency and aggressiveness was also seen in ovarian cancer, where glutamine is required to promote migration and invasiveness [32, 33].
We studied molecular mechanisms allowing COLO-357sh-p15/p16 cells to adapt to glutamine-low ammonia-high environment and the transcriptomic and metabolomic data suggested that p15/p16-deletion led to reduced glutaminolysis. This was evidenced by an increase in fractional labeling of [U-13C]-glucose in citrate and glutamate labeling but a decrease in succinate labeling. This suggests a drop in the relative contribution of glutaminolysis to TCA cycle anaplerosis supported by a decrease in GLUD1/2 and GLS2, along with an increase in GLUL (Fig. 5). The significance of GLUL in mediating the effects of p15/p16 silencing was demonstrated in our study where COLO-357sh-p15/p16 cells were more sensitive to GLUL silencing compared to COLO-357sh-NS (Fig. 6). These findings support the fact that aggressive phenotype enhanced by p15/p16 co-deletion is mediated by a reduction in glutamynolysis driven by overexpression of GLUL and a decrease in GLS2. Recently, a critical role of GLUL in PDAC progression and proliferation under metabolite limitation was reported. GLUL allows de novo glutamine synthesis by recycling excessive ammonia for nitrogen anabolism (e.g. biosynthesis of purines, pyrimidines, and asparagine) [26]. On the other hand, enhanced glutamynolysis was shown to enhance cancer progression. Thus, in colorectal cancer, PIK3CA mutations led to reprogramming of glutamine metabolism, resulting in increased conversion of glutamine to α-ketoglutarate, thus replenishing the TCA cycle to generate more ATP, thereby promoting proliferation and tumor growth [34]. Therefore, we conclude that reduction in glutamynolysis may promote cancer growth under cetain conditions, like in hepatocyte environment, and also may be cell-type dependent. Importantly, due to the importance to GLUL for cancer progression specific inhibitors of this enzymes were studied ([35]). However, the current inhibitors, including one of the most potent, methionine sulphoxide, have severe adverse effects and therefore, it raises the need to develop better specific inhibitors.
While CDKN2A/p16 and CDKN2B/p15 are commonly co-deleted in cancer, most studies reported to date focused on the role p16 [36]. Co-deletion of both proteins showed significantly increased effect on colony formation (Figs. S1–3), as well as toward sphere formation of COLO-357 cells in liver microenvironment (Fig. 2F and Fig. 3E) or colony formation in the presence of ammonia (Fig. S5C). The transcriptomic data indicated altered regulation of 271 genes upon co-deletion, whereas of the, only 66 genes were directly affected by each of the protein alone. Thus, the effect of co-deletion seems to be beyond the summation of the two separated genes and imply complex interactions between them. Further studies are required in order to decipher this complex regulatory machinery.
It is also important to take into concideration that while the platform used by us analyzed a limited set of cancer associated genes, other regulatory sequences, like micro RNAs, may reside within those genes and contribute to the cancerous phenotype. For example, additional regulatory elements, including miR-31, are encoded on chromosome 9, in proximity to the area encoding CDKN2A/B. miR-31 plays a tumor-dependent dual role and may serve as either an oncogene or as a tumor suppressor [37]. In PDAC it has been shown to harbor oncogenic activities [38] and thus, it is not expected that miR-31 deletion would contribute to the more aggressive phenotype observed in PDAC following deletion of CDKN2A/B. However, it is possible that other regulatory elements reside in this area and contribute to the effects observed by us.
Taken together, this study indicates that GAs play a role in mediating tropism of PDAC cells. One of these alterations, p15/p16 deletion, prone PDAC cells to form liver metastasis, mediated at least in part, by modulation of glutamine-ammonia metabolism. Liver metastases are a direct and common cause of death among PDAC patients, therefore, targeting p15/p16 activity, through cell cycle inhibition or through metabolic inhibitors could be a novel strategy for PDAC patients. While targeting this pathway may not offer a cure, it may slow disease progression in the liver and prolong life of PDAC patients.
