Abstract
RUNX3 is frequently inactivated by DNA hypermethylation in numerous cancers. Here, we show that RUNX3 has an important role in modulating apoptosis in immediate response to tumor necrosis factor-related apoptosis-including ligand (TRAIL). Importantly, no combined effect of TRAIL and RUNX3 was observed in non-cancerous cells. We investigated the expression of the death receptors (DRs) DR4 and DR5, which are related to TRAIL resistance. Overexpression of RUNX3 increased DR5 expression via induction of the reactive oxygen species (ROS)-endoplasmic reticulum (ER) stress-effector CHOP. Reduction of DR5 markedly decreased apoptosis enhanced by the combined therapy of TRAIL and RUNX3. Interestingly, RUNX3 induced reactive oxygen species production by inhibiting SOD3 transcription via binding to the Superoxide dismutase 3 (SOD3) promoter. Additionally, the combined effect of TRAIL and RUNX3 decreased tumor growth in xenograft models. Our results demonstrate a direct role for RUNX3 in TRAIL-induced apoptosis via activation of DR5 and provide further support for RUNX3 as an anti-tumor.
Similar content being viewed by others
Introduction
Tumor necrosis factor-related apoptosis-including ligand (TRAIL), a member of the tumor necrosis factor cytokine family, binds to death receptors, DR4 [1] and DR5 [2]. TRAIL causes cell death in various cancers through intrinsic and extrinsic death pathways but does not cause normal primary epithelial cell death [3, 4]. In the extrinsic death pathway, DR-induced cell death is initiated when TRAIL binds to DR4 and DR5, forming the death-inducing signaling complex (DISC) which leads to the cleavage of pro-caspase-8. Caspase-8 activation results in the formation of caspase-3 to initiate apoptosis. Approximately 70% of tumor cells are sensitive to TRAIL therapy. However, cotreatment with several chemotherapeutic agents is required to induce cytotoxic effects, as cancer cells are tolerant to TRAIL therapy. The systems of TRAIL resistance were reported previously [5,6,7,8], including increase of Fas-associated death domain-like IL-1-converting enzyme-inhibitory protein, decrease of DR4 and DR5, loss of apoptotic proteins such as Bax, Bak, PUMA, and BIM, and upregulation of the NK-κB and PI3K/AKT pathways [9,10,11,12]. Therefore, therapeutic strategies using a combination of genes, molecules, or agents to overcome TRAIL resistance are urgently needed.
The human runt-related transcription factor (RUNX) genes are homologous to the Drosophila genes, runt and lozenge [13, 14], encoding the α subunit of the Runt-domain transcription factor PEBP2/CBF [15]. RUNX family consists of RUNX1, RUNX2, and RUNX3. Among them, RUNX3 is a tumor suppressor and was shown to be downregulated in various cancers [16]. RUNX3 is inactivated by promoter hypermethylation, protein mislocalization, or loss of heterozygosity [17, 18]. RUNX3 was reported to inhibit cancer proliferation, the cell cycle, and metastasis [19, 20]. These reports highlight the importance of RUNX3 in regulating apoptosis and cell cycle genes. Therefore, we predicted that RUNX3 could improve drug sensitivity when treated with several chemotherapeutic agents.
The endoplasmic reticulum (ER) related to variety cellular processes, including protein synthesis, protein folding, and stress-sensing [21, 22]. One factor related to cell death by ER stress is the CCAAT/enhancer-binding protein homologous protein (CHOP), which is preferentially upregulated at the translational level upon eIF2 phosphorylation. Increase of CHOP protein following ER stress and unfolded protein response induce the expression of pro-apoptotic proteins [23,24,25,26,27]. Thus, various types of anti-cancer drugs or molecular regulators for modulating proteins related to ER stress reaction have been developed. Our study, we demonstrated the anti-cancer effects of RUNX3 in sensitizing TRAIL-tolerant colorectal cancer (CRC) cells to TRAIL-induced apoptosis through regulation of the ER stress reaction and apoptosis pathways.
We investigated whether RUNX3 sensitizes human CRC cells to TRAIL-mediated apoptosis. We observed that RUNX3 elevated TRAIL-mediated apoptosis by inducing DR5 expression and the caspase-dependent pathway. Furthermore, we found that RUNX3 has a critical role in regulating ROS via downregulation of extracellular superoxide dismutase 3 (SOD3) expression. Overall, our data suggest that a cotreatment of TRAIL and RUNX3 is an attractive strategy for cancer treatment.
Results
RUNX3 sensitizes TRAIL-induced cell death in human CRC cells
We confirmed the RUNX3 protein in tumor tissue of human CRC by immunohistochemistry (IHC) analysis. The level of RUNX3 was lower in tumors compared to in normal colon tissues (Fig. 1a). Overexpression of RUNX3 was not previously reported to induce anti-proliferative effects in CRC cells [19]. To study function of RUNX3 in TRAIL-mediated cell death, we performed RUNX3 overexpression and knockdown experiments. First, to assess the TRAIL sensitizing effects of RUNX3, we performed an MTT assay. Cell proliferation was not altered by RUNX3 expression (Fig. 1b, Supplementary Fig. 1A). TRAIL mediated the dose-dependent reduction in CRC cell proliferation (Fig. 1c). When RUNX3-overexpressing cells were exposed to TRAIL, cell death was significantly enhanced in TRAIL-resistant HT29, Colo205, and DLD-1 cell lines (Fig. 1e). However, cell death did not induce in normal epithelial primary colon cells (CCD-18Co) and normal lung cells (BEAS-2B) (Fig. 1d, Supplementary Fig. 1B). Additionally, cell death was significantly increased in HT29 cells at various concentrations (Supplementary Fig. 1C). Additionally, knockdown of RUNX3 decreased TRAIL-induced cell death (Fig. 1f). These results show that RUNX3 sensitizes CRC cells to TRAIL-mediated cell death.
