Abstract
Treatment outcomes widely vary for individuals diagnosed with major depressive disorder, implicating a need for deeper understanding of the biological mechanisms conferring a greater likelihood of response to a particular treatment. Our improved understanding of intrinsic brain networks underlying depression psychopathology via magnetic resonance imaging and other neuroimaging modalities has helped reveal novel and potentially clinically meaningful biological markers of response. And while we have made considerable progress in identifying such biomarkers over the last decade, particularly with larger, multisite trials, there are significant methodological and practical obstacles that need to be overcome to translate these markers into the clinic. The aim of this review is to review current literature on brain network structural and functional biomarkers of treatment response or selection in depression, with a specific focus on recent large, multisite trials reporting predictive accuracy of candidate biomarkers. Regarding pharmaco- and psychotherapy, we discuss candidate biomarkers, reporting that while we have identified candidate biomarkers of response to a single intervention, we need more trials that distinguish biomarkers between first-line treatments. Further, we discuss the ways prognostic neuroimaging may help to improve treatment outcomes to neuromodulation-based therapies, such as transcranial magnetic stimulation and deep brain stimulation. Lastly, we highlight obstacles and technical developments that may help to address the knowledge gaps in this area of research. Ultimately, integrating neuroimaging-derived biomarkers into clinical practice holds promise for enhancing treatment outcomes and advancing precision psychiatry strategies for depression management. By elucidating the neural predictors of treatment response and selection, we can move towards more individualized and effective depression interventions, ultimately improving patient outcomes and quality of life.
Similar content being viewed by others
Introduction
“‘Healing,’
Papa would tell me,
‘is not a science,
but the intuitive art
of wooing Nature.”
– W.H. Auden, The Art of Healing
Published in 1969, W.H. Auden’s dedication to his physician, Dr. David Protetch, was written on the brink of a paradigm shift for modern psychiatry and our understanding of major depressive disorder (MDD). Psychopharmacologist Joseph J. Schildkraut had recently published his seminal 1965 work on the catecholamine hypothesis of affective disorders [1], which set the stage for a biological understanding of MDD and second-generation pharmacotherapies over the next three decades. At nearly the same time, psychotherapist Gordon L. Paul wrote on the challenges of trial research in psychotherapy: “[i]n all its complexity, the question towards which all outcome research should ultimately be directed is the following: What treatment, by whom, is most effective for this individual with that specific problem, and under which set of circumstances?” [2, 3] Auden’s eulogy to his physician and friend poignantly underscores the tension between these two concepts: the generalizations drawn from neuropsychopharmacological research and the need to treat the individual. It is this tension that contemporary precision medicine and biological marker research in MDD seeks to resolve.
Today, MDD is diagnosed according to the DSM-5-TR when an individual possesses five of nine symptoms, including low mood, anhedonia, sleep and appetite disturbances, and suicidal ideation [4]. Consequently, numerous possible symptom combinations of MDD exist; for example, over 1000 unique symptom profiles were identified in 3703 outpatients with MDD [5]. Furthermore, the DSM-5-TR includes clinical specifiers, such as MDD with sleep disturbances, and previous editions include other specifiers like melancholic MDD [6]. Unfortunately, these specifiers do not currently yield homogenous MDD subtypes [7] nor differentiate antidepressant responses to many interventions, including pharmaco- [8,9,10,11,12] and psychotherapies [13,14,15]. Unsurprisingly, two-thirds of patients do not remit to their first course of pharmacological treatment, and roughly one-third develop treatment-resistance [16]. Such highly variable symptomatology suggests that antidepressant response for MDD is complex, and clinical characteristics alone are insufficient to predict response to a certain intervention or distinguish responses to different treatments [8,9,10,11,12,13,14,15].
Functional and structural neuroimaging can help to identify pre-treatment brain network-based characteristics of antidepressant response. Recent neuroimaging studies from large, clinical trials have advanced our understanding of biological markers—biomarkers—in brain networks implicated in MDD pathophysiology. Such markers may guide patients through first-line antidepressant treatments and for second-line treatments in a treatment-resistant population [17]. For the purposes of this review, we distinguish between two types of biomarkers of antidepressant response: prognostic and prescriptive [2]. Prognostic biomarkers are biological characteristics that differentiate dichotomous response and nonresponse or correlate with change on a continuous primary clinical outcome measure to a single treatment. Prescriptive biomarkers are biological characteristics that differentiate responses or improvements on a primary outcome measure between two or more treatments. While numerous studies report promising prognostic and, to a lesser extent, prescriptive biomarkers, there currently is a remarkable paucity of prospective clinical trials that are critically needed to bring neuroimaging biomarkers into clinical practice.
Although the current literature consists of a considerable amount of heterogeneity in the study design and analysis, thereby yielding numerous prognostic biomarkers [17,18,19], our narrative review describes recent advances in structural and functional brain network-based biological markers of treatment response and selection in MDD with a particular focus on reviewing candidate neuroimaging biomarkers with the highest quality of evidence such as large clinical trials. Therefore, we conducted a sensitive search for peer-reviewed papers in PubMed and Web of Science using synonyms of unipolar depression, neuroimaging/connectivity, and treatments (e.g., antidepressants, psychotherapy, and neuromodulation). We extensively discuss studies that employed task- and resting-state blood-oxygenation-level-dependent (BOLD) functional magnetic resonance imaging (fMRI), structural MRI (sMRI), and diffusion tensor imaging. Wherever relevant, we also discuss studies using arterial spin labeling (ASL) perfusion MRI, positron emission tomography (PET) and electroencephalogram (EEG). In the final section, we examine the obstacles hindering the clinical translation of current brain-based biomarkers and discuss aspects of study design and analysis worth considering for future biomarker research in MDD. Although we focus specifically on biomarkers in unipolar MDD, the challenges and future directions highlighted by this review are likely applicable to biomarker research in other psychiatric disorders. Ultimately, biomarker development efforts are urgently needed for people diagnosed with MDD as this disorder is characterized by overwhelming disease burden [20, 21] and a frustrating trial-and-error approach to treatment [22].
Brain networks related to treatment response and selection in MDD
Network neuroscience is an approach to characterize complex large-scale interactions between spatially distributed brain regions. In humans, neuroimaging techniques are used to gain insights into our structurally and functionally connected neurobiological systems. For one, fMRI captures correlated patterns of neural activity in widely separated brain regions that occur spontaneously or are evoked by stimuli. These interactions between different regions of the brain can be used to delineate several distinct large-scale intrinsic brain networks (IBN) serving various aspects of human cognition [23,24,25]. IBNs can be discerned during rest-state or while performing a cognitive task, and notably, IBNs have been hypothesized to reflect both monosynaptic and complex polysynaptic connections of the brain [26, 27]. By studying IBNs, we can understand how network-like brain organization either produces or constrains cognitive function. Likewise, we can compare how structural and functional brain networks are altered in psychiatric populations to elucidate pathophysiology and disease mechanisms. Pre-treatment inter-individual variability in IBN structure or function present in a particular clinical population may help to identify features predictive of response and/or inform treatment selection.
Large-scale structural networks provide an anatomical organization of the brain that underlies cognition. Briefly, brain areas or “nodes” are determined using anatomical parcellation techniques. For instance, a participant could undergo an sMRI scan, and the structural imaging data could be parcellated into different regions using standardized atlas (for review, see ref. [28]). We can characterize the morphological relationships between nodes using structural covariance, which uses measures like cortical thickness and gray matter volume to identify spatially distributed but morphologically similar nodes [29]. These nodes are structurally connected by the axons in the white matter. We can call these fiber tracts connecting each node as “edges”. Edges are identified in vivo using diffusion tensor [30, 31] and diffusion spectrum imaging [32, 33].
Our understanding of the structural organization of the brain helps to infer possible functional interactions within and across structural networks [23, 26, 34]. In other words, interconnected brain areas that work together for a particular set of cognitive functions are a functional network. The nodes of the functional network for a particular function are typically inferred by concurrent activation or deactivation [28]. The functional edges between each node represent the statistical relationship between brain regions’ activity using the time series data from fMRI, electroencephalography, and magnetoencephalography. The quantified statistical relationships between nodes can be undirected (e.g., correlation-based metrics, such as seed-to-voxel-based functional connectivity [35]) or directed/causal (e.g., Granger causality analysis [36] and dynamic causal modeling [37]).
On the other hand, large-scale functional networks can be discerned either during task-related or resting-state functional neuroimaging. Task-based paradigms measure how the brain responds to the cognitive processes in question while the resting-state paradigms measure the spontaneous activity of the brain in a task-free environment. Using functional neuroimaging paradigms, IBNs have been identified via hypothesis-driven and data-driven approaches. For instance, seed-based functional connectivity analysis extracts the time series of an a priori region of interest (ROI), which is then correlated with the activity in other brain regions to identify which areas might be functionally related to the seed region. A more data-driven approach like independent component analysis decomposes the fMRI signal into a set of statistically independent components, each representing a network of distinct patterns of brain activity [38].