Materials and methods
Clinical sample analysis
Comprehensive Genomic Profiling (CGP) was carried out at Foundation Medicine (Cambridge, MA) in a Clinical Laboratory Improvement Amendments (CLIA)-certified, CAP (College of American Pathologists)-accredited laboratory on PDAC all-comers during the course of routine clinical care. Approval for the study was obtained from the Western Institutional Review Board (Protocol No. 20152817). Hybrid capture was carried out on exons from up to 395 cancer-related genes (FoundationOne v3: 323; FoundationOne v5: 395) and select introns from up to 31 genes frequently rearranged in cancer (FoundationOne v3: 24; FoundationOne v5: 31) (Supplemental Tables 1 and 2). We assessed all known and likely pathogenic alterations across all classes of genomic alterations (GA) including short variant (SV), copy number (CN), and rearrangement (RE) alterations, as described previously [39]. Briefly, base substitutions were detected using a Bayesian method allowing the detection of somatic mutations at low MAF with increased sensitivity for mutations at hotspot sites. Indels were identified using a de Bruijn approach. Copy number events were detected by fitting a statistical copy-number model to normalized coverage and allele frequencies at all exons and ~3,500 genome-wide, single-nucleotide polymorphisms. Rearragnments were detected through an analysis of chimeric reads. Tumor mutational burden (TMB) was determined on 0.8–1.1 Mb (v3: 0.8; v5: 1.1) as described previously [40]. At the time of the analysis, the dataset consisted of 8,880 local-biopsied and 7,983 distantly metastatic PDAC samples in addition to 19,847 metastatic samples from other tumor types, including 7,869 biopsied from the liver (Table 2).
Plasmids
Detailed in Table S3.
Cells transfection and infection
PDAC cell lines, PANC-1, MIA-PaCa2, Capan-2 and COLO-357, were originally obtained from the ATCC (Manassas, VA) and authenticated with the DNA markers used by ATCC. Cells were grown in DMEM containing 10% fetal bovine serum (FBS). Constructs used for transfection are detailed in Table S3. Cells were transfected using Jet Pei (Polyplus Transfection, France). Lentiviral infection was conducted as described [31] and detailed in Supplementary.
Hepatocyte conditioned media
Primary human hepatocytes were purchased from Lonza Cell Culture Products (Basel, Switzerland) and cultured following the manufacturer’s instructions and detailed in Supplementary. Primary mouse hepatocytes were isolated as previously described [41]. Conditioned media (CM) was generated by collecting hepatocyte media after 48 h.
Generation of stably silenced cells
Generation of p15/p16 silenced cells (COLO-357sh-NS, COLO-357sh-p15, COLO-357sh-p16, and COLO-357sh-p15/p16): We stably infected COLO-357 cells with two clones of each shRNAs (Mission shRNA, Sigma) against p15 and p16 independently, and stably-silenced cells were generated as described in Supplementary data. GLUL silencing was performed in COLO-357sh-p15/p16 and COLO-357sh-NS cells by infecting cells with clones of each shRNAs (Mission shRNA, Sigma) as described in Supplementary data.
Metabolomic analysis
For labeling experiment, COLO-357sh-p15/p16 and COLO-357sh-NS cells were grown in DMEM without glucose, supplemented with 10% dialyzed fetal bovine serum (Tarom Applied Technologies) with [U-13C]-glucose at a final concentration 5 mM. Cells were grown in an incubator containing 5% CO2 and ambient oxygen at 37 °C and after 24 h metabolite extractions were conducted at 70–80% confluency. Intracellular metabolites were extracted by cold solution (−20 °C) composed of methanol, acetonitrile, and water (5:3:2) and extrcellular metabolites were extracted by 5:3 methanol-acetonitrile solution in the final proportion of 80:20 extraction solution to medium and collected for subsequent liquid chromatography mass spectrometry (LC-MS) analysis.