RUNX3 sensitizes TRAIL-induced cell death in human CRC cells. a Representative immunofluorescence image of human CRC specimens stained for RUNX3 at a magnification of ×20 and ×40. b Cell viabilities of colorectal cancer (CRC) cell lines transfected with pFlag-c1 RUNX3 were measured by an MTT assay for 24 h. c Cell viabilities of colorectal cancer (CRC) cell lines were measured by an MTT assay and treated with 0–100 ng/mL TRAIL for 6 h. d, e Normal colon cell line (CCD-18Co) and CRC cell lines transfected with pFlag-c1 RUNX3 treated with TRAIL 20 ng/mL. Cell viabilities were assessed by trypan blue dye staining. f SNU283 was transfected with RUNX3 shRNA and then treated with TRAIL 30 ng/mL. The cell viabilities were assessed by trypan blue dye staining. Data are expressed as the means of three independent experiments. **P < 0.01
RUNX3 enhances TRAIL-mediated apoptosis in CRC cells by activating DR and apoptotic pathways
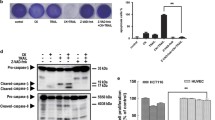
First, the combined efficacy of TRAIL and RUNX3 expression on HT29 cell morphology was studied under a microscope and confirmed to be altered compared to in HT29 control cells or RUNX3-overexpressing cells with TRAIL (Fig. 2a). The colony-forming ability was reduced in RUNX3-overexpressing cells with TRAIL compared to that when either was treated alone (Fig. 2b). Next, we performed Annexin V/propidium iodide (PI) staining with fluorescence-activated cell sorting (FACS) analysis to determine if apoptotic sensitivity was related to the enhanced expression of TRAIL induced by RUNX3. TRAIL-induced apoptosis in RUNX3-overexpressing cells was significantly increased (Fig. 2c). To confirm these, we tested the activation of pro-apoptotic proteins, such as cleaved caspase-8, caspase-9, caspase-3, and PARP. The levels of the cleaved forms of these proteins were increased under combined expression of RUNX3 and TRAIL (Fig. 2d). We pretreated the cells with a pan-caspase inhibitor, z-VAD-fmk, to find the role of caspase in the effects mediated by RUNX3-overexpressing cells with TRAIL. As expected, z-VAD-fmk significantly decreased the activities of caspase-8, 9, 3 and PARP mediated by RUNX3-overexpressing cells with TRAIL (Fig. 2e). RUNX3-hypermethylated CRC cells (HT29) were treated with a DNA methylation inhibitor, 5-Aza-dc, to find the efficacy of RUNX3 methylation on TRAIL. 5-Aza-dc induced significant upregulation of RUNX3 expression in HT29 cells and enhanced TRAIL-induced apoptosis (Fig. 2f). Therefore, the enhanced apoptosis caused by RUNX3-overexpressing cells with TRAIL was induced through both intrinsic and extrinsic pathways.
RUNX3 enhanced TRAIL-induced apoptosis in CRC cells by activating DR and mitochondrial apoptotic pathways. a HT29 cells were treated with 20 ng/mL TRAIL for 4 h, and the cell morphologies were examined by light microscopy (upper). Scale bar: 100 µM. The graphs represent the percentage of live cells using MTT solution (lower). b HT29 cells were treated with 20 ng/mL of TRAIL. After 14 days, cells were stained with crystal violet dye, and photographs of the colonies are shown (upper). The graphs represent the percentage of stained colonies (lower). c HT29 cells treated with TRAIL were stained with annexin V and propidium iodide and then analyzed via FACS. d The levels of cleaved-PARP and cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. e HT29 cells were pretreated with 25 µM z-VAD-fmk for 30 min and then treated with 20 ng/mL of TRAIL for 4 h. The levels of cleaved-PARP and cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. f HT29 cells were pretreated with 20 μM of 5-Aza-dc for 3 days and then treated with 20 ng/mL of TRAIL for 4 h. The levels of cleaved-PARP and RUNX3 were detected by western blotting. β-Actin was used as a loading control for each lane. Data are expressed as the means of three independent experiments. **P < 0.01
RUNX3 enhances TRAIL-induced apoptosis by increasing DR5 expression
To further study on the mechanisms of RUNX3-induced TRAIL sensitivity, we determined pro-apoptotic proteins, anti-apoptotic proteins, and DRs. We found that DR5 expression was markedly increased by RUNX3 overexpression in HT29 cells (Fig. 3a). Upregulation of DR5 by RUNX3 was also confirmed in Colo205 and DLD-1 cells (Supplementary Figure 2A). However, DR4 expression was unchanged in the other CRC cell lines (Fig. 3a and Supplementary Figure 2A). In previous studies, DR5 was downregulated in normal tissues compared to that in cancer tissues [28]. Because the expression of DR5 was very low in the normal cell line (CCD-18Co), RUNX3 caused only a slight change in DR5 levels (Supplementary Figure 2D). Additionally, the protein levels of Bim, Bid, Survivin, Bcl-2, Bcl-xL, and Mcl-1 were unaltered by RUNX3 overexpression (Fig. 3a). Induction of DR5 was confirmed by immunofluorescence (Fig. 3b) and FACS analysis (Fig. 3c and Supplementary Figure 2C). To confirm the importance of upregulated DR5 expression, we transfected pCDNA DR5 into HT29 cells, where overexpression of DR5 caused enhanced TRAIL-induced apoptosis (Fig. 3d). In contrast, knockdown by DR5 siRNA inhibited the RUNX3-sensitizing effect of TRAIL-mediated apoptosis (Fig. 3e, f). Also, we assessed whether TRAIL-mediated apoptosis enhanced by RUNX3 after DR4 knockdown by siRNA but observed no significant effect (Supplementary Figure 2E). These results show that RUNX3 enhances TRAIL-induced apoptosis by upregulating DR5.