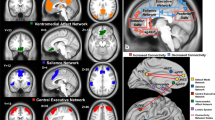
Given the fundamental role of IBNs in cognition and its close relation to structural and task-based functional networks, understanding interactions within and between intrinsic networks is key to elucidating the pathophysiology of psychiatric disorders and searching for network-based biomarkers based on mechanistic understanding of treatment response [17, 39]. While a recent study yielded inconclusive results regarding the identification of neuroimaging biomarkers [40], networks including but not limited to the default mode network (DMN), salience network (SN), central executive network (CEN), and ventromedial limbic/affective network (VMN) have emerged as focal points for potential biomarker discovery in the context of major depressive disorder [39, 41] (Fig. 1). We summarize key nodes within each IBN and their hypothesized functions in Table 1. Dysfunctional interactions within and between IBNs have been associated with various depressive symptoms such as increased negative emotional and cognitive bias [39, 42], rumination [43,44,45], impaired emotional regulation [46, 47], cognitive deficits [48], and anhedonia [49] (Box 1). Based on the current evidence, distinct connectivity patterns, notably within the salience and DMNs, might underlie multiple phenotypic expressions [50] rather than one-to-one relationships between brain networks and symptoms as previously hypothesized [51].
Resting-state functional connectivity (rsFC) map of default mode network (a), central executive network (b), salience network (c), and ventromedial affect network (d), generated using NeuroSynth [281]. Colormap displays brain regions that are correlated with the seed voxel indicated with crosshairs (MNI coordinates). Red and yellow represent the minimum and maximum Pearson correlations (r) respectively. The seeds are nodes of each network: posterior cingulate cortex (a), right dorsolateral prefrontal cortex (b), dorsal anterior cingulate cortex (c), and subgenual anterior cingulate cortex (d). The coordinates of the seed voxel were obtained from [282] and confirmed using the term-to-coordinate mapping in NeuroSynth [281].
Prognostic biomarkers of MDD treatment response
We will first consider prognostic biomarkers from major clinical trials for first-line antidepressant treatments, followed by trials for second-line treatments. Given that the psychopathology of heterogeneous MDD involves complex perturbations of large-scale brain networks, patients with the same MDD diagnosis may have varying aberrant structural and functional connectivity. This heterogeneity implies that patients with certain structural or functional networks might be more suitable for a particular treatment compared to other patients. As we summarize breakthroughs in using pre-treatment brain networks or early changes in the brain as prognostic biomarkers, we note that different factors such as types of study design, analysis methods, validation techniques, and sample size will influence the robustness and clinical translatability of the biomarker. For instance, studies that prospectively validated their biomarkers in large clinical trials and an independent dataset within the same report (i.e., using the same computational pipeline) will likely yield a reliable biomarker. While most current neuroimaging biomarkers have not yet undergone a successful prospective validation, we have made substantial progress in the past decade. Some prognostic biomarkers particularly for pharmacotherapy and neuromodulation have been validated in an independent dataset within the same study or at least replicated across different large clinical trials, while there are relatively less neuroimaging biomarkers predictive of psychotherapy response.
Pharmacotherapy
Since two-thirds of patients do not respond to their first antidepressant medication [16], many trials aim to improve prognostic biomarkers for commonly prescribed drugs like selective serotonin reuptake inhibitors (SSRIs; e.g., escitalopram and sertraline), and serotonin and norepinephrine reuptake inhibitors (SNRIs; e.g., venlafaxine). Early clinical studies identified promising neuroimaging biomarkers, which classified responders from non-responders above 80% [52, 53]. However, studies had a modest sample size (<30), which posed substantial challenges in applying these markers prospectively in a heterogeneous MDD population. Recognizing the need for robust biomarkers, we are making rapid progress in finding more reliable biomarkers using larger, multisite datasets. Very large clinical trials are the most common in pharmacotherapy studies. For instance, antidepressant drug trials with neuroimaging and N > 70 include International Study to Predict Optimized Treatment for Depression clinical trial (iSPOT-D) [54], Canadian Biomarker Integration Network in Depression (CAN-BIND) [55, 56], and Establishing Moderators and Biosignatures of Antidepressant Response for Clinical Care for Depression (EMBARC) [57]. The details of major clinical trials are summarized in Table 2.
Resting-state functional connectivity (rsFC) of the DMN has been frequently reported as candidate biomarkers to first-line antidepressants [58,59,60,61,62,63,64,65,66] across wide-ranging clinical trial settings, types of drugs, and analysis methodologies (e.g., seeds-based vs connectomic approach). For example, increased DMN intra-connectivity was found to be associated with responders or remitters in different clinical trials [58,59,60,61,62] such as iSPOT-D [58, 61] and EMBARC [60]. One example of strong evidence supporting increased pre-treatment DMN intra-connectivity as prognostic biomarker is by ref. [58]. In this study, they randomized 75 MDD participants to escitalopram, sertraline, or venlafaxine, and found that rsFC between the posterior cingulate cortex (PCC) and rostral anterior cingulate cortex (rACC)/medial prefrontal cortex (mPFC) predicted remission with greater than 75% accuracy [58]. Remitters had relatively intact PCC-rACC/mPFC rsFC that closely resembles that of healthy controls. While [58] used ROI-based analysis, ref. [61] used network-based statistics and similarly found that greater baseline functional connectivity within the DMN predicted treatment remission regardless of the type of medication [62]. However, this within-DMN rsFC is likely a treatment non-specific biomarker as it did not differentially predict remission by medication [58]. Consistent with the above findings, an early decrease in DMN intra-connectivity also predicted better sertraline response in the EMBARC study [67].
In addition to DMN intra-connectivity, between-network connectivity of the DMN could differentiate remission. Using a larger, multi-site sample [55, 56] of CAN-BIND-1 participants treated with open-label escitalopram (N = 129), ref. [65] found greater baseline SN anterior cingulate cortex (ACC)-DMN PCC rsFC among early remitters relative to non-remitters [65]. Furthermore, greater insular connectivity with anterior and posterior DMN nodes could differentiate early from late remitters. Remitters also had weaker intra-connectivity within the SN (left-right insula), and DMN (PCC-right superior temporal gyrus) [65]. Extending these findings, the connectomic approach uncovers potential brain connectivity beyond the canonical brain networks. For instance, remitters in the iSPOT-D study had greater inter-connectivity among DMN, somatomotor networks, attention networks, somatomotor networks, and fronto-parietal networks. Adding the neuroimaging connectome biomarkers to clinical variables significantly improved their cross-validated model accuracy from 61.5% to 68.8% [62]. High baseline connectivity between the DMN and other nodes, and low connectivity between the executive network and the rest of the brain predicted general treatment response to sertraline and placebo for 200 MDD patients who completed the EMBARC study [66]. Overall, there is strong evidence that increased DMN intra-connectivity and the inter-connectivity involving nodes of DMN could be potential prognostic biomarkers of pharmacotherapy response. However, it remains a challenge to resolve different inter-connectivity biomarkers found across different study designs and analysis pipelines. The need to address the heterogeneity across studies and subsequently prospectively validate these biomarkers will be the recurring theme throughout.
Nucleus accumbens (NAc) connectivity, an important area for reward neurocircuitry, has also been identified as a candidate biomarker given that SSRIs and augmentation therapies act on dopaminergic systems. One CAN-BIND report found that baseline NAc rsFC with nodes of the SN (ACC) and DMN (precuneus, PCC) correlated with anhedonia improvement for escitalopram non-responders who received adjunct aripiprazole, a dopamine D2 receptor agonist [68]. However, another study (N = 128 MDD) did not find any difference in the baseline NAc rsFC between responders and non-responders, but an increase in right NAc-right dorsal ACC (dACC) rsFC from pre- to post-treatment was able to differentiate responders from non-responders, regardless of antidepressant type [69]. So far, our understanding of the neural mechanism by which different drugs decrease anhedonia remains limited. Based on the current evidence, anhedonia improvement in MDD may have unique prognostic biomarkers that integrate the striatal regions related to reward processing with the DMN and SN.
Furthermore, baseline structural connectivity measures such as fractional anisotropy of predefined white matter tracts yield promising biomarkers [70,71,72]. For instance, the cingulum connects important nodes of DMN and SN within the cingulate cortex (e.g., PCC, dACC, and rACC) [73] while stria terminalis subserves the limbic/affective network (e.g., amygdala and hypothalamus) [74]. Their dysfunctional structural connectivity has been implicated in MDD [75,76,77] and treatment response [70]. Fractional anisotropy of the cingulum and stria terminalis could predict antidepressant treatment response up to 74% when combined with sociodemographic variables like age [70].
