Abstract
Proteomic signatures associated with clinical measures of more aggressive cancers could yield molecular clues as to disease drivers. Here, utilizing the Clinical Proteomic Tumor Analysis Consortium (CPTAC) mass-spectrometry-based proteomics datasets, we defined differentially expressed proteins and mRNAs associated with higher grade or higher stage, for each of seven cancer types (breast, colon, lung adenocarcinoma, clear cell renal, ovarian, uterine, and pediatric glioma), representing 794 patients. Widespread differential patterns of total proteins and phosphoproteins involved some common patterns shared between different cancer types. More proteins were associated with higher grade than higher stage. Most proteomic signatures predicted patient survival in independent transcriptomic datasets. The proteomic grade signatures, in particular, involved DNA copy number alterations. Pathways of interest were enriched within the grade-associated proteins across multiple cancer types, including pathways of altered metabolism, Warburg-like effects, and translation factors. Proteomic grade correlations identified protein kinases having functional impact in vitro in uterine endometrial cancer cells, including MAP3K2, MASTL, and TTK. The protein-level grade and stage associations for all proteins profiled—along with corresponding information on phosphorylation, pathways, mRNA expression, and copy alterations—represent a resource for identifying new potential targets. Proteomic analyses are often concordant with corresponding transcriptomic analyses, but with notable exceptions.
Similar content being viewed by others
Introduction
Mass spectrometry-based proteomics can provide a window into cancer biology not possible using other -omics technologies. Cancers arise from DNA damage, including single nucleotide variants or indels. However, mutation events for a particular gene in a given cancer type tend to be sparse and would not necessarily capture the downstream gene expression alterations initiated by different mutations in different genes [1]. While gene transcript or mRNA levels often correspond to total protein levels for a given gene, there is also widespread decoupling between proteins and their mRNAs, due to differences in translation [2,3,4,5]. Moreover, the stability, function, and activity of proteins are precisely regulated through posttranscriptional modifications, including the phosphorylation of kinases that regulate growth signaling pathways in cancers. By combining proteomic data with transcriptomic data, we can obtain a more comprehensive view of the complex regulatory mechanisms that underlie cancer. The National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) is a national effort to accelerate the understanding of the molecular basis of cancer through the application of large-scale proteomics. CPTAC-generated proteomics data for human tumors of several different cancer types [3,5,6,7,8], just recently provided to the research community, represent a resource for identifying protein correlates of interest, which may provide clues as to deregulated pathways.
Specific sets of differentially expressed proteins, or proteomic “signatures,” associated with clinical measures of advanced disease, could provide molecular clues as to the drivers of more aggressive cancers [9, 10]. Indicators of more aggressive disease, as used in the clinical setting, include grade and stage. Cancer grade is a histologic parameter assigning the degree of differentiation of the cancer cells, where high-grade cancers look poorly-differentiated and tend to grow and spread more quickly than low-grade cancers that look well-differentiated (resembling the tissue of origin) and tend to have a more indolent clinical behavior. Cancer stage is a clinical parameter indicating how extensively the tumor has spread outside of its site of origin. Patient survival or time to adverse event represents another indicator of aggressive disease. However, such information requires tracking patients over a long period, and such follow-up data are not present in the CPTAC cohorts, as freshly collected patient samples are required for proteomics research. Nevertheless, both grade and stage can predict patient survival to a high degree.
Our present study hypothesizes that grade and stage differences among cancer cases represent altered pathways manifested at the protein level, and that such data can also aid in the identification of functional targets. Pan-cancer molecular analyses seek to bring data from diverse cancer types together, to identify commonalities, differences, and emergent themes across tumor lineages [11,12,13,14,15]. Past pan-cancer efforts in proteomics, such as by The Cancer Genome Atlas (TCGA), have been limited to focused analysis of ~200 protein features as available by Reverse Phase Protein Array platform [11]. In contrast, mass spectrometry-based proteomics allows for profiling of thousands of proteins, representing a unique opportunity for CPTAC proteomics data. Our study follows a similar basic approach as that of a previous study examining grade-specific transcriptomic differences [15]. At the same time, here we expect some unique differences that would exist at the proteome level. We also utilize data on proteomic grade correlations to identify protein kinases having a functional impact in vitro in uterine endometrial cancer cells.
Results
Proteomic signatures of high grade or high stage cancers by cancer type
We sought to define differentially expressed proteins and mRNAs associated with higher grade or with higher stage, for each of seven cancer types (Table 1 and Supplementary Data 1): breast invasive carcinoma (n = 105 cases), colon adenocarcinoma (n = 97), lung adenocarcinoma (lung AD, n = 111), clear cell renal cell carcinoma (renal, n = 110), ovarian serous carcinoma (n = 169), uterine corpus endometrial carcinoma (n = 100), and pediatric glioma (n = 102). Mass spectrometry-based proteomic data were provided by CPTAC, with mRNA data on these cases provided using expression arrays (ovarian) or RNA-sequencing. Both total protein and phosphoprotein features were profiled, involving 14586 total proteins (by unique gene) and 44763 phosphoproteins detected across the various projects, where for each cancer type the average number of total proteins detected was ~9700. Histologic grade information was available for lung AD, renal, ovarian, uterine, and glioma cancer types. Clinical or pathological stage information was available for breast, colon, lung AD, renal, ovarian, and uterine cancer types (as pediatric gliomas are not typically staged but instead are classified using World Health Organization guidelines [16]). To facilitate access to CPTAC proteomic results by the general biomedical research community, we integrated CPTAC data with the UALCAN data portal [17], allowing users to query proteins of interest for associations with grade or stage (http://ualcan.path.uab.edu/). To determine differential levels for a given gene or protein according to increasing tumor grade or stage, we took the Pearson’s correlation between the log-transformed gene-level molecular values and the grade or stage as translated into a numerical value (e.g., translating grade categories such as “G1”, “G2”, and “G3” into 1, 2, and 3, respectively).