RUNX3 enhanced TRAIL-induced apoptosis by increasing DR5 expression. a Protein expression levels of BAX, BIM, NOXA, PUMA, p53, BID (pro-apoptotic protein), survivin, BCL2, Mcl-1, XIAP (anti-apoptotic protein), DR4 (death receptor), and DR5 were determined by western blotting. b Immunofluorescence of DR5 and RUNX3 was detected by confocal laser-scanning microscopy (original magnification: ×40). Scale bar: 10 µM. c HT29 and HT29 pFlag-C1 RUNX3 analyzed by FACS to determine the expression of DR5. d HT29 cells were transfected with pCNDA DR5. Cells were treated with 20 ng/mL TRAIL for 4 h. Cell viabilities were assessed by trypan blue dye staining. e HT29 and HT29-RUNX3-overexpressing cells were transfected with control siRNA or DR5 siRNA. The levels of cleaved-PARP, cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. f HT29 and HT29-RUNX3-overexpressing cells were transfected with control siRNA or DR5 siRNA. HT29 cells treated with 20 ng/mL TRAIL were stained with annexin V and propidium iodide, and then analyzed via FACS. Data are expressed as the means of three independent experiments. **P < 0.01
Overexpression of RUNX3 induces activation of the ROS-ER stress-CHOP pathway
To investigate how RUNX3 regulates DR5 expression, we evaluated DR5 mRNA levels by real-time PCR. Overexpression of RUNX3 elevated both the mRNA level and protein levels of DR5 (Fig. 4a and Supplementary Figure 2B). The DR4 mRNA was increased by Runx3 overexpression, but the effect may not be significant (Fig. 4a). These data show that RUNX3 regulates the activation of DR5 at the transcription level. Because CHOP is known to act upstream of DR5 activation [29], we tested whether RUNX3 regulates CHOP and ER stress proteins. We found that RUNX3 overexpression induced DR5 via IRE1α-JNK-CHOP (Fig. 4b). We also confirmed that transient RUNX3 overexpression increased ER stress via IRE1α-JNK-CHOP (Supplementary Figure 3B). To verify whether ER stress caused by overexpression of RUNX3 was induced via IRE1α, we inhibited IRE1α with IRE1α siRNA. We observed a reduction in TRAIL-induced apoptosis following inhibition of IRE1α in RUNX3-overexpressing cells (Fig. 4c). Because ROS are known to cause ER stress [30], we postulated that RUNX3-mediated ROS can lead to enhanced ER stress. We found that RUNX3 significantly increased ROS generation using DCF-DA (Fig. 4d), which was confirmed by immunofluorescence analysis (Fig. 4e). To further investigate whether ROS generation by RUNX3 affects TRAIL-induced apoptosis, we pretreated the cells with 1 mM ROS scavenger, N-acetyl-l-cysteine (NAC), for 1 h. Apoptotic proteins, whose expression was enhanced by RUNX3-overexpressing cells with TRAIL, were inhibited by NAC (Fig. 4f), indicating that ROS acts a direct role in the susceptibility to TRAIL. Additionally, we pretreated the cells with 20 µM JNK inhibitor for 1 h. Apoptosis enhanced by the RUNX3-overexpressing cells with TRAIL was decreased by the JNK inhibitor (Supplementary Figure 3C). Additionally, knockdown of CHOP by siRNA decreased apoptosis in RUNX3-overexpressing cells with TRAIL (Fig. 4g). Our data represent that the ROS induced by RUNX3 enhanced TRAIL-induced apoptosis.
Overexpression of RUNX3-induced activation of the ROS-ER stress-CHOP pathway. a mRNA levels of DR5 and DR4 were measured by real-time PCR. Expression was normalized to that of GAPDH. b Protein expression levels of p-IRE1α, IRE1α, XBP1S, p-eIF2α, eIF2α, ATF6, GRP94, Bip, Chop, and p-JNK (ER stress) were determined by western blotting. c HT29 cells were transfected with control siRNA or IRE1α siRNA. The levels of p-IRE1α, IRE1α, p-JNK, DR5, CHOP, cleaved-PARP, cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. d ROS levels were measured by DCFH-DA and analyzed by flow cytometry. e To measure ROS generation, the immunofluorescence of DCFH-DA was detected by confocal laser-scanning microscopy (original magnification: ×40). Scale bar: 10 µM. f HT29 cells were pretreated for 1 h with NAC and then treated for 4 h with TRAIL. The levels of DR5, Chop, cleaved-PARP and cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. g HT29 cells were transfected with control siRNA or CHOP siRNA. The levels of DR5, CHOP, cleaved-PARP, cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. Data are expressed as the means of three independent experiments. **P < 0.01
RUNX3 induces ROS generation via transcriptional inhibition of SOD3
To investigate how RUNX3 induces ROS generation, we measured the levels of ROS-related factors. Overexpression of RUNX3 did not change SOD1, SOD2, catalase, NADPH oxidase 2, and NADPH 4 levels, while SOD3 levels were significantly reduced (Fig. 5a). Additionally, RUNX3-overexpressing cells showed relatively low SOD3 expression in CRC cell lines (Supplementary Figure 4B). Consistent with the results at the protein level, the mRNA level of SOD3 was also decreased (Fig. 5b, Supplementary Figure 4C), whereas the mRNA level of SOD3 in RUNX3-knockdown cells was increased (Fig. 5c and Supplementary Figure 4D). The mRNA levels of SOD1 and SOD2 did not change in RUNX3-overexpressing cells (Supplementary Figure 4A). These results indicate that the transcriptional level of SOD3 was regulated by RUNX3. In agreement with these finding, overexpression of RUNX3 inhibited SOD3, as observed in immunofluorescence analysis (Fig. 5d). Because RUNX3 affected both the SOD3 protein and mRNA levels, we predicted that RUNX3 transcriptionally inhibits SOD3 expression. Within the promoter region (−3500–100 bp upstream), we found two RUNX3-binding sites (Fig. 5e). To evaluate the binding of RUNX3 within the SOD3 promoter, we achieved a chromatin immunoprecipitation (ChIP) assay, which demonstrated that RUNX3 binds to RUNX3BS1 and RUNX3BS2 in RUNX3-overexpressing cells (Fig. 5f). This indicates that binding of RUNX3 within the SOD3 promoter inhibited SOD3 transcription. To further confirm the function of SOD3, we transfected SOD3 siRNA into HT29 cells. Knockdown of SOD3 enhanced TRAIL-mediated increase of caspase-3, caspase-9, and caspase-8 as well as PARP cleavage (Fig. 5g). Additionally, increased TRAIL-mediated apoptosis was verified by FACS analysis in SOD3 knockdown cells (Supplementary Figure 4E). Overexpression of SOD3 inhibited the sensitivity of RUNX3-overexpressing cells for TRAIL-induced apoptosis (Fig. 5h). These data suggest that RUNX3 synergistically promotes TRAIL-induced apoptosis by downregulating SOD3 expression.