Combining structural and functional data may also improve prediction accuracies. Tian et al. [78] combined structural and functional data (N = 106) from an open-label escitalopram trial and found whole-brain biomarkers that predicted 69–72% accuracy [78]. Recently, multi-modal connectomes trained on 184 MDD patients in a naturalistic antidepressant monotherapy setting (SSRI or SNRI) could predict post-treatment depression severity of the external validation dataset (N = 26 MDD) at 76.92% accuracy, as compared to using structural connectome (73.1%) and functional connectome (65.38%) alone [79]. Newer ways are being developed to quantify combined structural and functional connectome (e.g., anatomically weighted functional connectivity [80]), which may bolster prediction accuracy. Neuroimaging data has also been incorporated into a multimodal dataset that combined clinical, behavioral, and molecular biomarkers [81], but its predictive value in this study was limited (accuracy: 57%).
So far, neuroimaging studies have reported prognostic biomarkers either by using group-level statistics to measure the differences between responders and non-responders or by evaluating predictive accuracy of their machine learning models or classifiers. Particularly for those studies evaluating predictive accuracy, different choices of preprocessing steps, first-level statistical modeling, and machine learning methodologies could yield drastically different prediction accuracies. For example, there are different approaches to performing anatomical parcellations, quantifying functional connectivity, identifying the most predictive connectomic features, and classifying responders and non-responders. Using CAN-BIND data, ref. [82] tested the performance of 240 different models using either the baseline or the post-treatment resting-state connectome (N = 144 MDD) and none were capable of predicting response above chance [82]. Comparing different machine learning classifiers on a pre-treatment sMRI dataset for an open-label escitalopram trial (N = 79 MDD), ref. [83] found cross-validated accuracies for predicting treatment response ranging from 0.46 (support vector machine) to 0.62 (random forest) [83]. However, the classifiers trained on their dataset were not generalizable to an independent dataset, albeit for a different SSRI (sertraline). These findings contradict smaller single-site studies reporting >80% accuracy [52, 53] as well as the single-site iSPOT-D study [58, 61]. Interestingly, ref. [82] reported that the change in functional connectivity from baseline to week 2 showed a predicted accuracy of up to 69.6% for the best-performing model [82]. This aligns with quantitative electroencephalography studies indicating that early changes predict antidepressant response at >70% accuracy [84, 85]. Collecting neuroimaging data at baseline and early on during the treatment may have some clinical utility, as it may capture early changes in the brain that might be predictive of a delayed symptomatic change [86].
Thus far, we have made substantial progress in finding baseline intrinsic functional connectivity as prognostic network-based biomarkers for treatment response to antidepressant pharmacotherapy. Interactions within DMN and inter-connectivity between DMN and the rest of the brain appeared to be biomarkers that emerged from both seeds-based and connectomic approaches, while the reward neurocircuitry might play a role in predicting anhedonia response. Recent studies have shown that combining structural and functional connectomes, and novel computational techniques could improve the predictive value of neuroimaging biomarkers. However, it is not clear which of these biomarkers is truly suitable for real-world prediction of antidepressant response. Future studies should rigorously validate the accuracy of our current biomarkers using a larger independent dataset to capture the heterogeneity of MDD.
Psychotherapy
Psychotherapy is a first-line psychological treatment for MDD with comparable efficacy to pharmacotherapy [87,88,89], and includes cognitive behavioral therapy (CBT) and interpersonal therapy. Like pharmacotherapy, 45% of MDD patients do not respond to CBT [89, 90]. Yet, there is relatively less research on prognostic biomarkers of psychotherapy treatment response compared to that of pharmacotherapy [91].
Prognostic studies for psychotherapy in MDD have predominantly involved task-based fMRI during emotional processing or reward learning. The rationale for these tasks stems from the hypothesized mechanistic action of psychotherapy. For instance, the development of cognitive-based therapies for depression is geared towards identifying and correcting dysfunctional thinking patterns implicated in processing negative emotional information and reward-based learning [92,93,94]. From a brain network perspective, these cognitive processes relate to the dual-process model of emotional regulation: prefrontal emotional regulation regions inhibit limbic nodes implicated in emotional reactivity [91, 95, 96]. Specifically, nodes of the CEN (e.g., dorsolateral prefrontal cortex or DLPFC) inhibit limbic/affective regions (amygdala) via the SN (dACC), and via VMN and DMN nodes implicated in emotional appraisal/evaluation like the orbitofrontal prefrontal cortex (OFC), subgenual ACC (sgACC), and rACC [97,98,99,100]. Thus, for MDD participants, limbic areas such as the amygdala are hyperactivated in response to negative stimuli [101,102,103,104] likely due to decreased top-down control from dorsal prefrontal regions [105, 106].
Based on this model, it is believed that pharmacotherapy and psychotherapy have distinct mechanistic actions. Antidepressant drugs act via decreasing amygdala hyperactivity directly while psychotherapy increases PFC hypoactivity, which has a top-down effect on normalizing amygdala dysfunction [107]. Thus, we would expect amygdala hyperactivity to be a prognostic marker for response to both psychotherapy and pharmacotherapy because both treatments ultimately normalize amygdala activity. While converging evidence suggests amygdala hyperactivation during negative emotional reactivity to be a prognostic biomarker of pharmacotherapy response [108, 109], such trends are mixed for psychotherapy. Two studies have reported baseline amygdala hyperactivity during negative emotional processing [110] or reward learning [111] as predictive of response to CBT. However, most task-based studies report nonsignificant amygdala findings [112,113,114,115] including a follow-up study [116] of Siegle et al. [110].
Alternatively, we would expect areas related to emotion regulation and evaluation to be a possible biomarker for psychotherapy. A few fMRI studies found that baseline hypoactivity in sgACC [110, 116] and dACC [112] during emotional processing tasks predicted psychotherapy response. Similarly, a PET study found that baseline sgACC hyperactivity sgACC at rest predicted non-response to CBT [117]. However, a recent CBT task-fMRI study (N = 32 MDD) did not find any baseline emotion regulation or emotional reactivity-related brain activity significantly associated with depression improvement [115]. Although it could be that more research with a larger sample size is needed to resolve the conflicting literature, newer models [118, 119] have moved beyond a simplistic binary interaction between the prefrontal cortex and limbic regions to better account for treatment mechanisms and biomarkers.
To date, there is relatively less research on rsFC predictors of psychotherapy treatment response. Investigating four canonical resting-state networks (DMN, SN, dorsal attention network, CEN), ref. [120] found that increased baseline right insula (SN)–middle temporal gyrus (MTG/DMN) rsFC was predictive of improvements in anhedonia following behavioral activation treatment, which is a psychotherapy aimed at improving engagement with rewarding stimuli [120]. However, this biomarker did not predict improvements in overall depression severity. Interestingly, this rsFC was hypoconnected in all MDD participants relative to controls, but individuals with relatively normalized right insula–MTG connectivity had worse baseline anhedonia and better response to treatment [120]. Although this study used resting-state fMRI, MDD participants excessively recruit MTG during the generation of negative affect as a compensatory mechanism for inefficient activation of dACC and supplementary motor area during the down-regulation of emotions [121]. Increased insula-MTG rsFC may reflect maladaptive functional compensation. Thus, these findings suggest that those who exhibit compensatory mechanism might best respond to psychotherapy. These findings also support the notion that certain treatments may be best suited to address subgroups of patients exhibiting similar symptomatotic and rsFC profiles (i.e., behavioral activation treatment for individuals presenting with high anhedonia and normalized MTG rsFC).
Other psychotherapy biomarkers are consistent with the idea that psychotherapy addresses maladaptive rumination and top-down emotional regulation. Straub et al. [122] found that higher baseline amygdala-left DLPFC, amygdala/left anterior insula, and right sgACC-right DLPFC rsFC predicted greater CBT improvement [122]. Thus far, current literature suggests a few potential prognostic biomarkers for psychotherapy response possibly involving (i) increased rsFC between SN (insula) and DMN (MTG), (ii) insula hypermetabolism at rest, (iii) hyperactivity in sgACC and dACC (emotional regulation system) in response to negative emotional processing. However, future research should explore data-driven and connectomic approaches using a larger sample size to uncover depressive brain states that are responsive to psychotherapy. Particularly with resting-state studies, it is unclear if early brain changes can be used as prognostic biomarkers for psychotherapy. It is also unknown whether these neuroimaging biomarkers are general to every psychotherapy or simply cognitive/behavioral therapy, or with improvements in specific symptoms. Lastly, since psychotherapy may reduce the risk of relapse or recurrence [123, 124], future research should address whether these biomarkers can predict long-term remission and relapses.