For most cancer types studied, we found on the order of hundreds of total proteins and of phosphoproteins to be differentially expressed with higher grade or with higher stage at a nominal significance level of p < 0.01 (Pearson’s, Figs. 1a and Figs. S1 and S2 and Supplementary Data 2–6). Except for stage correlates in the instance of breast and colon cancers, the numbers of nominally significant proteins exceeded that expected in a randomized proteomic dataset for which no relation between grade or stage and protein expression would be expected (Fig. S3). Protein correlates of grade in ovarian cancer were also notably low. Even in instances of nominally significant proteins not exceeding or only moderately exceeding chance expectations, the nominally significant proteins may still contain molecular information. Such information would represent real biological differences, which may be revealed by downstream analyses, as described below. For example, we could also find widespread patterns involving gene transcripts or mRNAs associated with grade or stage (Fig. 1b), and, for all cancer types, there were significant overlapping gene features between the total protein signatures and the mRNA signatures (Fig. 1c). Most phosphoprotein signatures also overlapped significantly with their corresponding total protein signatures (Fig. S2b). At the same time, an appreciable fraction of proteins in the proteomic signatures—on the order of half—were not included in the corresponding mRNA signature (Fig. 1c). Except for ovarian cancer, for each cancer type, the global proteomic and mRNA patterns associated with grade were respectively broadly similar to proteomic and mRNA patterns associated with stage (Fig. 1d, taking the t-statistics for all profiled features for the respective grade/stage associations and then correlating them to each other).
a For each of the indicated cancer types, numbers of top differentially expressed proteins (p < 0.01, Pearson’s using log-transformed data), associated with higher cancer grade (left) or with higher cancer stage (right). Numbers of unique genes tested are indicated for each comparison (involving proteins for which measurements were made in over half of samples profiled). b Similar to part a, but for differentially expressed mRNAs. Numbers of unique genes tested are indicated for each comparison (involving mRNAs with available data for which proteins were considered in part a). c Overlapping top protein and mRNA features (p < 0.01 for protein and p < 0.05 for mRNA, based on shared gene) by comparisons according to grade (left) or according to stage (right). P values for significance of overlap by one-sided Fisher’s exact tests. d For each cancer type, overall concordance or discordance in differential gene feature patterns between grade (columns) and stage (rows) comparisons for both protein data (left) and mRNA data (right). For all gene features examined, correlations between t-statistic by grade comparison versus t-statistic by stage comparison were computed (based on all profiled features). R-values by Pearson’s are shown. Red square denotes overall concordance between grade and stage results for the given cancer type, and blue square denotes discordance. Breast, Breast invasive carcinoma; Colon, Colon adenocarcinoma; Lung AD, Lung adenocarcinoma; Renal, Clear cell renal cell carcinoma; Ovarian, Ovarian serous carcinoma; Uterine, Uterine corpus endometrial carcinoma; Glioma, Pediatric glioma. See also Figs. S1, S2 and S3 and Supplementary Data 1–6.
Proteomic signatures predict patient survival
As another indication of the biological information contained in our proteomic signatures of grade or stage, the mRNAs represented by the signatures for a given cancer type could significantly predict patient survival in that cancer type. For each cancer type, we examined an independent dataset of primary tumor sample transcriptomic profiles of that cancer type, for which sufficient patient follow-up information was available (taken from previously published datasets, see Supplementary Methods). We applied each proteomic signature of grade or stage (from Fig. 1a) to the corresponding independent dataset, to score patient profiles based on overall similarities of the mRNA patterns with the direction of change associated with grade (Fig. 2a) or stage (Fig. 2b) at the protein level. Lung AD grade and stage proteomic signatures [18], renal grade and stage signatures [9], uterine grade and stage signatures [19], ovarian stage signature [20,21,22,23,24,25], breast stage signature [26], and pediatric glioma grade signature could all stratify patients of the corresponding cancer type to high-, low-, and intermediate-risk groups representing significant differences in outcome. The ovarian grade and colon stage [27] proteomic signatures could not successfully stratify patients in this way (Figs. 2 and S4a). For the other signatures, in most instances, the actual differences in patient outcomes were not enough to be able to potentially guide treatment decisions, as all stratified groups would have a risk of death. However, for the pediatric glioma signature, the low-risk group of patients (2 high grade and 79 low grade, out of 281 cases) had no adverse events, and the intermediate-risk group consisted mostly of low-grade cases (66 out of 80, Supplementary Data 7). As another reflection of the proteomic signatures containing information on disease aggressiveness, significant numbers of proteins within most of the signatures were for genes individually associated with patient survival (Fig. S4b, c and Supplementary Data 7). Transcriptomic signatures of stage or grade, as well as the intersection of the above proteomic and transcriptomic signatures, yielded similar levels of prognostic power as the proteomic signatures (Figs. S5 and S6).