RUNX3 induces ROS generation via transcriptional inhibition of SOD3. a The levels of SOD1, SOD2, SOD3, catalase, NOX2, and NOX4 were detected by western blotting. β-Actin was used as a loading control for each lane. b, c The mRNA level of SOD3 was measured by real-time PCR. The expression was normalized to that of GAPDH. d Immunofluorescence of SOD3 was detected by confocal laser-scanning microscopy (original magnification: ×40). Scale bar: 10 µM. e The two predicted RUNX3-binding sites in the SOD3 promoter presented as an illustration and as DNA sequence (from −3500 to +100). f A chromatin immunoprecipitation (ChIP) assay was performed to confirm direct binding of RUNX3 to the SOD3 promoter. g HT29 cells were transfected with control siRNA or SOD3 siRNA. The levels of SOD3, cleaved-PARP, cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. h HT29 cells were transfected with Myc-DDK SOD3. The levels of Myc, DR5, CHOP, cleaved-PARP, cleaved-caspase3, cleaved-caspase9, and cleaved-caspase8 were detected by western blotting. β-Actin was used as a loading control for each lane. Data are expressed as the means of three independent experiments. **P < 0.01, *P < 0.05
RUNX3 overexpression enhances TRAIL-induced apoptosis in vivo
HT29-FLAG- and HT29-RUNX3-overexpressing cells were subcutaneously injected into nude mice and treated with 4 ng/kg TRAIL every two days. Treatment with RUNX3-overexpressing cells with TRAIL significantly decreased tumor growth compared to in the control, control + TRAIL, and RUNX3 groups (Fig. 6a, b). Tumor weight was also lowest in RUNX3-overexpressing cells with TRAIL group compared to that in the other groups (Fig. 6c). We performed IHC to measure the expression of RUNX3, DR5, and SOD3. Consistent with the in vitro results, DR5 expression was increased in RUNX3-overexpressing groups, while SOD3 expression was decreased in these groups (Fig. 6d). Lastly, we performed a TUNEL assay to detect cell apoptosis. Samples from RUNX3-overexpressing cells with TRAIL group contained significantly more apoptotic cells than those from other groups (Fig. 6e). These data show that RUNX3 synergistically increased TRAIL-mediated cell apoptosis in vivo.
RUNX3 enhanced TRAIL-induced apoptosis in vivo. a HT29 cells were implanted subcutaneously in nude mice and then tumor growth was evaluated by fluorescence intensity after 3 weeks of treatment with TRAIL at a dose of 4 ng/kg every 2 days (n = 6). b Representative tumor volume from xenograft nude mice. Tumor dimensions were measured every two days. c The tumor weights were measured at the termination of the experiments. d Immunohistochemical (IHC) staining revealed the expression of RUNX3, DR5, and SOD3 in tumors from xenograft mice of the two groups at a magnification of ×20. Scale bar, 50 μm. Graphs represent the expression of RUNX3, DR5, and SOD3 by IHC staining (right). e The tumors were subjected to a TUNEL assay and DAPI was used to visualize the nucleus (left). The percentage of TUNEL-positive cells was counted and plotted as a histogram (right). Scale bar, 20 μm. f Schematic for the working model of TRAIL-induced apoptosis by RUNX3. The data are expressed as the means of three independent experiments. **P < 0.01
Discussion
Because TRAIL and agonistic antibodies against DRs are safe and tumor-specific, these proteins have been widely developed as therapeutic molecules and compared to other tumor necrosis factor family members. However, because of the resistance of tumors to these molecules and their insufficient effects, it has become necessary to study their effects when combined treatment with other chemotherapeutic agents or adjuvant therapies. Therefore, therapies that elevate the efficacy of TRAIL and overcome TRAIL resistance are needed.
CRC cells have been reported to be resistant to TRAIL [5, 31]. The high concentration of 500 ng/mL of TRAIL used to treat CRC cells shows no effect [32]. Surprisingly, the present study revealed greater downregulation of human RUNX3 in CRC tissues and related cell lines than in normal colon tissues and human primary CRC. Notably, the IHC data suggested that downregulation of RUNX3 expression is clinically related to carcinogenesis and the TRAIL resistance of CRC. Epidemiological studies of RUNX3 in CRC have not clearly revealed the functional participation of RUNX3 in tumor suppression [33, 34]. Recently, RUNX3 was reported to function in pro-apoptosis. RUNX3 has a pivotal role in lung normal epithelial cell apoptosis induced by influenza A virus infection and Toll-like receptor-dependent interferon-gamma by activating DR-related apoptosis pathways [35, 36]. Whereas, some reports show that RUNX3 plays an oncogenic functions in skin cancers, including basal cell carcinoma [37, 38]. Our previous report demonstrated that RUNX3 inhibits metastasis and invasion act as a tumor suppressor in CRC [19]. Although the RUNX3 have double-edged sword, depending on the type of cancer in a particular area, our study is based on the patient data and CRC cell lines, and suggests that RUNX3 have a role of tumor suppressor.
In this study, we investigated the association between RUNX3 expression and the TRAIL response in various CRC cell lines. We observed restored TRAIL sensitivity following methylated RUNX3 in CRC cells that had been treated with the demethylation reagent. Furthermore, we found that activation of DR5 was involved in regulating RUNX3 in CRC cells. To further determine the functional role of RUNX3, we evaluated apoptosis-related proteins (anti- or pro-apoptotic) by western blotting; these proteins were found to have novel functions in carcinogenesis. Recently, anti-apoptotic proteins responsible for TRAIL resistance, such as survivin and Mcl-1, were reported to associate with Bcl-2 or Bcl-xL members [39, 40]. Overexpressing RUNX3/TRAIL in CRC cell lines resulted in significantly enhanced cell death, which was not observed in siRUNX3- and/or TRAIL-treated CRC cells; this observation further supports the finding that RUNX3 functions as a pro-apoptotic protein, particularly in TRAIL-resistant cancers. This was also supported by transient overexpression of RUNX3 in CRC cell lines, suggesting that RUNX3 sensitizes the apoptotic machinery to TRAIL-induced death signals. Our study using a mouse xenograft model confirmed that RUNX3 overexpression combined with TRAIL leads to cell death, strongly supporting the role of RUNX3 as a pro-apoptotic protein. Mechanistically, RUNX3 significantly increased the protein levels of pro-apoptotic proteins, including NOXA and Bax, and upregulated DR5 expression. Additionally, overexpression of RUNX3/TRAIL induced apoptosis, but not of human primary epithelial colon cells, indicating that RUNX3 overexpression combined with TRAIL is a promising therapy for cancers showing resistance to TRAIL therapy.