Neuromodulation and second-line treatments
Treatment-resistant depression is typically defined as non-response to two or more trials of pharmaco- or psychotherapies [125, 126], and affects 35% of people diagnosed with MDD [127]. Neuromodulation therapies are indicated for those who do not respond to or cannot tolerate first-line treatments. These treatments modulate brain activity using the delivery of a stimulus such as electrical currents in electroconvulsive therapy (ECT), magnetic pulses in repetitive transcranial magnetic stimulation (rTMS) targeting the DLPFC [128], or electrical stimuli via implanted electrode leads in deep brain stimulation (DBS). The neuromodulatory effects of these treatments can locally affect the stimulation site and further propagate to other remote areas, targeting specific IBN implicated in MDD psychopathology [129]. Treatment responses to neuromodulation-based treatments vary greatly across individuals [130]. Searching for neuroimaging biomarkers, particularly for neuromodulation-based treatments, will guide the development of neuromodulation techniques that optimally engage with brain regions or circuits implicated in MDD based on identified biomarkers, and generate individualized care. This targeted and personalized approach increases the likelihood of therapeutic efficacy while minimizing potential side effects.
Pre-treatment sgACC connectivity is implicated in responses to numerous neuromodulation-based treatments. For DLPFC-rTMS, baseline hyperconnectivity between the sgACC and DMN at rest, but not between DMN and CEN, predicted response 10Hz-rTMS over the left DLPFC [131, 132]. Of recent interest, greater anticorrelation between the DLPFC stimulation site and sgACC is associated with better outcomes to treatment [133,134,135]. However, one study reported that stronger baseline sgACC and right DLPFC connectivity predicted non-response to high-frequency rTMS over the left DLPFC [136]. Other groups have indicated that this biomarker may be sensitive to modeling choices and may have a relatively modest effect size [137]. Future studies are suggesting that symptom-specific manipulations of brain activity via rTMS may yield more robust and personalized care using this intervention [138, 139].
Similarly implicating the sgACC, DBS for MDD targets this region, which alters blood flow locally at the stimulation site, but also in other remote areas within the limbic and prefrontal networks that are functionally connected to sgACC [140,141,142]. This normalization of sgACC activity in TRD is associated with a reduction in depressive symptoms [140, 141]. Given this proposed mechanism, DBS response is associated with the white matter tracts and structural connectivity of the stimulation site [143]. Individuals with DBS leads implanted near white matter tracts connecting the sgACC to the mPFC, rACC, dACC, and subcortical nuclei responded best to treatment. In a prospective, open-label, follow-up study, ref. [143] used this knowledge to select the optimal stimulation site using tractography, finding that 82% of patients responded one year post-surgery [143]. This response rate is higher than previous trials from the same group [144], demonstrating the potential of this marker in personalizing care for this population.
Despite the stimulation non-specificity of ECT, this treatment also implicates sgACC structure and function. Greater baseline sgACC volume predicts improved outcomes to ECT [145]. At rest, greater variability in sgACC activity is correlated with ECT response, which decreased after treatment [146]. Another study found that sequential changes in sgACC, DLPFC, and amygdala rsFC correlated with ECT improvement [147]. Furthermore, a classifier trained on a multimodal dataset with features from sMRI, BOLD rs-fMRI, and arterial spin-labeled fMRI predicted ECT response with a balanced accuracy of 68% [148]. Although the sample size in this study was modest (N = 46), the classification models consistently highlighted left DLPFC and sgACC, as well as the connectivity between motor and temporal networks around electrodes used in electroconvulsive therapy (ECT), as predictive features.
In addition to sgACC, connectivity within and between different IBNs, such as DMN and CEN, has been implicated in non-invasive brain stimulation response [149,150,151,152,153,154]. For instance, within-DMN hypoconnectivity and connectivity between the DMN and CEN are associated with ECT response [151]. Van Waarde et al. [152] used a classifier trained on either resting-state fMRI or sMRI to predict ECT remission in TRD [152]. Their approach identified two functional networks that predicted remission at >80%: one network involving the mPFC, DLPFC, OFC, and PCC, and another centered on the dACC, DLPFC, sensorimotor cortex, parahippocampal gyrus, and midbrain. Notably, structural data was not significant predictors, contradicting other studies that found structural biomarkers such as hippocampal subfields [155], striatum [156], and sgACC [145] to be predictive of ECT outcomes.
One limitation to identifying biomarkers of treatment response is that MDD is a symptomatically heterogeneous condition. Therefore, the connectomic signatures of MDD widely vary across individuals, making it potentially advantageous to incorporate biological subtypes of depression with the predictive connectomic biomarkers into classifiers for predicting response. Using this approach, ref. [157] found that classifiers based on both resting-state connective features and biotype diagnosis can accurately predict MDD responders to rTMS over the mPFC (90%), which significantly outperformed classifiers based on the connectomic features (79%) or clinical features (64%) alone [157]. They found that areas across DMN (PCC, mPFC), CEN (left DLPFC), and limbic/reward system (amygdala, OFC) had the strongest discriminant effects [157]. These methods were recently updated to address limitations related to overfitting, feature selection, and multisite variability in a sample of MDD patients. We found that symptom-RSFC and subtypes were not only stable and generalizable in unseen data using an updated approach, but also stratified individuals treated with rTMS targeting the DLPFC by response [158]. Another approach is to cluster symptom-response maps and use individual depression symptoms since most studies used total depression scores [133,134,135, 159]. The symptom-response maps are the correlations between the symptom changes and the expected rsFC map for each patient’s stimulation site. The connectivity of each stimulation site is usually derived from a large functional connectome dataset of healthy participants [160, 161] to improve the reliability of the connectomic biomarkers [159]. Preliminary evidence in retrospective datasets suggests that a connectomic approach to defining regions functionally related to stimulation sites may help identify distinct targets for different clusters of depressive symptoms [162].
Prescriptive biomarkers of MDD treatment selection
Much progress has been made in finding prognostic biomarkers that predict treatment response or remission in MDD. Many of those studies, however, have used reported prognostic biomarkers based on a single intervention prescribed open-label or relative to a placebo [65, 82, 163, 164]. While such prognostic biomarkers lay the foundation for understanding responsive depressive brain states specific to those treatments, they are limited in their clinical utility of guiding whether such biomarkers can differentially predict response better than other treatments. Relatively few neuroimaging studies compared more than two differing treatment options to search for prescriptive biomarkers that can predict the optimal type of intervention. However, many of those comparative studies were only able to find treatment non-specific, prognostic biomarkers, rather than prescriptive biomarkers, for the treatment options in question. We summarize below advances toward finding prescriptive neuroimaging biomarkers for MDD.
Earlier studies involving modest sample sizes found baseline neuroimaging predictors between two different classes of antidepressant medication [165, 166]. For instance, ref. [166] found that bupropion responders (n = 6) showed cerebellar hypermetabolism, whereas venlafaxine responders (n = 7) exhibited bilateral temporal and basal ganglia hypometabolism. However, these findings have been challenging to replicate. Even among larger randomized controlled trials for pharmacotherapy, prescriptive biomarkers have been difficult to identify. For example, studies from the iSPOT-D trial sought to identify whether rsFC can be used as prescriptive biomarkers for optimizing treatment selection to escitalopram, sertraline, or venlafaxine-XR [58, 69]. Both seed-based [58] and connectomic [61] approaches did not yield any prescriptive biomarkers. Given the mechanism of antidepressant drugs is to modulate monoaminergic neurotransmission [167], identifying neuroimaging biomarkers with sufficient predictive value and specificity to individual classes of antidepressant medications remains a significant challenge.
On the other hand, the EMBARC study yielded fMRI and EEG biomarkers that differentially predicted sertraline and placebo. Using the seed-based functional connectivity analysis, Chin Fatt et al. [60] found that higher DMN-ECN interconnectivity predicted better sertraline and worse placebo response [60]. Higher interconnectivity of hippocampus with VMN/ECN/attention network, and limbic network with SN and somatomotor network predicted better placebo and worse sertraline response. The ASL perfusion study did not find opposite-direction predictions but found several regions in the DMN, SN, VWN, and limbic network similar to the rsFC study that predicted treatment-specific responses [168]. And while we would expect that early changes in perfusion patterns might yield potential biomarkers similar to that of rsFCs [82] since cerebral blood flow has a higher signal-to-noise ratio than resting-state fMRI, a recent perfusion study did not find any perfusion patterns predictive of sertraline response [169]. However, ASL may emerge as an important modality when considered with other neuroimaging biomarkers since ASL data contributed the most to the recent multimodal MRI biomarker predicting sertraline-specific response [170].