a For each of the indicated cancer types (Lung adenocarcinoma, Clear cell renal cell carcinoma, Ovarian serous carcinoma, Uterine corpus endometrial carcinoma, Pediatric glioma), the corresponding proteomic signature (“sig.”) of higher grade (from Fig. 1a) was applied to an independent set of primary tumor sample mRNA profiles of the same cancer type [9, 18, 19, 45] (using datasets for which sufficient patient follow-up data were available). Across the sample mRNA profiles in the independent set, the grade signature similarity scores (t-statistic as derived from the “t-score” metric [9, 45, 46]) were correlated with patient survival. P values by log-rank test (comparing patients with top third or middle third or bottom third of signature scores) or by univariate Cox, as indicated. b Similar to part a, but for proteomic signatures of higher stage (Lung adenocarcinoma, Clear cell renal cell carcinoma, Ovarian serous carcinoma, Uterine corpus endometrial carcinoma, Breast invasive carcinoma) [9, 18, 19, 45]. See also Figs. S4, S5, and S6 and Supplementary Data 7.
Proteomic patterns shared or not shared across cancer types
We found some differential protein expression patterns associated with grade or stage shared across multiple cancer types. Still, at the same time, each cancer type showed a proteomic signature that was distinctive from those of the other cancer types. In comparing the respective proteomic signatures of grade from each cancer type (from Fig. 1a) to each other, we observed significant levels of gene set overlap (Fig. 3a), in particular between lung AD and renal cancers, between lung AD and glioma, between lung AD and uterine, and between renal and glioma. For the proteomic signatures of stage (from Fig. 1a), only lung AD and renal signatures significantly overlapped (Fig. 3a). A set of 1056 total proteins associated significantly with grade (p < 0.01) with the same direction of change for two or more cancer types (Fig. 3b and Supplementary Data 2), revealing many shared patterns between lung AD, renal, and glioma cancer types in particular. Most of the 1056 proteins had differential expression patterns represented at the mRNA level. However, for a subset of 142 proteins, we found no significance (p < 0.05) at the mRNA level for any of the involved cancer types. We examined protein and mRNA patterns in our data for a set of genes previously identified as part of a transcriptome-based meta-signature of undifferentiated cancer [15] (i.e., mRNAs associated with high-grade cancers). We found that this mRNA-based signature was highly reflected in the mRNA patterns and mostly reflected but to a somewhat lesser extent in the protein patterns (Fig. 3c). In contrast to the grade associations, fewer total proteins, 60, were significantly associated with stage (p < 0.01) with the same direction of change for two or more cancer types (Fig. 3d and Supplementary Data 3), mostly reflecting shared patterns between lung AD and renal cancers. As compared to the above regarding total proteins, phosphoproteins associated with stage or grade showed similar patterns of overlap between cancer types (Fig. S7).
a For both the proteins over-expressed or under-expressed with higher grade for at least one cancer type (left, proteins from Fig. 1a) and the proteins over-expressed or under-expressed with higher stage for at least one cancer type (right, proteins from Fig. 1a), the numbers of overlapping proteins between any two cancer types are indicated, along with the corresponding significances of overlap (using color map, p values by one-sided Fisher’s exact test). b Heat map of differential t-statistics (Pearson’s on log-transformed data), by cancer type, comparing higher grade versus lower grade (red, higher expression with higher grade; white, not significant with p > 0.05), for 1056 proteins significant for two or more cancer types (p < 0.01). Differential t-statistics by grade for the mRNA corresponding to the 1056 proteins are also shown. Proteins significantly over-expressed (p < 0.01) with higher grade for three or more cancer types are indicated by name. c For both protein and mRNA, differential t-statistics by grade are shown for a set of genes previously identified as part of a transcriptome-based meta-signature of undifferentiated cancer [15]. d Similar to part b, but for a set of 60 proteins significant for two or more cancer types (p < 0.01) when comparing higher stage versus lower stage cancers. See also Fig. S7.
Proteomic signatures involve copy number alterations
We hypothesized that some of the proteins associated with grade or stage might reflect somatic copy number alterations (CNAs) in the cancer genome. We collected CNA data on each of the cancer types in our study, and for each cancer type, we compared the proteins associated with grade or stage with the genes showing higher or lower copy numbers with increasing grade or stage. For both the proteomic and transcriptomic grade signatures (from Fig. 1a, b), significant numbers of genes showed CNA changes in the same direction (Figs. 4a, S8, and Supplementary Data 8). In particular, proteins having lower expression with higher grade involved genes more frequently lost with higher grade. In contrast, gene set overlap involving the proteomic stage signatures and CNA patterns were not significant or not as markedly significant (Fig. 4a).