ER stress is triggered when unfolded or misfolded proteins were aggregated in the ER lumen. ER stress increases the expression level of DRs through the ATF4-CHOP [41,42,43] or ATF6-CHOP signaling pathways [44, 45]. Specifically, ER stress is activated by oxidative agents modulate the level of survival or apoptotic proteins [46]. In agreement with these observations, we identified that ER stress mediated by RUNX3 significantly increases DR5 by inducing the IRE1α/JNK/CHOP pathway and upregulates NOXA in TRAIL-resistant CRC cell lines.
In a previous study, we studied a close association between ER stress and DR5 activation after TRAIL treatment [5, 31]. Additionally, thapsigargin [47], tunicamycin [48], and MG132 [49], which are ER stress inducers, have been confirmed to increase DR5 in numerous cancer cells. Regulation of DR5 by CHOP provides association between ER stress and DR5. Our data are consistent with previous finding that CHOP binds to within DR5 promoter and increases its expression [5, 50, 51]. We investigated whether RUNX3 directly regulates DR5 transcription. Expression of DR4 and DR5 did not change in RUNX3-knockdown cells (Supplementary Figure 3A). As a result, we confirmed that RUNX3 controls the activation of DR5 via CHOP. It appears likely that RUNX3 induces ER stress. We found that important upstream pathway related to the sensitive of TRAIL by RUNX3 is ROS. RUNX3 induced ROS quenching by NAC, anti-oxidant, which attenuated the effects of RUNX3 on inducing CHOP and DR5. Additionally, quenching of ROS attenuated TRAIL-induced apoptotic cell death by RUNX3. Our findings agree with those of previous studies showing that RUNX3 promotes accumulation of ROS and PKC-θ activity for DR5 activation [52].
The increase in DR5 levels is of specific interest for selectively targeting DR5, but not DR4, to treat cancer in humans. Recently, several studies reported that DR5 has potential pro-metastatic functions in cancer [41, 53,54,55,56]. Thus, upregulation of DR5 by RUNX3 suggests its potential in therapy by enhancing the sensitivity to TRAIL. Our data demonstrate that DR5 is important for enhancing the sensitivity of TRAIL. Knockdown of DR5 decreased the TRAIL sensitivity enhanced by RUNX3. In previous studies, ROS was shown to induce ER stress in various cancers [57]. Therefore, we hypothesized that RUNX3-induced ER stress could be induced by ROS, which is also involved in RUNX3 overexpression and apoptosis. ROS have pivotal role in cell death and are associated with overcoming TRAIL resistance by inducing DR5 [29, 58]. ROS are generated by the mitochondria and NADPH oxidases (NOX enzymes) [59], which are known to produce large amounts of superoxide. SOD is an enzyme that catalyzes the dismutation of the superoxide (O2−) radical into oxygen or hydrogen peroxide. Thus, SODs prevent the generation of highly aggressive ROS. SOD1 is located in the cytosol, SOD2 in the mitochondria, and SOD3 is extracellular [60]. We evaluated the expression of NOX and SODs to investigate how RUNX3 regulates ROS. Overexpression of RUNX3 decreased SOD3 protein level, although the level of other factors did not alter, while the RNA level of SOD3 was also downregulated. We also found that RUNX3 regulates the transcription of SOD3 by binding to two sites, which we identified within the SOD3 promoter, to inhibit its expression.
In summary, the results of our study demonstrate that decreased SOD3 expression by RUNX3 increased ROS induction. And then, upregulation of DR5 via the ROS-ER stress-CHOP pathway enhanced TRAIL-induced apoptosis (Fig. 6f).
Our study reveals the mechanism of how RUNX3 sensitizes CRC cells to TRAIL-mediated cell death and enables to overcome TRAIL resistance. Future examinations should aim to investigate the clinical potential of combined treatment with RUNX3 and TRAIL in CRC patients.
Materials and methods
Patients and tissue specimens
A total of 197 cases of colon cancers were collected from the Korea University Guro Hospital tissue bank between 2000 and 2006. The patients with colon carcinoma were 109 males and 88 females. This protocol was approved by the Institutional Review Board of Guro Hospital (KUGH 12110).
Cell culture and stable cell lines
Human colon cancer cells were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI 1640 medium or McCoy’s 5 A medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (HyClone, Logan, UT, USA) and 1% antibiotic-Antimycotic (Katy, TX, USA). Human colon cells (CCD18CO) and human lung cells (BEAS-2B) were purchased from American Type Culture Collection. For RUNX3 stable overexpression, we cloned RUNX3 between the HindIII and BamHI restriction enzyme sites in the pFlag-c1 vector. We then transfected the cells with pFlag-c1 or pFlag-c1 RUNX3 using Lipofectamine 2000 (Invitrogen) and selected transfectants with puromycin.
Reagents and antibodies
Anti-DR5 (AF631) and TRAIL were purchased from R&D Systems (Minneapolis, MN, USA). NAC, 5-aza-2′-deoxycytidine, and STF-083010 were purchased from Sigma (Sigma, St. Louis, MO, USA). Anti-BAX (sc-20067), anti-Bcl-xL (sc-8392), anti-Bcl-2 (sc-509), anti-DR4 (sc-6823), anti-CHOP (sc-7531), anti-SOD3 (sc-271170), anti-Myc (sc-788), and Protein G PLUS-Agarose (sc-2002) were from Santa Cruz Biotechnology (Dallas, TX, USA); Anti-Noxa (#14766), anti-Bim (#2819), anti-Puma (#4976), anti-XIAP (#2042), anti-Bid (#2002), anti-Mcl-1 (#4572), anti-Survivin (#2808), anti-cleaved PARP (#9541), anti-caspase-8 (#4927), anti-caspase-3 (#9662), anti-caspase-9 (#9508), anti-phospho JNK (#9251), anti-XBP1s (#12782), anti-eIF2α (#9722), anti-phospho eIF2α (#3597), anti-ATF6 (#65880), anti-GRP94 (#2104), anti-BIP (#3177), anti-IRE1α (#3294), anti-SOD1 (#2770), and anti-SOD2 (#13194) were from Cell Signaling Technology (Danvers, MA, USA); anti-catalase (ab76110), anti-RUNX3 (ab40278), anti-DR5 (ab8416), anti-phospho IRE1 α, anti-NOX2 (ab129068), and anti-NOX4 (ab133303) were from Abcam (Cambridge, UK); anti-Flip (AXL-804-961-0100) was from Enzo Life Sciences (Farmingdale, NY, USA); anti-actin antibody (A5316) was from Sigma. The secondary antibodies, anti-mouse IgG horseradish peroxidase and anti-rabbit IgG horseradish peroxidase, were from Cell Signaling Technology.