Furthermore, an initial attempt to find connectomic prescriptive biomarkers using the same EMBARC dataset did not succeed [66]. However, ref. [171] used individual-specific rsFC from the EMBARC dataset and successfully found connectomic signatures that were specific to either sertraline (e.g., DMN-somatomotor network connectivity) or the placebo arm (e.g., somatomotor network and visual network connectivity) [171]. In other words, models trained on the sertraline arm did not predict placebo treatment outcome (r = 0.00, p = 0.97) and vice-versa (r = 0.08, p = 0.55). Furthermore, in comparison to using the raw rsFC data, the individualization of FCs improved the prediction power and changed connectivity weights of important nodes of DMN (middle/inferior temporal cortex) and SN (insula) in sertraline; the left superior temporal cortex and right middle cingulate cortex in the placebo [171]. These regions were also the most predictive of sertraline and placebo response respectively, suggesting that individualized precision functional mapping might be more sensitive to subtle differences in varying connectomic features that might contain critical information for predicting treatment selection. Additionally, EEG represents a more accessible modality that could potentially inform treatment selection. Similar to fMRI, most EEG studies could not find reproducible, prescriptive biomarkers [172,173,174] partly because EEG faces the challenge of signal smearing [175], the risk of overfitting high-dimensional EEG [176], and optimizing feature identification [177]. However, a recent EMBARC study developed a novel rs-EEG computational model and found the latent signatures from alpha frequency range rs-EEG data that were specific to either sertraline or placebo [178]. Similarly, the CAN-BIND group was able to classify open-label escitalopram response using baseline and treatment week 2 EEG recordings with roughly 80% accuracy [179]; this finding is currently being prospectively validated (NCT05017311). Lastly, EEG-based biomarkers have been prospectively replicated in patients treated with agomelatine/ALTO-300 [180]. These results point to EEG as an accessible, and potentially effective tool to identifying and validating biomarkers of existing and novel treatments.
Another potential avenue for treatment selection is to compare two different interventions with highly divergent hypothesized mechanisms: pharmacotherapy and psychotherapy. The most compelling biomarkers thus far come from the PReDICT study, where participants were randomized to either CBT or pharmacotherapy [181]. Dunlop et al. [182] found baseline sgACC rsFC with the left frontal operculum, left ventromedial prefrontal cortex, and dorsal midbrain as prescriptive biomarkers for CBT and pharmacotherapy treatment selection [182]. They then summed up the mean functional connectivity between SCC and each of the three regions. Positively summed rsFC accurately predicted CBT remission (78%) and pharmacotherapy non-remission (75%) while negatively summed rsFC accurately predicted CBT non-remission (89%) and pharmacotherapy remission (72%) [182]. These findings suggest that baseline sgACC rsFC could potentially be used to recommend CBT or pharmacotherapy, but more replication and prospective validation must be done before implementing this biomarker in real-world psychiatric care.
In addition to the rsFC, resting-state metabolic activity measured using PET has also been explored. In a randomized controlled trial of CBT and escitalopram (n = 38), ref. [183] found six regions in the SN (right anterior insula), DMN (right inferior temporal cortex and precuneus), limbic network (left amygdala), and somatomotor network (left premotor cortex and right motor cortex) as potential prescriptive biomarkers [183]. Of note, the right anterior insula showed the highest discriminative ability, with insula hypermetabolism being associated with escitalopram remission and CBT non-remission, and insula hypometabolism being associated with CBT remission and escitalopram non-remission [183]. This finding, along with studies showing heterogenous insula metabolism profiles among MDD participants [184, 185], led to the hypothesis that anterior insula metabolism might be a viable treatment selection biomarker [119] due to its role in initiating network switching between DMN and CEN [39]. However, when the clinical utility of the prescriptive biomarker was prospectively tested by using the anterior insula metabolism to assign MDD participants to either CBT or escitalopram, ref. [186] did not find anterior insula beneficial in predicting remission rates (38%) [186]. The findings emphasize the importance of prospectively validating current biomarkers and integrating our understanding of large-scale brain networks in depression into the development of novel network-based biomarkers.
Despite the extensive body of research dedicated to understanding the etiology, pathophysiology, and treatment mechanism of MDD, the identification of neuroimaging biomarkers capable of guiding personalized treatment approaches is yet to be successful. Based on the current literature, potential prescriptive biomarkers for predicting different classes of pharmacotherapy, or between pharmacotherapy and psychotherapy could be based on interactions among DMN (e.g., lateral temporal cortex, precuneus), SN (anterior insula), and VMN (e.g., sgACC). However, many of the biomarkers identified for the same intervention but in different clinical trials consist of many overlapping brain regions within the well-known IBN implicated in MDD, likely reflecting the need to account for heterogeneity in the MDD participants, study design, eligibility criteria, operationalization of treatment outcomes, and computational methodologies in analyzing the data and calculating the predictive utility of biomarkers. These challenges are extensively discussed in the next section below.
Future directions and clinical implications
The reviewed studies demonstrate that significant strides have been made over the last decade toward unraveling the complex underpinnings of antidepressant response, with the strong potential of informing personalized treatment strategies. However, challenges remain on the path to clinical translation. For example, the therapeutic window defining treatment response can vary considerably from one biomarker study to the next (discussed in [17]). Response and remission rates also differ slightly depending on the therapeutic window considered, with longer trials having higher rates [187]. Therefore, it is unsurprising that biomarkers of response qualitatively and substantially differ when investigating 6-week or 12-week outcomes [17]. Another area of concern is publication bias, which further limits our ability to independently validate and prospectively use biomarkers. Open data, and pre-registered, freely-available method will be paramount to furthering our confidence in efforts to translate findings from the scanner into the clinic [188, 189].
In this section, we highlight considerations on study design, methodological, and implementation that, if addressed in future work, will improve our understanding of and aid in implementing biomarkers of treatment response and selection.
Clinical trial design
Study design can significantly influence response and remission rates to antidepressants. For example, response and remission rates are higher in open-label relative to placebo-controlled trials [187], likely due to expectancy effects [190]. Furthermore, the choice of intervention impacts willingness to participate in research and therefore may bias participant selection, as individuals are willing to participate in head-to-head or placebo-controlled studies using psychotherapy over head-to-head drug trials [191]. It is therefore likely that these aspects of study design could impact the identification and validation of biomarkers of response.
Researchers have used a variety of different study designs in an effort to identify biomarkers of antidepressant response. For studies whose primary aim was to evaluate biological modulators of response, some groups, such as EMBARC [57], used a placebo-controlled study of a single SSRI, while others, like PReDICT [181] and iSPOT-D [54], randomized participants into one of three active treatment arms. Others, including EMBARC and CAN-BIND-1 [55], have used a staged (mono- and adjunctive therapy) approach, which more closely mirrors the trial-and-error approach to antidepressant treatment and other studies evaluating efficacy [22]. Heterogeneity in study design could negatively impact our ability to pool studies for meta-analysis [192], potentially limiting the generalizability of findings. Reassuringly and in contradiction to this notion, recent efforts pooling CAN-BIND, PReDICT, EMBARC, and other trial data have identified sMRI dimensions of MDD that stratified response to SSRIs and placebo [193]. Regardless, idiosyncrasies in study design will need to be carefully considered in upcoming efforts to pool studies in larger consortia including the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium [194, 195] and the COORDINATE-MDD Consortium [196].
Moving forward, research is needed to clarify the impact of trial design on biological markers of treatment response, and different study designs may be better equipped to answer specific questions on the robustness or utility of candidate biomarkers. For one, the goal of placebo-controlled trials is to unravel the treatment-specific biomarkers, while controlling for psychophysiological factors contributing to the placebo response [197]. These comparisons between biomarkers of active and placebo are helpful both from a mechanistic and prognostic perspective. That said, placebo responses are high but variable across a variety of different interventions [198], and will be present in prospective, real-world translation efforts. Some have argued that the placebo response could be harnessed to bolster treatment efficacy or refine treatment options, which would be particularly relevant to biomarkers research [197]. Indeed, both biomarkers of antidepressant and analgesic placebos are associated with networks pertinent to MDD, including the SN and DMN [171, 199], so more research is needed to understand the clinical utility of these markers.
Another area needing more investigation is research comparing two or more active treatments to better understand predictors of specific interventions and aid in treatment selection. While this has shown helpful in disentangling biomarkers of psycho- vs. pharmacotherapy [117, 182, 183] and has ethical benefits by randomizing patients to receive at least one efficacious treatment, there are relatively few direct comparisons of predictors to specific drugs. Furthermore, studies investigating active interventions have randomized participants into arms irrespective of patient preference. This line of research suggests that patient preference does not influence overall treatment efficacy but does impact dropout rates [200, 201], and we do not yet know whether or how patient preference impacts biomarkers of response/selection. Partially randomized patient preference trials could improve the internal and external validity of biomarkers by assigning patients their preferred treatment, which would emulate real-world treatments while retaining response and remission rates.