a For both the proteins altered with higher grade for at least one cancer type (left) and the proteins altered with higher stage (right), the numbers of overlapping gene features with protein expression and copy number changes in the same direction (using Pearson’s p < 0.01 for each comparison), along with the corresponding significances of overlap (using color map, p values by one-sided Fisher’s exact test). TCGA LUAD [18] CNA data do not include grade information. b For the sets of overlapping genes showing both protein under-expression and lower copy numbers with higher grade for renal, ovarian, uterine, or glioma cancer types, significantly enriched cytoband regions (FDR < 1% [42], one-sided Fisher’s exact test). c For the sets of overlapping genes showing both protein under-expression and lower copy numbers with higher grade for renal, ovarian, or uterine cancer types, heat maps represent t-statistics according to increasing grade (blue, lower with higher grade) for differential protein expression, differential mRNA expression, and differential CNA, as well as CNA patterns (blue, copy loss) by cancer type. d Taking all of the genes in the 13q14-q22 region (region associated in part b with both copy loss and protein under-expression in high grade renal cancers), overall patient survival in TCGA KIRC [9] patient cohort is assessed for cases with copy loss on average for these genes (left) and for cases with low (bottom 5%) average mRNA expression (middle). Overall patient survival is also assessed for low average mRNA expression of 13q14-q22 genes in an independent clear cell renal cell carcinoma dataset [28] (right). P values by log-rank test. e In the TCGA UCEC [19] patient cohort, overall survival is assessed for cases with copy loss on average for all 15q15 genes (left), for cases with copy loss on average for all 15q24 genes (middle), and for cases with low average mRNA expression of genes (bottom 20%) in 15q15 or 15q24 cytoband regions. P values by log-rank test. Copy loss for parts (d and e) defined as average log2 (tumor/normal) less than −0.4. See also Fig. S8.
Focusing on the genes showing both protein under-expression and lower copy numbers with higher grade in renal, ovarian, uterine, or glioma cancer types, we could identify significantly enriched cytoband regions associated with each cancer type (Fig. 4b). (Our focus here was on those cancer types for which CNA data with grade information were available.) Genomic regions associated with copy loss and corresponding protein changes with higher grade included 9q21-q34, 13q14-q22, 14q11-q32, and 18q14-q21 for renal cancers; 16q22-q23 for ovarian cancer; 15q15-q24 for uterine cancers; and 10q26 and 11p15 for pediatric gliomas (Fig. 4b, c). The above regions showed loss at the copy number level at a higher frequency in higher grade versus lower grade cancers for the given type (Fig. 4c). For clear cell renal cell carcinoma, lower average levels of genes in the 13q14-q22 region—either CNA or mRNA—were associated with worse patient outcomes in two separate patient cohorts (Fig. 4d) [9, 28]. Similarly, lower average levels of 15q15 genes and 15q24 genes were associated with worse patient outcomes in uterine corpus endometrial carcinoma [19] (Fig. 4e).
Proteomic signatures represent common pathways across cancer types
The information contained within the proteomic signatures can represent altered pathways or functional gene categories. In terms of functional gene categories, we found significantly enriched Gene Ontology (GO) annotation terms for each proteomic signature of grade or stage (Figs. 5a and S9 and Supplementary Data 9 and 10, signatures taken from Fig. 1a). However, we found markedly fewer significant GO terms for the proteomic stage signatures as compared to the corresponding grade signatures (Fig. S9). GO terms significant for the proteomic signatures for multiple cancers included “extracellular exosome,” “immune system process,” “response to stress,” “cadherin binding,” “cell cycle process,” and “translational initiation” (Fig. 5a). GO terms significant for one or more proteomic signatures were often but not always significant in the corresponding mRNA-based signatures, and vice versa. GO terms significant for proteomic but not transcriptomic signatures of grade included “Golgi subcompartment,” “MHC protein binding,” and “ribosome” for renal cancers, and “p53 binding” and “euchromatin” for gliomas (Fig. S9a). GO terms significant for transcriptomic but not proteomic signatures of grade included “T cell receptor signaling pathway,” “regulation of Wnt signaling pathway,” and “protein ubiquitination” for renal cancers, and “tricarboxylic acid cycle” and “electron transport chain” for gliomas (Fig. S9b).
a Selected significantly enriched Gene Ontology (GO) terms, involving the top set of proteins or mRNAs either over-expressed (left) or under-expressed (right) with higher grade for each cancer type represented. Differential gene features selected using p < 0.01 by Pearson’s (Fig. 1a, b). Enrichment p values by one-sided Fisher’s exact test. b Significance of enrichment (by one-sided Fisher’s exact test) for wikiPathway [29] gene sets with the respective sets of proteins and mRNAs over-expressed (p < 0.01, Pearson’s) with tumor grade for each cancer type represented. The set of pathways represented were significant (FDR < 10% [42]) within the over-expressed proteins for at least one cancer type. c Heat map of differential protein t-statistics (Pearson’s on log-transformed data), by cancer type, comparing higher grade versus lower grade (red, higher expression with higher grade; white, not significant with p > 0.05), for proteins in the wikiPathway “Metabolic reprogramming in cancer” (full name “Metabolic reprogramming in colon cancer”). d Pathway diagram representing core metabolic pathways (“Metabolic reprogramming in cancer”), with differential protein and mRNA expression patterns represented, correlating expression with increasing tumor stage for Lung AD, Renal, and Glioma cancer types. RNA features are indicated using italics. Phosphoprotein features are indicated by residue. Red denotes significantly higher expression with higher grade and blue denotes significantly lower expression. e Heat map of differential protein and mRNA t-statistics (Pearson’s on log-transformed data), by cancer type, comparing higher grade versus lower grade, for genes in the wikiPathway “Translation factors”. See also Fig. S9 and Supplementary Data 9–11.