Immunoblotting
Cells were prepared and western blotting was performed as previously described [61]. Signals were detected on X-ray film. Protein sizes were determined using GangNam-Stain protein ladder (Cat. 24052).
Survival assay
Cell proliferation was measured using MTS reagent. Cells were seeded and then treated. Cells were incubated with EZ-Cytox kit MTS reagent (DoGen) for 4 h at 37 °C. Absorbance at 550 nm was measured using a microplate reader.
Apoptosis assay (flow cytometry)
HT29 cells and HT29-RUNX3 cells were untreated or treated with TRAIL for 4 h. The cells were mixed with7 μL FITC reagent and 1.25 μL Annexin V (BioBud, Seoul, Korea, Cat. LS-02-100) and incubated for 30 min at 4 °C in the dark. The cells were immediately analyzed by flow cytometry.
Methylation specific PCR (MSP)
Genomic DNA was extracted by the DNeasy kit (Qiagen, Hilden, Germany). Genomic DNA was modified by bisulfite using a Methylamp™ DNA modification kit (Epigentek, Pittsburgh, PA, USA) according to the manufacturer’s instructions. PCR was performed hot-started at 95 °C 5 min. this was followed by 40 cycles at 95 °C for 30 s, 65 °C (unmethylated) or 71 °C (methylated) for 30 s and 72 °C for 40 s, and final elongation at 72 °C for 10 min. Primer sequences for the unmethylated reaction were 5′-TTATGAGGGGTGGTTGTATGTGGG-3′ (sense) and 5′- AAAACAACCAACACAAACACCTCC-3′ (anti-sense), and primer sequences for the methylated reaction were 5′-TTACGAGGGGCGGTCGTACGCGGG-3′ (sense) and 5’-AAAACGACCGACGCGAACGCCTCC-3′ (anti-sense).
Immunofluorescence staining
The expression of DR5 and SOD3 were observed by fluorescence microscopy as previously described [61].
Flow cytometric analysis for DR5
Cells were stained with phycoerythrin-conjugated anti-DR5 (clone DJR2-4, BioLegend, San Diego, CA, USA) on ice for 20 min in the dark. Stained cells were washed with cell staining buffer by centrifugation at 350 × g for 5 min. The stained cells were analyzed by flow cytometry.
Small interfering RNA (siRNA)
DR5 siRNA, DR4 siRNA, CHOP siRNA, IRE1α siRNA, and SOD3 siRNA were purchased from Santa Cruz Biotechnology. Cells were transfected with siRNA using Lipofectamine RNAiMax reagent (Invitrogen) according to the manufacturer’s instructions.
ROS measurement (DCF-DA assay)
ROS levels were measured using dichloro-dihydro-fluorescein diacetate (DCF-DA). Cells were incubated for 30 min with 20 µM DCF-DA. The stained cells were fixed with 3.7% formaldehyde for 15 min at room temperature. Fluorescence intensity was measured using a flow cytometer or fluorescence microscopy.
ChIP (chromatin immunoprecipitation) assay
Overall, 1.5 × 106 HT29 cells and HT29-RUNX3 cells were seeded. ChIP assay was performed as previously described [61]. The DNA was analyzed by PCR using the following specific primers: RUNX3BS1 forward, 5′ GGT ACA GCA GAG GCT CTT AGT AAA 3′; reverse, 5′ ATG CTT GTG GTC CAA ACT ACT GAG 3′; RUNX3BS2 forward, 5′ CCA CCA CAC CTG GTT GAT TT 3′; reverse, 5′ AAA TAA TGG GCC AGA CAC AG 3′.
Animal xenograft experiment
Animal experiments were conducted in accordance with animal care guidelines approved by the Korea University Institutional Animal Care and Use Committee. Four-week-old female BALB/c nude mice were obtained from Orient Bio (Seongnam-si, Korea) and housed in a pathogen-free environment. The animals had a period of adaptation for 1 week before study and had free access to food and water. HT29 cells (2 × 106) in 100 µL of PBS were subcutaneously implanted into BALB/c nude mice. The tumor size was measured every two days. Tumor volume calculations were obtained using the formula V = (width2 × length)/2 after caliper measurements.
Immunohistochemistry staining (IHC) and assessment
Tissue were prepared and IHC was performed as previously described [61]. The antibodies, clones, catalogs, and dilutions are listed in Table 1 and IHC scoring is listed in Table 2.
Statistical analysis
Statistical analysis was performed using GraphPad InStat 6 software (GraphPad Software, Inc., La Jolla, CA, USA). The results were expressed as the mean of arbitrary values ± SEM. Statistics were analyzed by using one-way analysis of variance with GraphPad InStat 6. One-way analysis of variance followed by Turkey post hoc tests were performed in all statistical analysis. To determine the significance between two groups, an unpaired t-test was used, where a P-value of <0.05 was considered significant.
References
Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, et al. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–3.
Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997;16:5386–97.
Yang F, Tay KH, Dong L, Thorne RF, Jiang CC, Yang E, et al. Cystatin B inhibition of TRAIL-induced apoptosis is associated with the protection of FLIP(L) from degradation by the E3 ligase itch in human melanoma cells. Cell Death Differ. 2010;17:1354–67.
Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782–98.
Na YJ, Lee DH, Kim JL, Kim BR, Park SH, Jo MJ, et al. Cyclopamine sensitizes TRAIL-resistant gastric cancer cells to TRAIL-induced apoptosis via endoplasmic reticulum stress-mediated increase of death receptor 5 and survivin degradation. Int J Biochem Cell Biol. 2017;89:147–56.
Lee DH, Rhee JG, Lee YJ. Reactive oxygen species up-regulate p53 and Puma; a possible mechanism for apoptosis during combined treatment with TRAIL and wogonin. Br J Pharmacol. 2009;157:1189–202.