All of the metabolite samples were stored at −80 °C for at least 2 h. Protein free metabolite extractions were prepared by spinning the samples at 20,000 × g for 20 min at 4 °C twice. Samples were subsequently analyzed using liquid chromatography mass spectrometry (LC-MS) method. Samples were chromatographically separated on a SeQuant ZIC-pHILIC column (2.1 × 150 mm, 5 mμ, EMD Millipore) with an HPLC system (Ultimate 3000 Dionex LC system, Thermo Fisher Scientific, Inc., USA). Flow rate was set to 0.2 mL min-1, column compartment was set to 30 °C, and autosampler tray maintained 4 °C. Mobile phase A consisted of 20 mM ammonium carbonate and 0.01% (v/v) ammonium hydroxide. Mobile Phase B was 100% acetonitrile. The mobile phase linear gradient (%B) was as follows: 0 min 80%, 15.0 min 20%, 15.1 min 80%, 23.0 min 80%. Mass spectrometry detection was performed using a Q Exactive Hybrid Quadrupole Orbitrap high-resolution mass spectrometer with an electrospray ionization source (ESI, Thermo Fisher Scientific, Inc., USA). Ionization source parameters were following: sheath gas 25 units, auxiliary gas 3 units, spray voltage 3.3 and 3.8 kV in negative and positive ionization mode respectively, capillary temperature 325 °C, S-lens RF level 65, auxiliary gas temperature 200 °C. Metabolites were analyzed in the range 72–1080 m/z. Retention time of metabolites in the chromatogram were identified by corresponding pure chemical standards. Data were analyzed using the MAVEN software suite [42] Metabolite labeling patterns were adjusted for natural 13C abundance and for enrichment impurity of labeled substrate.
RNAseq
Cells were seeded in triplicates for 24 h. Total RNA extraction, library preparation and sequencing were conducted as previously described [43] and detailed in the Supplementary data.
Quantitative RT-PCR
Total RNA isolation and cDNA synthesis were perfomed as previously described [31]. Primers were synthesized by IDT (Coralville, IA) and are listed in Table S4. Quantitative RT-PCR (qRT-PCR) was used to determine mRNA level as previously described [31].
Western blot
Cells were harvested, lysed, and the total protein was extracted as previously described [31]. Lysates were resolved on 10% SDS-PAGE and immunoblotted with the indicated antibodies (Table S5).
Colony assay
Cells were cultured in a 6-well plate, 500 cells per well, in triplacates, for two weeks. Media was replaced twice a week, then cells were fixed, stained and quantitated as previously described [31]. Each experiment was repeated at least three times.
Growth assay
Cells were plated in a 96-well plate, 3000 cells per well, ten wells per treatment. For overexpression experiments, the cells were transfected with either an empty vector (pCDNA3), or indicated constructs. After 24 h, medium was changed to the appropriate media (e.g., regular media or CM) or control media (depending on the experiment) and incubated at 37 °C, 5% CO2 for 24 h. Methylene blue assay was conducted as previously described [31] and normalized to viability before treatment. Each experiment was repeated at least three times.
Viability assay
Cells (3000 per well) were plated, ten wells per treatment and after 24 h, medium was changed to the appropriate media (e.g., regular media or CM) or control media (depending on the experiment) and incubated at 37 °C, 5% CO2 for 24 h. Cells’ viability was assessed using Realtime-Glow MT assay (Promega, Wisconsin) following manufacturer instructions. Each experiment was repeated at least three times.
Invasion assay
Cells were plated in triplicates, into the upper side of matrigel coated 24 transwell inserts, with pore sizes of 8 µm (Corning, New York), in media without FBS, whereas the lower chamber contained media with 10% FBS. After 48 h, the upper side of the apical chamber was scraped gently with cotton swabs to remove non-invading cells, and invading cells were fixed and stained with crystal violet. Each experiment was repeated at least three times.
Migration assays
Transwell: Cells were plated in triplicates as for invasion assay, only the inserts were not covered with matrigel and the assay was terminated after 24 h.
Wound healing assay: Cells were grown to confluency in six-well plates, in triplicates, and a day later cells were treated with mitomycin C (Sigma; 40 μg/ml), scraped in a straight line and photographed at indicated time points. Each experiment was repeated at least three times.
3D sphere formation assays
PDAC cells, 1000 per well, were seeded in GravityPLUS plates (Insphero, Switzerland), 10 repeats per condition, and after five days spheres were transferred to GravityTRAP. Viability of sphere cells was determined after seven days, using ATPlite 1 step 3D (PerkinELMER, Massachusetts) and spheres were photographed at the end of the experiment. For CM experiments, PDAC cells were seeded as above. After three days, spheres were transferred to GravityTRAP and medium was changed to m-CM or control media. Cell viability was assessed using Realtime-Glow MT assay (Promega, Wisconsin) and spheres were photographed at the end of the experiment, 3 days later. Each experiment was repeated at least three times.