In terms of represented pathways, we searched the wikiPathway [29] gene sets with the respective sets of proteins/phosphoproteins and mRNAs over-expressed (p < 0.01, Pearson’s, from Fig. 1a, b) with tumor grade for each cancer type (Supplementary Data 11). Out of 417 pathways considered, 49 were significant by one-sided Fisher’s exact test with FDR <10% for at least one cancer type (Fig. 5b). Pathways significant by protein analysis were often significant by corresponding mRNA analysis, but with some exceptions. One example pathway of particular interest, significant for four of the five cancer types, involved core metabolic pathways (Fig. 5c), including glycolysis, lipid synthesis, Krebs cycle, and Warburg effect (Fig. 5d). For the above pathways, different cancer types had higher expression of different sets of proteins with higher grade, and differences at the protein level were often reflected at the mRNA level though not always. Another example pathway of interest involved translation factors, significantly associated at the protein level with higher grade for four out of five cancer types yet significant at the mRNA level for only two of these four cancer types (Fig. 5b, e).
Proteomics reveals kinases having functional impact
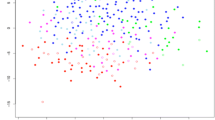
Proteomic correlates of stage or grade would include proteins having a functional impact in cancer cells. To identify such proteins, we focused on protein kinases, as these would represent stronger candidates for therapeutic targets [30]. As each cancer type showed a distinctive proteomic signature of higher grade or stage (Fig. 3), we chose to examine the uterine data for potential targets for functional studies in uterine endometrial cell lines. Taking a set of 347 protein kinases with available uterine data, we compared the protein-level associations with higher tumor grade, against the corresponding mRNA associations (Fig. 6a), paying particular attention to the proteins associated with grade in more than one cancer type. Of the 347 kinases, 37 associated (p < 0.05, Pearson’s) with higher grade in uterine cancer, and 20 of these proteins were associated with grade in three or more CPTAC cancer types (including uterine). From these 20 kinases, we selected four for functional studies—MAP3K2, MASTL, SCYL1, and TTK—although other selection criteria and cutoffs could reveal other candidates. Of these four kinases, three had a corresponding mRNA association with grade, while MAP3K2 did not (Fig. 6a). We surveyed the expression of these kinases at the mRNA and protein level in five different malignant human uterine cell lines, with expression detected for all four kinases by qRT-PCR and Western blot (Figs. 6b and S10). We transfected Ishikawa and HEC-1-A cell lines with non-targeting siRNA (siNT) or siRNA targeting TTK (siTTK), MASTL (siMASTL), MAP3K2 (siMAP3K2), and SCYL1 (siSCYL1). After 48 h of siRNA treatment, the transfected cells were lifted and seeded for cell viability assays (schematically depicted in Fig. S11a). In parallel, we evaluated transfected cells for knock-down efficiency by qRT-PCR and Western blot (Fig. S11b–e).
a Proteomic identification of significantly expressed kinases in uterine endometrial cancer. For 347 protein kinases with available data, protein abundance correlates with tumor grade are plotted against corresponding mRNA expression correlates (Pearson’s correlation). Genes associated with grade in three or more CPTAC cancer types (including uterine) are indicated by red data points. Proteins selected for functional studies (MAP3K2, SCYL1, MASTL, TTK) are labeled by name. b Western blot analysis of basal protein expression in malignant uterine cell lines. c Time-course evaluation of cell viability in Ishikawa cells following siRNA-mediated depletion of kinases. Data normalized to luminescence signal in control transfection (siNT) at day 1 (Mean ± SEM, n = 4 biological replicates, 2-way ANOVA, with Dunnett’s multiple comparisons post-test). d Time-course evaluation of cell viability of HEC-1-A following siRNA-mediated depletion of kinases. Data normalized to luminescence in control transfection (siNT) at day 1 (Mean ± SEM n = 4 biological replicates, 2-way ANOVA, with Dunnett’s multiple comparisons post-test). e Representative brightfield images at 0 and 72 h post-scratch in Ishikawa cells following siRNA-mediated depletion of kinases. Size-bar = 200 µm. f Quantification of 3 biological replicates for wound-healing in Ishikawa cells at 72 h (Mean ± SEM, One-way ANOVA with Dunnett’s multiple comparisons post-test). g Brightfield images of representative wounds at 0 and 72 h post-scratch in HEC-1-A cells following siRNA-mediated depletion of kinases. h Quantification of 3 biological replicates for wound-healing in HEC-1-A cells at 72 h (Mean ± SEM). RLU, relative luminescence units. For parts (c, d, f, and h), p values are represented as follows: 0.033 (*), 0.002(**), and <0.001 (***). See also Figs. S10, S11.