Lee DH, Kim DW, Jung CH, Lee YJ, Park D. Gingerol sensitizes TRAIL-induced apoptotic cell death of glioblastoma cells. Toxicol Appl Pharmacol. 2014;279:253–65.
Shi K, Xue J, Fang Y, Bi H, Gao S, Yang D, et al. Inorganic Kernel-reconstituted lipoprotein biomimetic nanovehicles enable efficient targeting “Trojan Horse” delivery of STAT3-decoy oligonucleotide for overcoming TRAIL resistance. Theranostics. 2017;7:4480–97.
Chawla-Sarkar M, Bae SI, Reu FJ, Jacobs BS, Lindner DJ, Borden EC. Downregulation of Bcl-2, FLIP or IAPs (XIAP and survivin) by siRNAs sensitizes resistant melanoma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2004;11:915–23.
Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–7.
Kandasamy K, Srivastava RK. Role of the phosphatidylinositol 3’-kinase/PTEN/Akt kinase pathway in tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in non-small cell lung cancer cells. Cancer Res. 2002;62:4929–37.
Franco AV, Zhang XD, Van Berkel E, Sanders JE, Zhang XY, Thomas WD, et al. The role of NF-kappa B in TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of melanoma cells. J Immunol. 2001;166:5337–45.
Daga A, Karlovich CA, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–205.
Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–13.
Ito Y. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells. 1999;4:685–96.
Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–24.
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65:7743–50.
Waki T, Tamura G, Sato M, Terashima M, Nishizuka S, Motoyama T. Promoter methylation status of DAP-kinase and RUNX3 genes in neoplastic and non-neoplastic gastric epithelia. Cancer Sci. 2003;94:360–4.
Kim BR, Kang MH, Kim JL, Na YJ, Park SH, Lee SI, et al. RUNX3 inhibits the metastasis and angiogenesis of colorectal cancer. Oncol Rep. 2016;36:2601–8.
Chi XZ, Yang JO, Lee KY, Ito K, Sakakura C, Li QL, et al. RUNX3 suppresses gastric epithelial cell growth by inducingp21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol Cell Biol. 2005;25:8097–107.
Zhang K. Integration of ER stress, oxidative stress and the inflammatory response in health and disease. Int J Clin Exp Med. 2010;3:33–40.
Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29.
Glab JA, Doerflinger M, Nedeva C, Jose I, Mbogo GW, Paton JC, et al. DR5 and caspase-8 are dispensable in ER stress-induced apoptosis. Cell Death Differ. 2017;24:944–50.
Jiang Y, Chen X, Fan M, Li H, Zhu W, Chen X, et al. TRAIL facilitates cytokine expression and macrophage migration during hypoxia/reoxygenation via ER stress-dependent NF-kappaB pathway. Mol Immunol. 2017;82:123–36.
He K, Zheng X, Li M, Zhang L, Yu J. mTOR inhibitors induce apoptosis in colon cancer cells via CHOP-dependent DR5 induction on 4E-BP1 dephosphorylation. Oncogene. 2016;35:148–57.
Zlotorynski E. Apoptosis. DR5 unfolds ER stress. Nat Rev Mol Cell Biol. 2014;15:498–9.
Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, et al. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101.
Ichikawa K, Liu W, Zhao L, Wang Z, Liu D, Ohtsuka T, et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat Med. 2001;7:954–60.
Prasad S, Kim JH, Gupta SC, Aggarwal BB. Targeting death receptors for TRAIL by agents designed by Mother Nature. Trends Pharmacol Sci. 2014;35:520–36.
Boyce M, Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 2006;13:363–73.
Lee DH, Sung KS, Guo ZS, Kwon WT, Bartlett DL, Oh SC, et al. TRAIL-induced caspase activation is a prerequisite for activation of the endoplasmic reticulum stress-induced signal transduction pathways. J Cell Biochem. 2016;117:1078–91.
Toscano F, Fajoui ZE, Gay F, Lalaoui N, Parmentier B, Chayvialle JA, et al. P53-mediated upregulation of DcR1 impairs oxaliplatin/TRAIL-induced synergistic anti-tumour potential in colon cancer cells. Oncogene. 2008;27:4161–71.
Kodach LL, Jacobs RJ, Heijmans J, van Noesel CJ, Langers AM, Verspaget HW, et al. The role of EZH2 and DNA methylation in the silencing of the tumour suppressor RUNX3 in colorectal cancer. Carcinogenesis. 2010;31:1567–75.
Ku JL, Kang SB, Shin YK, Kang HC, Hong SH, Kim IJ, et al. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004;23:6736–42.
Gan H, Hao Q, Idell S, Tang H. Transcription factor Runx3 is induced by influenza A virus and double-strand RNA and mediates airway epithelial cell apoptosis. Sci Rep. 2015;5:17916.
Gan H, Hao Q, Idell S, Tang H. Interferon-gamma promotes double-stranded RNA-induced TLR3-dependent apoptosis via upregulation of transcription factor Runx3 in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2016;311:L1101–L12.
Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS, Kang SG, et al. Expression of RUNX3 in skin cancers. Clin Exp Dermatol. 2011;36:769–74.
Lotem J, Levanon D, Negreanu V, Bauer O, Hantisteanu S, Dicken J, et al. Runx3 at the interface of immunity, inflammation and cancer. Biochim Biophys Acta. 2015;1855:131–43.
Park SH, Lee DH, Kim JL, Kim BR, Na YJ, Jo MJ, et al. Metformin enhances TRAIL-induced apoptosis by Mcl-1 degradation via Mule in colorectal cancer cells. Oncotarget. 2016;7:59503–18.
Harashima N, Takenaga K, Akimoto M, Harada M. HIF-2alpha dictates the susceptibility of pancreatic cancer cells to TRAIL by regulating survivin expression. Oncotarget. 2017;8:42887–900.
Dufour F, Rattier T, Constantinescu AA, Zischler L, Morle A, Ben Mabrouk H, et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress. Oncotarget. 2017;8:9974–85.
Iurlaro R, Puschel F, Leon-Annicchiarico CL, O’Connor H, Martin SJ, Palou-Gramon D, et al. Glucose deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol Cell Biol. 2017;37:e00479–16.