UCSC cancer genomics browser analysis
The heat map and correlation between CDKN2A, CDKN2B and GLUL in the same patient cohort were constructed by data mining in the Cancer Genome Atlas (TCGA) pancreatic, breast, colorectal cancers and GDC TCGA protstate cancer using the UCSC Xena browser (http://xena.ucsc.edu/).
Statistical analysis
Data was analyzed using the GraphPad Prism software: T test, One or two Way ANOVA with multiple comparisons Bonferroni post hoc analysis and considered significant at P-values *≤ 0.05, **≤ 0.01 and ***≤ 0.001. Bar graphs represented mean and standard deviation (SD) across multiple independent experimental repeats.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: A Cancer J Clin. 2012;62:10–29.
Pokorny AMJ, Chin VT, Nagrial AM, Yip D, Chantrill LA. Metastatic pancreatic ductal adenocarcinoma: diagnosis and treatment with a view to the future. Intern Med J. 2018;48:637–44.
Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140–1.
Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, et al. Metastatic patterns in adenocarcinoma. Cancer. 2006;106:1624–33.
Weissmueller S, Manchado E, Saborowski M, Morris JP,4th, Wagenblast E, Davis CA. et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell. 2014;157:382–94.
Yachida S, White CM, Naito Y, Zhong Y, Brosnan JA, Macgregor-Das AM, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res. 2012;18:6339–47.
Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49:358–66.
Connor AA, Denroche RE, Jang GH, Lemire M, Zhang A, Chan-Seng-Yue M, et al. Integration of genomic and transcriptional features in pancreatic cancer reveals increased cell cycle progression in metastases. Cancer Cell. 2019;35:267–82.e7.
Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–7.
Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, Haigis MC. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science. 2017;358:941–6.
Hatting M, Tavares CDJ, Sharabi K, Rines AK, Puigserver P. Insulin regulation of gluconeogenesis. Ann NY Acad Sci. 2018;1411:21–35.
Kumar A, Cherukumilli M, Mahmoudpour SH, Brand K, Bandapalli OR. ShRNA-mediated knock-down of CXCL8 inhibits tumor growth in colorectal liver metastasis. Biochem Biophys Res Commun. 2018;500:731–7.
Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32.
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–40.
Naumann M, Savitskaia N, Eilert C, Schramm A, Kalthoff H, Schmiegel W. Frequent codeletion of p16/MTS1 and p15/MTS2 and genetic alterations in p16/MTS1 in pancreatic tumors. Gastroenterology. 1996;110:1215–24.
Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–6.
Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–35.
Morgan RT, Woods LK, Moore GE, Quinn LA, McGavran L, Gordon SG. Human cell line (COLO 357) of metastatic pancreatic adenocarcinoma. Int J Cancer. 1980;25:591–8.
Sipos B, Moser S, Kalthoff H, Torok V, Lohr M, Kloppel G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: towards the establishment of an in vitro research platform. Virchows Arch: Int J Pathol. 2003;442:444–52.
Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7.
Häussinger D, Graf D, Weiergräber OH. Glutamine and cell signaling in liver. J Nutr. 2001;131:2509S–14S. discussion 23S-4S
Walker V. Ammonia metabolism and hyperammonemic disorders. Adv Clin Chem. 2014;67:73–150.
Watford M. Glutamine and glutamate metabolism across the liver sinusoid. J Nutr. 2000;130:983s–7s.
Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer. 2017;3:169–80.
Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31.
Bott AJ, Shen J, Tonelli C, Zhan L, Sivaram N, Jiang YP, et al. Glutamine anabolism plays a critical role in pancreatic cancer by coupling carbon and nitrogen metabolism. Cell Rep. 2019;29:1287–98. e6.
Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, et al. Minimal functional driver gene heterogeneity among untreated metastases. Science. 2018;361:1033–7.
Tanaka M, Mihaljevic AL, Probst P, Heckler M, Klaiber U, Heger U, et al. Meta-analysis of recurrence pattern after resection for pancreatic cancer. Br J Surg. 2019;106:1590–601.