To evaluate the potential role of the selected kinases in malignant processes of sustained proliferation and activated invasion [31], we evaluated cell viability and 2-D migration (wound-healing) in Ishikawa and HEC-1-A cell lines following siRNA-mediated depletion (Fig. 6c–h). Ishikawa cells treated with siSCYL1 demonstrated increased viability compared to control cells treated with siNT. In contrast, treatment with siMASTL significantly decreased cell viability five days after seeding (Fig. 6c). Noticeably, siTTK treatment potently suppressed Ishikawa cell viability beginning at day 3 (Fig. 6c). In HEC-1-A cells, both siTTK and siMASTL treatments robustly and significantly decreased cell viability. siMAP3K2 led to a slight but significant increase in HEC-1-A cell viability on the last day of analysis (Fig. 6d). To assess the candidate kinases’ contribution to cell migration, we measured the wound-healing capacity of Ishikawa and HEC-1-A cells 72 h after siRNA treatment (Fig. 6e–h). Compared to siNT-treated Ishikawa cells, cells treated with siTTK demonstrated a significant reduction in wound closure (60.64% ± 11.46 vs. 14.11% ± 12.32) (Fig. 6e, f), indicating the critical contribution of this kinase to the migratory potential of the Ishikawa endometrial cancer cell line. HEC-1-A cells on the other hand, showed a significant reduction in wound-closure capacity compared to siNT after treatment with siMAP3K2 (75.78% ± 9.36 vs. 43.66 ± 5.34), siMASTL (75.78% ± 9.36 vs. 28.44 ± 9.22) and siTTK (75.78% ± 9.36 vs. 29.58 ± 4.13). These results suggest that compared to Ishikawa cells (representative of grade I), the migratory potential of HEC-1-A cells (grade II) is more sensitive to siRNA knock-down of MAP3K2 and MASTL. Notably, although cell viability was not affected by siMAP3K2 treatment in HEC-1-A cells (Fig. 6d), a significant reduction in wound-healing capacity was observed, suggesting a unique role for MAP3K2 in cell migration.
Discussion
Using CPTAC proteomic data, we explored protein features and associated pathways of aggressive cancers, which associations may cut across multiple cancer types at the protein level in addition to the mRNA level. Results from protein analysis were often concordant with corresponding mRNA analysis. However, proteomic patterns were often not observable in transcriptomic patterns or vice versa. This aspect demonstrates the need for proteomics in capturing biological information that may not be present within transcriptomics data, where the vast majority of gene expression profiling studies to date have dealt with only the transcriptome. Interestingly, we found more proteins differentially expressed with higher grade than with higher stage, suggesting that grade as a clinical measure could be capturing more in terms of biological differences. Simultaneously, significant associations for both grade and stage signatures were identified in downstream analyses integrating protein results with other modalities such as pathway associations, demonstrating the signatures as enriched for real biological correlations. Our results are consistent with previous studies involving multiple cancer types identifying widespread transcription patterns associated with tumor grade [15, 32, 33]. The associations of a subset of the proteomic grade signatures with CNA patterns may represent proteins that are selectively amplified or lost in the more aggressive cancers. Numerous pathways of interest are associated with the grade- or stage-associated proteins, including pathways of altered metabolism and Warburg-like effects. This particular finding substantiates previous findings from other studies utilizing mRNA or protein analysis of clear cell renal cell carcinoma [9, 34] and lung adenocarcinoma [35].
Gene-level and protein-level results of the present study, for all proteins profiled, are provided as supplementary data, with the intention that these would provide a resource to the research community. Supplementary data tables offer not only differential statistics by grade or stage for each protein across multiple cancer types, but also corresponding differential patterns at the mRNA or phosphoprotein levels, corresponding differential gain or loss at the copy number level, and corresponding patient survival associations in independent cohorts. By integrating differential protein expression with the other modalities offered here, one may be more confident of a given association or obtain more information as to what the association may represent. Besides the supplementary data, we have added the CPTAC datasets to the user-friendly UALCAN data portal [17], facilitating differential analyses by protein. These data would facilitate exploration for potential and novel targets for therapy. Also, genes studied extensively in the laboratory setting, using experimental models that may not entirely reflect human tumors, may be examined in our results, thereby reinforcing the gene’s potential relevance to disease in the patient setting. The potential of these proteomic signatures to form the basis of a molecular assay to guide clinical decision making represents future work. However, this would be conceivable given analogous efforts with mRNA-based testing [36].
Results from our functional studies indicate that identifying overexpressed kinases using proteomic analysis of tumor tissues is a useful and tractable approach for the identification of novel therapeutic drug targets. The experimental results provide a proof-of-concept regarding the resource value of our compiled results. We observed impacts on either cell viability or 2-D migration in uterine endometrial cancer cells or both for kinases MAP3K2, MASTL, and TTK. The kinase MAP3K2 (MEKK2) activates the MEK5/ERK5 cell signaling pathway and is thought to play an important role in tumor growth and metastasis [37]. MASTL, or microtubule-associated serine/threonine kinase like, is an important mitotic kinase that regulates mitotic progression of normal or transformed cells by blocking tumor suppressor protein phosphatase 2 A (PP2A) activity, where MASTL deregulation has been detected in multiple cancer types and associated with aggressive clinicopathological features [38]. The spindle assembly checkpoint kinase TTK (Mps1) is a key regulator of chromosome segregation, with functional roles demonstrated for multiple cancer types [39]. Inhibitors exist for MAP3K2, MASTL, and TTK [37,38,39]. Previously studied in other cancer types, these kinases may represent therapeutic targets for uterine endometrial cancer in particular. Additional proteins of interest remained to be uncovered and explored for uterine and other cancer types, using the CPTAC proteomic datasets.
Methods
Additional details are provided in Supplementary Materials
Molecular profiling datasets
Results are based in part upon data generated by CPTAC, by TCGA Research Network (http://cancergenome.nih.gov/), and by the Children’s Brain Tumor Tissue Consortium (CBTTC). CPTAC [40] generated the mass spectrometry-based proteomic data used in this publication. Molecular profiling data were generated through informed consent as part of CPTAC efforts and analyzed per CPTAC data use guidelines and restrictions. We obtained processed protein expression data from the supplementary tables of the associated CPTAC publications or from the CPTAC Data Portal (https://cptac-data-portal.georgetown.edu/cptacPublic/) [41].