Study population
Another factor to consider is study population. To date, most large biomarker trials recruit adults diagnosed with unipolar MDD [54,55,56,57], with fewer biomarker trials occurring in adolescence and in later life. Studies within the unipolar space vary considerably on inclusion/exclusion criteria, particularly on factors like antidepressant treatment history, comorbid mental illness, and a depression severity cutoff [202, 203]. Nearly all studies exclude individuals with a high suicide risk, but the methods to determine risk differ (e.g., clinician judgement, score on scales acquired during screening). While these criteria help to ensure a homogenous sample/population, comorbid mental illnesses are common [204, 205] and, therefore, limit the external validity of biomarkers. Given that individuals suffering from comorbid mental illness experience more MDD recurrence, greater treatment resistance, and slower recovery than those without [206, 207], future studies should “lean in” than “shy away” from this aspect of clinical heterogeneity to maximize external validity.
Furthermore, more inclusive study criteria may help converge our biomarker study efforts across mental illnesses, especially for therapies that are indicated for multiple disorders other than unipolar MDD. For example, researchers have reported convergent mechanisms and predictors of psychotherapy and pharmacotherapy response in mood and anxiety disorders despite strong between-study heterogeneity [91, 208]. Future research should consider convergent and divergent biomarkers of treatment response considering the high comorbidity of mental illness and the transdiagnostic indications of many psychotropic interventions.
Defining treatment response
Defining treatment response, the dependent variable, is not trivial. Studies frequently acquire a clinician-rated or self-reported depression severity scale, like the Hamilton Rating Scale for Depression [209] or Beck Depression Inventory [210], pre-treatment, at regular intervals during treatment, and post-treatment to track improvement. Selecting which scale and how to assess response (i.e., as a continuous or dichotomous variable), as previously discussed [17], varies considerably across trials, limiting our ability to generalize findings.
Further complicating this issue, these scales often evaluate multiple symptom dimensions [211]. This likely negatively impacts our ability to detect biomarkers of response and distinct biomarkers of specific treatments, when evaluating the change in total score on these scales due to heterogenous baseline symptom profiles and symptom-specific change. Confirming this notion, specific pre-treatment symptom clusters are associated with differential response to both first- and second-line treatments [158, 212]. Furthermore, researchers have shown that different interventions yield distinct patterns of symptom-specific response [213,214,215]. For example, ref. [216] found that individuals treated with escitalopram or duloxetine experienced greater improvements in melancholic symptoms relative to those treated with psychotherapy [216]. Second-line treatments for MDD may also impact specific symptom dimensions, including ketamine [217] and rTMS [162]. Given that specific symptom domains have distinct underlying biology [157, 158, 218], it is possible that change in these domains are a fruitful way forward to identifying treatment-specific and robust biomarkers.
How else might we characterize response? Incorporating multiple scales may be one path forward, particularly if they assess symptoms not fully realized in one scale (e.g., anhedonia in the Hamilton Depression Rating Scale [209]). Alternatively, quality of life and functional impairment scales are commonly acquired in biomarkers studies but typically considered a secondary outcome measure. Scales such as the Sheehan Disability Scale [219] could also yield important biomarkers of clinically meaningful improvement, especially if aggregated across many trials. Early improvements in functional impairment predict later depressive response [220, 221], and structural and functional connectivity related to functional impairment in MDD implicates nodes of depression-related networks [222, 223]. Another consideration is identifying biomarkers predicting distinct trajectories of treatment response, which will consider all available timepoints. Linear growth mixture modeling is one method that stratifies subgroups of participants by trajectory, which has shown high prediction accuracy for response to psychotherapy in late-life depression [224] and has been used to characterize response to psychotherapy and pharmacotherapy in adults [225, 226]. Early work incorporating trajectories with neuroimaging is promising [227, 228].
Neuroimaging and modeling considerations
Another nontrivial consideration is selecting the neuroimaging paradigm and acquisition parameters. First, there is considerable variability in biomarker modality across studies (e.g., sMRI, resting-state and/or task-based fMRI), again limiting our ability to generalize findings. Resting-state and sMRI are popular options, as they are relatively simple to acquire and do not rely on task compliance or performance. However, interpreting functional connectivity unconstrained by a task is complicated by the fact that individuals may experience the resting-state scan differently, and this variability is inadequately accounted for [229]. Further, task-based fMRI may be better suited to characterizing inter-individual differences in behavior over resting-state. In a nondepressed sample, task-based functional connectivity is superior at characterizing variability in a number of cognitive domains, including reading comprehension [230], attention [231], behavioral inhibition [232], and fluid intelligence [233, 234]. In MDD, tasks that have been used to distinguish antidepressant response have included tasks related to emotional reactivity and social cognition [109, 112], appetitive and avoidance behaviors [111, 235,236,237], emotion regulation [115, 238, 239], and working memory/cognitive control [240,241,242]. Another untapped area is passive movie watching, which outperforms resting-state fMRI in predicting individual differences in cognition and emotional traits [243] without risking omitting data due to task performance. Lastly, alternative modalities, such as ASL or EEG, may yield robust or scalable biomarkers of treatment response or selection. It is therefore possible that such modalities or using task-based or naturalistic viewing (movie watching) may be preferred functional methods in future biomarker efforts.
Due to the inherently low signal-to-noise ratio of fMRI data [244], it also remains a challenge to use individual data rather than the averaged group-level imaging data to create individual-specific functional connectomes for a personalized targeting approach. Newer scan acquisition parameters, such as multi-echo fMRI, may also yield novel insights. Multi-echo fMRI acquires multiple echoes, imaging readouts, per volume, which help distinguish blood oxygen-level dependent signal from nonneuronal noise [245,246,247]. Optimally combining and denoising these multiple echoes [248] has been shown to improve the signal-to-noise ratio and reliability of findings [249,250,251], and improve the interpretability of task-based fMRI results [252, 253]. Multi-echo may also improve test-retest reliability of precision functional mapping [254, 255], a method modeling individualized rsFC maps that can provide clinically meaningful variation in cognition and behavior [256,257,258]. Further, machine learning approaches should carefully consider individual cases where the model fails. Greene and colleagues [259] recently demonstrated, in healthy samples, that misclassification in models predicting individual heterogeneity in behavior using fMRI is non-random, related to specific phenotypic profiles and generalizable across datasets [259]. Future biomarker studies should similarly assess model misclassification cases to better understand the robustness and refine predictive models. Other considerations include a larger sample size, evaluating biomarkers using an independent cohort, and validating the predictive accuracy across different models (e.g., parcellations) and algorithms.
Another important factor is how to model brain-based measures. There is substantial variation across studies in preprocessing methods, and statistical analyses, and these modeling decisions should be carefully considered. Systematically testing these factors is one employed solution, particularly for specific resting-state preprocessing steps (e.g., global signal regression) and parcellation selection [135, 260]. Large, whole-brain univariate correlations have been criticized due to limitations in statistical power, multiple comparisons, and reproducibility [261]. Multivariate whole-brain associations might be one way forward to developing predictive models without requiring tens of thousands of individuals in the sample [262], and we recently modeled the impact of sample size on the robustness of multivariate methods in a depressed sample [158]. Another proposed approach is to identify biomarker regions exhibiting atypical structure or function relative to nondepressed controls using normative modeling [263]. The rationale for this approach lies in the notion that patient heterogeneity should be linked to biological dysfunction [50, 51]. However, it is possible brain-based features correlated with symptom heterogeneity and antidepressant response need not be atypical relative to controls [120, 158, 264]. Alternative methods to characterize structure or function in a patient sample relative to controls, such as brain aging [265] or approaches that exploit larger control databases [162], may also help to distinguish prescriptive markers of response.
Conclusion
To conclude, significant progress has been made over the last decade to identify biologically based predictors of antidepressant treatments. That said, inability to replicate candidate biomarkers and considerable variability in model performance by study design, preprocessing, and analysis remains a significant challenge in this field [81, 82, 186]. Studies optimally designed to compare treatment-specific biomarkers, with pre-registered methods, will be paramount to uncovering rigorous prescriptive markers of response.