Kietzmann T, Roth U, Freimann S, Jungermann K. Arterial oxygen partial pressures reduce the insulin-dependent induction of the perivenously located glucokinase in rat hepatocyte cultures: mimicry of arterial oxygen pressures by H2O2. Biochemical J. 1997;321:17–20.
Häussinger D, Schliess F. Glutamine metabolism and signaling in the liver. Front Biosci. 2007;12:371–91.
Zinger L, Merenbakh-Lamin K, Klein A, Elazar A, Journo S, Boldes T, et al. Ligand-binding domain-activating mutations of ESR1 rewire cellular metabolism of breast cancer cells. Clin Cancer Res. 2019;25:2900–14.
Yuan L, Sheng X, Willson AK, Roque DR, Stine JE, Guo H, et al. Glutamine promotes ovarian cancer cell proliferation through the mTOR/S6 pathway. Endocr-Relat Cancer. 2015;22:577–91.
Yang L, Moss T, Mangala LS, Marini J, Zhao H, Wahlig S, et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol Syst Biol. 2014;10:728.
Hao Y, Samuels Y, Li Q, Krokowski D, Guan BJ, Wang C, et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun. 2016;7:11971.
Kim GW, Lee DH, Jeon YH, Yoo J, Kim SY, Lee SW, et al. Glutamine synthetase as a therapeutic target for cancer treatment. Int J Mol Sci. 2021;22:1701–18.
Hayashi N, Sugimoto Y, Tsuchiya E, Ogawa M, Nakamura Y. Somatic mutations of the MTS (multiple tumor suppressor) 1/CDK4l (cyclin-dependent kinase-4 inhibitor) gene in human primary non-small cell lung carcinomas. Biochemical biophysical Res Commun. 1994;202:1426–30.
Yu T, Ma P, Wu D, Shu Y, Gao W. Functions and mechanisms of microRNA-31 in human cancers. Biomedicine Pharmacother = Biomedecine pharmacotherapie. 2018;108:1162–9.
Kent OA, Mendell JT, Rottapel R. Transcriptional regulation of miR-31 by oncogenic KRAS mediates metastatic phenotypes by repressing RASA1. Mol Cancer Res. 2016;14:267–77.
Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31.
Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34.
Edwards M, Houseman L, Phillips IR, Shephard EA. Isolation of mouse hepatocytes. Methods Mol Biol. 2013;987:283–93.
Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–26.
Boldes T, Merenbakh-Lamin K, Journo S, Shachar E, Lipson D, Yeheskel A, et al. R269C variant of ESR1: high prevalence and differential function in a subset of pancreatic cancers. BMC Cancer. 2020;20:531.
Acknowledgements
This work was financially supported by the Israel Science Foundation to I.W. (grant no. 1320/14); the Israel Cancer Association to I.W. (grant no. 20160053); The Margaret Stultz foundation for Pancreatic Cancer Research, the Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel; and TASMC excellence fund. The funding sources had no implications on the study design, collections, analysis, and interpretation of data or writing the manuscript. Israel Cancer Association, ISF, Pancreatic Cancer Foundation
Author information
Authors and Affiliations
Contributions
SJ- Collected the data, performed the analysis of in vitro assays, and wrote the paper. AKG- Collected the data, performed the analysis of in vitro assays. ES- Conceived and designed the analysis, collected the data, contributed data and analysis tools, performed analysis, and participated in writing of the paper. LZ- Collected the data and performed analysis. MPC- Contributed data and analysis tools, performed analysis of RNA sequencing, and participated in writing. BS- Collected data, performed analysis and participated in writing. DS- Performed analysis of RNA sequencing. SF- Collected data and performed analysis. TS- Designed and performed analysis and participated in writing. TR- Conceived and designed the analysis, collected the data, performed analysis, and wrote the paper. IW- Conceived and designed the analysis, collected the data, performed analysis, and wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Journo, S., Goldberg, A.K., Sokol, E.S. et al. Genomic alterations drive metastases formation in pancreatic ductal adenocarcinoma cancer: deciphering the role of CDKN2A and CDKN2B in mediating liver tropism. Oncogene 41, 1468–1481 (2022). https://doi.org/10.1038/s41388-022-02184-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02184-2
- Springer Nature Limited