Statistical analysis
All p values were two-sided unless otherwise specified. We performed all tests using log2-transformed expression values. To determine differential levels for a given gene or protein according to increasing tumor grade or stage, we took the Pearson’s correlation between the log-transformed gene-level molecular values and the grade or stage as translated into a numerical value. When converting tumor grade into a numerical variable, grade categories such as “G1”, “G2”, and “G3”, for example, were translated as 1, 2, and 3, respectively, for differential analyses. When converting clinical or pathological stage into a numerical variable, stage categories such as “1 A”, “1B”, “II”, “III”, and “IV”, for example, were translated as 1, 1, 2, 3, and 4, respectively, for differential analyses. For a given cancer type, we considered for differential analyses only proteins and mRNAs for which the protein was detected in over half of the cancer cases by mass spectrometry. For each cancer type, we selected for downstream analyses the set of differential genes or proteins with Pearson’s p value < 0.01. The FDR that may be associated with each differential gene or protein set was estimated by both Story and Tibshirani method [42] and by permutation testing. Even in instances of nominally significant proteins not exceeding or only moderately exceeding chance expectations by FDR, the nominally significant proteins may still contain molecular information that would represent real biological differences. Therefore, we used the nominally significant p < 0.01 cutoff for downstream analyses, in which most of the protein lists examined showed significant enrichment patterns involving mRNA expression, patient survival, results from other cancer types, copy number alterations, and pathways. We evaluated enrichment of GO annotation terms [43] and wikiPathways [29] within sets of differentially expressed genes using SigTerms software [44] and one-sided Fisher’s exact tests, with FDRs estimated using the method of Storey and Tibshirini [42].
Functional studies
Cell lines were routinely monitored for mycoplasma contamination and grown in sterile conditions and validated by Short Tandem Repeat DNA fingerprinting test. SMARTPool siRNAs were obtained from Dharmacon, Inc. Cells were transfected with siRNAs using the Lipofectamine RNAiMAX Transfection Reagent per manufacturer’s instructions. For cell viability assays, cells were lifted 48 h after siRNA transfection and seeded at a density of 5000 cells per well in BioLite 96 well plates (Thermo Scientific). A second preparation of siRNA transfection media was applied to the cells. Cell growth was monitored daily following the second transfection treatment using the CellTiter-GLO® Luminescent cell viability assay (Promega, G7570) per manufacturer’s instructions. The experiments were performed three times, and plotted as mean ± SEM. For wound healing assays, cells were lifted 48 h after siRNA transfection and seeded 2.0 × 105 cell/mL in 12-well plates. A second preparation of siRNA transfection media was applied to the cells. Two perpendicular wounds were made through the center of the well using a sterile P200 pipette tip 24 h after seeding. To measure wound healing, the same regions of each well were photographed using an Axiovert A1 Tissue Culture microscope attached to an Axiocam 208 camera (Zeiss). The area of the scratch in each image was measured using the Zeiss Blue microscopy software and averaged across the 4 areas measured. The experiments were performed three times, and plotted as mean ± SEM Data was analyzed using an ordinary one-way ANOVA and Dunnett’s multiple comparisons test.
References
Zhang Y, Kwok-Shing NgP, Kucherlapati M, Chen F, Liu Y, Tsang Y, et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. 2017;31:820–32.
Chen G, Gharib T, Huang C, Taylor J, Misek D, Kardia S, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteom. 2002;1:304–13.
Mertins P, Mani D, Ruggles K, Gillette M, Clauser K, Wang P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62.
Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387.
Zhang H, Liu T, Zhang Z, Payne S, Zhang B, McDermott J, et al. Integrated proteogenomic characterization of human high-grade serous ovarian. Cancer Cell. 2016;166:755–65.
Dou Y, Kawaler E, Cui Zhou D, Gritsenko M, Huang C, Blumenberg L, et al. Proteogenomic characterization of endometrial carcinoma. Cell. 2020;180:729–48.
Gillette M, Satpathy S, Cao S, Dhanasekaran S, Vasaikar S, Krug K, et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. 2020;182:200–25.
Vasaikar S, Huang C, Wang X, Petyuk V, Savage S, Wen B, et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177:1035–1049. e1019.
The_Cancer_Genome_Atlas_Research_Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49.
Yuan Y, Van Allen E, Omberg L, Wagle N, Amin-Mansour A, Sokolov A, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol. 2014;32:644–52.
Akbani R, Ng P, Werner H, Shahmoradgoli M, Zhang F, Ju Z. et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887.
Cancer_Genome_Atlas_Research_Network, Weinstein J, Collisson E, Mills G, Shaw K, Ozenberger B, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–20.
Chen F, Zhang Y, Gibbons D, Deneen B, Kwiatkowski D, Ittmann M, et al. Pan-cancer molecular classes transcending tumor lineage across 32 cancer types, multiple data platforms, and over 10,000 cases. Clin Cancer Res. 2018;24:2182–93.
Chen F, Chandrashekar D, Varambally S, Creighton C. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun. 2019;10:5679.
Rhodes D, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–14.
Louis D, Perry A, Burger P, Ellison D, Reifenberger G, von Deimling A, et al. International Society Of Neuropathology-Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24:429–35.
Chandrashekar D, Bashel B, Balasubramanya S, Creighton C, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58.
Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, et al. Multiplatform-based molecular subtypes of non-small cell lung cancer. Oncogene. 2016;36:1384–93.