Even once replicable and robust markers are found, scalability and equitable access to care are significant impediments to translating any neuroimaging biomarker on a large scale [266]. For example, Canada has one of the lowest MRI scanners per capita and longest wait times amongst high income countries [267, 268]. In the United States, delays in acquiring an MRI are associated with individuals with public insurance, who identify as female, or live in a low socio-economic neighborhood [268]. It will be imperative to advocate for equitable access to care as neuroimaging biomarkers are integrating into clinical practice. Further, identifying analogue and more accessible markers of response, perhaps using molecular, genetic or ecological momentary assessment measures [81, 269] may help improve the scalability of markers of response to first-line interventions. It could be that MRI-based biomarkers in psychiatry, at least at first, are reserved for second-line treatments like DBS where pre-surgical tractography can be used to individualize care.
Despite these challenges, there are exciting prospective studies currently recruiting. For example, Optimized Predictive Treatment In Medications for Unipolar Major Depression (OPTIMUM-D; NCT05017311) is a follow-up to CAN-BIND, and seeks to test a previously identified prescriptive biomarker [270]. In the study, participants will be assigned to one of two arms. The active, personalized arm will receive open-label escitalopram and placebo-controlled brexpiprazole; active brexpiprazole or placebo will be assigned using a predictive algorithm. The placebo arm will be randomized to receive escitalopram/placebo or escitalopram/brexpiprazole. Using a similar trial design, Biomarker-guided rTMS for Treatment Resistant Depression (BioTMS; NCT04041479) will assign different rTMS targets (DLPFC or mPFC) based on rsFC-based subtype assignment in treatment-resistant depression [271]. These studies, if successful, will be monumental to reducing the frustration of treatment, by either enabling patients to bypass monotherapy trial-and-error and proceed with adjunctive treatment, or personalizing stimulation sites based on co-occurring symptoms and rsFC. Time will tell if these studies are indeed successful, but they are certainly on the right track to integrating neuroimaging data to help inform the “intuitive art of wooing Nature” in the context of treating MDD.
References
Schildkraut JJ. The catecholamine hypothesis of affective disorders: a review of supporting evidence. Am J Psychiatry. 1965;122:509–22.
Cohen ZD, DeRubeis RJ. Treatment selection in depression. Annu Rev Clin Psychol. 2018;14:209–36.
Paul GL. Strategy of outcome research in psychotherapy. J Consult Psychol. 1967;31:109–18.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders. DSM-5-TR. Washington, DC: American Psychiatric Association Publishing; 2022.
Fried EI, Nesse RM. Depression is not a consistent syndrome: An investigation of unique symptom patterns in the STAR*D study. J Affect Disord. 2015;172:96–102.
American Psychiatric Association, American Psychiatric Association, editors. Diagnostic and statistical manual of mental disorders: DSM-IV-TR. 4th ed. Washington, DC: American Psychiatric Association; 2000.
Lorenzo-Luaces L, Buss JF, Fried EI. Heterogeneity in major depression and its melancholic and atypical specifiers: a secondary analysis of STAR*D. BMC Psychiatry. 2021;21:454.
Kennedy SH, Lam RW, McIntyre RS, Tourjman SV, Bhat V, Blier P, et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 clinical guidelines for the management of adults with major depressive disorder: section 3. Pharmacological Treatments. Can J Psychiatry. 2016;61:540–60.
Uher R. Genes, environment, and individual differences in responding to treatment for depression. Harv Rev Psychiatry. 2011;19:109–24.
Arnow BA, Blasey C, Williams LM, Palmer DM, Rekshan W, Schatzberg AF, et al. Depression subtypes in predicting antidepressant response: a report from the iSPOT-D Trial. Am J Psychiatry. 2015;172:743–50.
Stewart JW, McGrath PJ, Fava M, Wisniewski SR, Zisook S, Cook I, et al. Do atypical features affect outcome in depressed outpatients treated with citalopram? Int J Neuropsychopharmacol. 2010;13:15–30.
McGrath PJ, Khan AY, Trivedi MH, Stewart JW, Morris DW, Wisniewski SR, et al. Response to a selective serotonin reuptake inhibitor (citalopram) in major depressive disorder with melancholic features: a STAR*D report. J Clin Psychiatry. 2008;69:1847–55.
Simon GE, Perlis RH. Personalized medicine for depression: can we match patients with treatments? Am J Psychiatry. 2010;167:1445–55.
Driessen E, Hollon SD. Cognitive behavioral therapy for mood disorders: efficacy, moderators and mediators. Psychiatr Clin North Am. 2010;33:537–55.
Parikh SV, Quilty LC, Ravitz P, Rosenbluth M, Pavlova B, Grigoriadis S, et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 clinical guidelines for the management of adults with major depressive disorder: section 2. Psychological treatments. Can J Psychiatry. 2016;61:524–39.
Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry. 2006;163:1905–17.
Dunlop K, Talishinsky A, Liston C. Intrinsic brain network biomarkers of antidepressant response: a review. Curr Psychiatry Rep. 2019;21:87.
Tura A, Goya-Maldonado R. Brain connectivity in major depressive disorder: a precision component of treatment modalities? Transl Psychiatry. 2023;13:196.
Gerlach AR, Karim HT, Peciña M, Ajilore O, Taylor WD, Butters MA, et al. MRI predictors of pharmacotherapy response in major depressive disorder. Neuroimage Clin. 2022;36:103157.
GBD 2019 Mental Disorders Collaborators. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry. 2022;9:137–50.
Greenberg PE, Fournier A-A, Sisitsky T, Simes M, Berman R, Koenigsberg SH, et al. The economic burden of adults with major depressive disorder in the United States (2010 and 2018). PharmacoEconomics. 2021;39:653–65.
Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, et al. Sequenced treatment alternatives to relieve depression (STAR*D): rationale and design. Control Clin Trials. 2004;25:119–42.
Sporns O. The human connectome: a complex network. Ann N. Y Acad Sci. 2011;1224:109–25.
Kelly AMC, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. Competition between functional brain networks mediates behavioral variability. NeuroImage. 2008;39:527–37.
Shannon BJ, Raichle ME, Snyder AZ, Fair DA, Mills KL, Zhang D, et al. Premotor functional connectivity predicts impulsivity in juvenile offenders. Proc Natl Acad Sci USA. 2011;108:11241–5.
Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78.
Hermundstad AM, Bassett DS, Brown KS, Aminoff EM, Clewett D, Freeman S, et al. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc Natl Acad Sci USA. 2013;110:6169–74.
Bressler SL, Menon V. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn Sci. 2010;14:277–90.
Spreng RN, Turner GR. Structural covariance of the default network in healthy and pathological aging. J Neurosci. 2013;33:15226–34.
Oishi K, Zilles K, Amunts K, Faria A, Jiang H, Li X, et al. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. Neuroimage. 2008;43:447–57.
Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, et al. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex. 2009;19:524–36.
Schmahmann JD, Pandya DN, Wang R, Dai G, D’Arceuil HE, de Crespigny AJ, et al. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 2007;130:630–53.
Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159.
Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, et al. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA. 2009;106:2035–40.
Hampson M, Peterson BS, Skudlarski P, Gatenby JC, Gore JC. Detection of functional connectivity using temporal correlations in MR images. Hum Brain Mapp. 2002;15:247–62.
Roebroeck A, Formisano E, Goebel R. Mapping directed influence over the brain using Granger causality and fMRI. Neuroimage. 2005;25:230–42.
Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19:1273–302.
Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, et al. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 2006;103:13848–53.
Menon V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci. 2011;15:483–506.
Winter NR, Leenings R, Ernsting J, Sarink K, Fisch L, Emden D, et al. Quantifying deviations of brain structure and function in major depressive disorder across neuroimaging modalities. JAMA Psychiatry. 2022;79:879.
Dutta A, McKie S, Deakin JFW. Resting state networks in major depressive disorder. Psychiatry Res Neuroimaging. 2014;224:139–51.
Kaiser RH, Andrews-Hanna JR, Spielberg JM, Warren SL, Sutton BP, Miller GA, et al. Distracted and down: neural mechanisms of affective interference in subclinical depression. Soc Cogn Affect Neurosci. 2015;10:654–63.
Cooney RE, Joormann J, Eugène F, Dennis EL, Gotlib IH. Neural correlates of rumination in depression. Cogn Affect Behav Neurosci. 2010;10:470–8.
Burkhouse KL, Jacobs RH, Peters AT, Ajilore O, Watkins ER, Langenecker SA. Neural correlates of rumination in adolescents with remitted major depressive disorder and healthy controls. Cogn Affect Behav Neurosci. 2017;17:394–405.
Lemogne C, Le Bastard G, Mayberg H, Volle E, Bergouignan L, Lehéricy S, et al. In search of the depressive self: extended medial prefrontal network during self-referential processing in major depression. Soc Cogn Affect Neurosci. 2009;4:305–12.
Sheline YI, Price JL, Yan Z, Mintun MA. Resting-state functional MRI in depression unmasks increased connectivity between networks via the dorsal nexus. Proc Natl Acad Sci USA. 2010;107:11020–5.