Li T, Su L, Lei Y, Liu X, Zhang Y, Liu X. DDIT3 and KAT2A proteins regulate TNFRSF10A and TNFRSF10B expression in endoplasmic reticulum stress-mediated apoptosis in human lung cancer cells. J Biol Chem. 2015;290:11108–18.
Tian X, Ye J, Alonso-Basanta M, Hahn SM, Koumenis C, Dorsey JF. Modulation of CCAAT/enhancer binding protein homologous protein (CHOP)-dependent DR5 expression by nelfinavir sensitizes glioblastoma multiforme cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J Biol Chem. 2011;286:29408–16.
Liu Z, Shi Q, Song X, Wang Y, Wang Y, Song E, et al. Activating transcription factor 4 (ATF4)-ATF3-C/EBP homologous protein (CHOP) cascade shows an essential role in the ER stress-induced sensitization of tetrachlorobenzoquinone-challenged PC12 cells to ROS-mediated apoptosis via death receptor 5 (DR5) signaling. Chem Res Toxicol. 2016;29:1510–8.
Kapur A, Felder M, Fass L, Kaur J, Czarnecki A, Rathi K, et al. Modulation of oxidative stress and subsequent induction of apoptosis and endoplasmic reticulum stress allows citral to decrease cancer cell proliferation. Sci Rep. 2016;6:27530.
Ma Z, Fan C, Yang Y, Di S, Hu W, Li T, et al. Thapsigargin sensitizes human esophageal cancer to TRAIL-induced apoptosis via AMPK activation. Sci Rep. 2016;6:35196.
Guha P, Kaptan E, Gade P, Kalvakolanu DV, Ahmed H. Tunicamycin induced endoplasmic reticulum stress promotes apoptosis of prostate cancer cells by activating mTORC1. Oncotarget. 2017;8:68191–207.
Park HS, Jun do Y, Han CR, Woo HJ, Kim YH. Proteasome inhibitor MG132-induced apoptosis via ER stress-mediated apoptotic pathway and its potentiation by protein tyrosine kinase p56lck in human Jurkat T cells. Biochem Pharmacol. 2011;82:1110–25.
He L, Jang JH, Choi HG, Lee SM, Nan MH, Jeong SJ, et al. Oligomycin A enhances apoptotic effect of TRAIL through CHOP-mediated death receptor 5 expression. Mol Carcinog. 2013;52:85–93.
Lim JH, Park JW, Choi KS, Park YB, Kwon TK. Rottlerin induces apoptosis via death receptor 5 (DR5) upregulation through CHOP-dependent and PKC delta-independent mechanism in human malignant tumor cells. Carcinogenesis. 2009;30:729–36.
Giambra V, Jenkins CR, Wang H, Lam SH, Shevchuk OO, Nemirovsky O, et al. NOTCH1 promotes T cell leukemia-initiating activity by RUNX-mediated regulation of PKC-theta and reactive oxygen species. Nat Med. 2012;18:1693–8.
Hartwig T, Montinaro A, von Karstedt S, Sevko A, Surinova S, Chakravarthy A, et al. The TRAIL-induced cancer secretome promotes a tumor-supportive immune microenvironment via CCR2. Mol Cell. 2017;65:730–42 e5.
Pal S, Amin PJ, Sainis KB, Shankar BS. Potential role of TRAIL in metastasis of mutant KRAS expressing lung adenocarcinoma. Cancer Microenviron. 2016;9:77–84.
Fritsche H, Heilmann T, Tower RJ, Hauser C, von Au A, El-Sheikh D, et al. TRAIL-R2 promotes skeletal metastasis in a breast cancer xenograft mouse model. Oncotarget. 2015;6:9502–16.
von Karstedt S, Conti A, Nobis M, Montinaro A, Hartwig T, Lemke J, et al. Cancer cell-autonomous TRAIL-R signaling promotes KRAS-driven cancer progression, invasion, and metastasis. Cancer Cell. 2015;27:561–73.
Kadara H, Lacroix L, Lotan D, Lotan R. Induction of endoplasmic reticulum stress by the pro-apoptotic retinoid N-(4-hydroxyphenyl)retinamide via a reactive oxygen species-dependent mechanism in human head and neck cancer cells. Cancer Biol Ther. 2007;6:705–11.
Jung EM, Lim JH, Lee TJ, Park JW, Choi KS, Kwon TK. Curcumin sensitizes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis through reactive oxygen species-mediated upregulation of death receptor 5 (DR5). Carcinogenesis. 2005;26:1905–13.
Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–9.
Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal. 2011;15:1583–606.
Kim BR, Jeong YA, Na YJ, Park SH, Jo MJ, Kim JL, et al. Genipin suppresses colorectal cancer cells by inhibiting the Sonic Hedgehog pathway. Oncotarget. 2017;8:101952–64.
Acknowledgements
This work was supported by a National Research Foundation (NRF) of Korea grant funded the Korean government (MSIP) [NRF-2017R1A6A3A11030765] and was supported by Korea University Grant and was supported by project for cooperative R&D between Industry, Academy, and Research Institute funded Korea ministry of SMES and Startups in 2018 [Grants No. Co558375].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
All experiments were approved by the Ethics Committee of Korea University.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, B.R., Park, S.H., Jeong, Y.A. et al. RUNX3 enhances TRAIL-induced apoptosis by upregulating DR5 in colorectal cancer. Oncogene 38, 3903–3918 (2019). https://doi.org/10.1038/s41388-019-0693-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0693-x
- Springer Nature Limited
This article is cited by
-
Rolapitant treats lung cancer by targeting deubiquitinase OTUD3
Cell Communication and Signaling (2024)
-
Emerging mechanisms of the unfolded protein response in therapeutic resistance: from chemotherapy to Immunotherapy
Cell Communication and Signaling (2024)
-
Ubiquitylation of RUNX3 by RNA-binding ubiquitin ligase MEX3C promotes tumorigenesis in lung adenocarcinoma
Journal of Translational Medicine (2024)
-
RETRACTED ARTICLE: RUNX3-regulated circRNA METTL3 inhibits colorectal cancer proliferation and metastasis via miR-107/PER3 axis
Cell Death & Disease (2022)
-
TRAIL promotes epithelial-to-mesenchymal transition by inducing PD-L1 expression in esophageal squamous cell carcinomas
Journal of Experimental & Clinical Cancer Research (2021)