Cancer_Genome_Atlas_Research_Network, Kandoth C, Schultz N, Cherniack A, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73.
Bonome T, Lee J, Park D, Radonovich M, Pise-Masison C, Brady J, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–12.
Crijns A, Fehrmann R, de Jong S, Gerbens F, Meersma G, Klip H, et al. Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med. 2009;6:e24.
Denkert C, Budczies J, Darb-Esfahani S, Györffy B, Sehouli J, Könsgen D, et al. A prognostic gene expression index in ovarian cancer - validation across different independent data sets. J Pathol. 2009;218:273–80.
Dressman H, Berchuck A, Chan G, Zhai J, Bild A, Sayer R, et al. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J Clin Oncol. 2007;25:517–25.
Tothill R, Tinker A, George J, Brown R, Fox S, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208.
Yoshihara K, Tsunoda T, Shigemizu D, Fujiwara H, Hatae M, Fujiwara H. et al. High-risk ovarian cancer based on 126-gene expression signature is uniquely characterized by downregulation of antigen presentation pathway. Clin Cancer Res. 2012;18:1374–85.
Pereira B, Chin S, Rueda O, Vollan H, Provenzano E, Bardwell H. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
Jorissen R, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–51.
Zhao H, Ljungberg B, Grankvist K, Rasmuson T, Tibshirani R, Brooks J. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3:e13.
Slenter D, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46:D661–7.
Sridhar R, Hanson-Painton O, Cooper D. Protein kinases as therapeutic targets. Pharm Res. 2000;17:1345–53.
Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100:57–70.
Creighton C, Beer D, Hanash S. Gene expression patterns define pathways correlated with loss of differentiation in lung adenocarcinomas. FEBS Lett. 2003;540:167–70.
Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–46.
Clark D, Dhanasekaran S, Petralia F, Pan J, Song X, Hu Y, et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–83.
Chen G, Gharib T, Wang H, Huang C, Kuick R, Thomas D, et al. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci USA. 2003;100:13537–42.
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26.
Ahmad S, St Hilaire V, Dandepally S, Johnson G, Williams A, Scott J. Discovery and characterization of an iminocoumarin scaffold as an inhibitor of MEKK2 (MAP3K2). Biochem Biophys Res Commun. 2018;496:205–11.
Fatima I, Singh A, Dhawan P. MASTL: A novel therapeutic target for Cancer Malignancy. Cancer Med. 2020;9:6322–9.
Libouban M, de Roos J, Uitdehaag J, Willemsen-Seegers N, Mainardi S, Dylus J, et al. Stable aneuploid tumors cells are more sensitive to TTK inhibition than chromosomally unstable cell lines. Oncotarget. 2017;8:38309–25.
Ellis M, Gillette M, Carr S, Paulovich A, Smith R, Rodland K, et al. Connecting genomic alterations to cancer biology with proteomics: the NCI clinical proteomic tumor analysis consortium. Cancer Disco. 2013;3:1108–12.
Edwards N, Oberti M, Thangudu R, Cai S, McGarvey P, Jacob S, et al. The CPTAC data portal: a resource for cancer proteomics research. J Proteome Res. 2015;14:2707–13.
Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–5.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29.
Creighton C, Nagaraja A, Hanash S, Matzuk M, Gunaratne P. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–96.
Cancer_Genome_Atlas_Research_Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474: 609–15.
Creighton C, Hernandez-Herrera A, Jacobsen A, Levine D, Mankoo P, Schultz N, et al. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS ONE. 2012;7:e34546.
Acknowledgements
This research was conducted using data made available by The Clinical Proteomic Tumor Analysis Consortium (CPTAC), The Children’s Brain Tumor Tissue Consortium (CBTTC), and The Cancer Genome Atlas (TCGA) Consortium. This work was supported by National Institutes of Health (NIH) grants P30CA125123 (CJC), P20CA221729 (MMM), R00HD096057 (DM), and a Core Facility Support Award from the Cancer Prevention Research Institute of Texas (RP160805 (MMM)). Diana Monsivais holds a PDEP Award from the Burroughs Wellcome Fund. We thank Jonathan Kurie for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: CJC, DM, YV; Methodology: CJC, FC, YZ, DM, YV; Investigation: CJC, FC, YZ, RPM, MES, DM, YV, JCF, MMM; Formal Analysis: CJC, FC, YZ, DSC, JCF; Data Curation: CJC, SV, DSC; Visualization; CJC; Writing: CJC, DM, YV; Manuscript Review: FC, YZ, DSC, RPM, MES; Supervision: CJC, SV, DM, MMM.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Monsivais, D., Vasquez, Y.M., Chen, F. et al. Mass-spectrometry-based proteomic correlates of grade and stage reveal pathways and kinases associated with aggressive human cancers. Oncogene 40, 2081–2095 (2021). https://doi.org/10.1038/s41388-021-01681-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01681-0
- Springer Nature Limited
This article is cited by
-
An essential gene signature of breast cancer metastasis reveals targetable pathways
Breast Cancer Research (2024)
-
Rummagene: massive mining of gene sets from supporting materials of biomedical research publications
Communications Biology (2024)
-
APOB is a potential prognostic biomarker in hepatocellular carcinoma
Discover Oncology (2024)
-
Global impact of somatic structural variation on the cancer proteome
Nature Communications (2023)
-
The N6-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway
Molecular Cancer (2022)