Beck AT. The evolution of the cognitive model of depression and its neurobiological correlates. Am J Psychiatry. 2008;165:969–77.
Snyder HR. Major depressive disorder is associated with broad impairments on neuropsychological measures of executive function: a meta-analysis and review. Psychol Bull. 2013;139:81–132.
Cooper JA, Arulpragasam AR, Treadway MT. Anhedonia in depression: biological mechanisms and computational models. Curr Opin Behav Sci. 2018;22:128–35.
Goldstein-Piekarski AN, Ball TM, Samara Z, Staveland BR, Keller AS, Fleming SL, et al. Mapping neural circuit biotypes to symptoms and behavioral dimensions of depression and anxiety. Biol Psychiatry. 2022;91:561–71.
Williams LM. Precision psychiatry: a neural circuit taxonomy for depression and anxiety. Lancet Psychiatry. 2016;3:472–80.
Cohen SE, Zantvoord JB, Wezenberg BN, Bockting CLH, Van Wingen GA. Magnetic resonance imaging for individual prediction of treatment response in major depressive disorder: a systematic review and meta-analysis. Transl Psychiatry. 2021;11:168.
Gao S, Calhoun VD, Sui J. Machine learning in major depression: from classification to treatment outcome prediction. CNS Neurosci Ther. 2018;24:1037–52.
Williams LM, Rush AJ, Koslow SH, Wisniewski SR, Cooper NJ, Nemeroff CB, et al. International Study to Predict Optimized Treatment for Depression (iSPOT-D), a randomized clinical trial: rationale and protocol. Trials. 2011;12:4.
Lam RW, Milev R, Rotzinger S, Andreazza AC, Blier P, Brenner C, et al. Discovering biomarkers for antidepressant response: protocol from the Canadian biomarker integration network in depression (CAN-BIND) and clinical characteristics of the first patient cohort. BMC Psychiatry. 2016;16:105.
Kennedy SH, Lam RW, Rotzinger S, Milev RV, Blier P, Downar J, et al. Symptomatic and functional outcomes and early prediction of response to Escitalopram Monotherapy and sequential adjunctive aripiprazole therapy in patients with major depressive disorder: a CAN-BIND-1 report. J Clin Psychiatry. 2019;80:18m12202.
Trivedi MH, McGrath PJ, Fava M, Parsey RV, Kurian BT, Phillips ML, et al. Establishing moderators and biosignatures of antidepressant response in clinical care (EMBARC): rationale and design. J Psychiatr Res. 2016;78:11–23.
Goldstein-Piekarski AN, Staveland BR, Ball TM, Yesavage J, Korgaonkar MS, Williams LM. Intrinsic functional connectivity predicts remission on antidepressants: a randomized controlled trial to identify clinically applicable imaging biomarkers. Transl Psychiatry. 2018;8:57.
Ye Y, Wang C, Lan X, Li W, Fu L, Zhang F, et al. Baseline patterns of resting functional connectivity within posterior default-mode intranetwork associated with remission to antidepressants in major depressive disorder. NeuroImage Clin. 2022;36:103230.
Chin Fatt CR, Jha MK, Cooper CM, Fonzo G, South C, Grannemann B, et al. Effect of intrinsic patterns of functional brain connectivity in moderating antidepressant treatment response in major depression. AJP. 2020;177:143–54.
Korgaonkar MS, Goldstein-Piekarski AN, Fornito A, Williams LM. Intrinsic connectomes are a predictive biomarker of remission in major depressive disorder. Mol Psychiatry. 2020;25:1537–49.
DeMaster D, Godlewska BR, Liang M, Vannucci M, Bockmann T, Cao B, et al. Effective connectivity between resting-state networks in depression. J Affect Disord. 2022;307:79–86.
Wu H, Liu R, Zhou J, Feng L, Wang Y, Chen X, et al. Prediction of remission among patients with a major depressive disorder based on the resting-state functional connectivity of emotion regulation networks. Transl Psychiatry. 2022;12:391.
Martens MAG, Filippini N, Harmer CJ, Godlewska BR. Resting state functional connectivity patterns as biomarkers of treatment response to escitalopram in patients with major depressive disorder. Psychopharmacology. 2022;239:3447–60.
Van Der Wijk G, Harris JK, Hassel S, Davis AD, Zamyadi M, Arnott SR, et al. Baseline functional connectivity in resting state networks associated with depression and remission status after 16 weeks of pharmacotherapy: a CAN-BIND report. Cereb Cortex. 2022;32:1223–43.
Fan S, Nemati S, Akiki TJ, Roscoe J, Averill CL, Fouda S, et al. Pretreatment brain connectome fingerprint predicts treatment response in major depressive disorder. Chronic Stress. 2020;4:247054702098472.
Nemati S, Akiki TJ, Roscoe J, Ju Y, Averill CL, Fouda S, et al. A unique brain connectome fingerprint predates and predicts response to antidepressants. iScience. 2020;23:100800.
Vaccarino SR, Wang S, Rizvi SJ, Lou W, Hassel S, MacQueen GM, et al. Functional neuroimaging biomarkers of anhedonia response to escitalopram plus adjunct aripiprazole treatment for major depressive disorder. BJPsych Open. 2024;10:e18.
Fischer AS, Holt-Gosselin B, Fleming SL, Hack LM, Ball TM, Schatzberg AF, et al. Intrinsic reward circuit connectivity profiles underlying symptom and quality of life outcomes following antidepressant medication: a report from the iSPOT-D trial. Neuropsychopharmacol. 2021;46:809–19.
Korgaonkar MS, Williams LM, Song YJ, Usherwood T, Grieve SM. Diffusion tensor imaging predictors of treatment outcomes in major depressive disorder. Br J Psychiatry. 2014;205:321–8.
Grieve SM, Korgaonkar MS, Gordon E, Williams LM, Rush AJ. Prediction of nonremission to antidepressant therapy using diffusion tensor imaging. J Clin Psychiatry. 2016;77:e436–43.
Davis AD, Hassel S, Arnott SR, Harris J, Lam RW, Milev R, et al. White matter indices of medication response in major depression: a diffusion tensor imaging study. Biol Psychiatry Cogn Neurosci Neuroimaging. 2019;4:913–24.
Bubb EJ, Metzler-Baddeley C, Aggleton JP. The cingulum bundle: anatomy, function, and dysfunction. Neurosci Biobehav Rev. 2018;92:104–27.
Mori S, Kageyama Y, Hou Z, Aggarwal M, Patel J, Brown T, et al. Elucidation of white matter tracts of the human amygdala by detailed comparison between high-resolution postmortem magnetic resonance imaging and histology. Front Neuroanat. 2017;11:16.
Murphy ML, Frodl T. Meta-analysis of diffusion tensor imaging studies shows altered fractional anisotropy occurring in distinct brain areas in association with depression. Biol Mood Anxiety Disord. 2011;1:3.
Korgaonkar MS, Cooper NJ, Williams LM, Grieve SM. Mapping inter-regional connectivity of the entire cortex to characterize major depressive disorder: a whole-brain diffusion tensor imaging tractography study. Neuroreport. 2012;23:566–71.
Cole J, Chaddock CA, Farmer AE, Aitchison KJ, Simmons A, McGuffin P, et al. White matter abnormalities and illness severity in major depressive disorder. Br J Psychiatry. 2012;201:33–9.
Tian S, Sun Y, Shao J, Zhang S, Mo Z, Liu X, et al. Predicting escitalopram monotherapy response in depression: the role of anterior cingulate cortex. Hum Brain Mapp. 2020;41:1249–60.
Wang X, Xue L, Shao J, Dai Z, Hua L, Yan R, et al. Distinct MRI-based functional and structural connectivity for antidepressant response prediction in major depressive disorder. Clin Neurophysiol. 2024;160:19–27.
Ayyash S, Davis AD, Alders GL, MacQueen G, Strother SC, Hassel S, et al. Assessing remission in major depressive disorder using a functional-structural data fusion pipeline: a CAN-BIND-1 study. IBRO Neurosci Rep. 2024;16:135–46.
Sajjadian M, Uher R, Ho K, Hassel S, Milev R, Frey BN, et al. Prediction of depression treatment outcome from multimodal data: a CAN-BIND-1 report. Psychol Med. 2023;53:5374–84.
Harris JK, Hassel S, Davis AD, Zamyadi M, Arnott SR, Milev R, et al. Predicting escitalopram treatment response from pre-treatment and early response resting state fMRI in a multi-site sample: A CAN-BIND-1 report. NeuroImage Clin. 2022;35:103120.
Beliveau V, Hedeboe E, Fisher PM, Dam VH, Jørgensen MB, Frokjaer VG, et al. Generalizability of treatment outcome prediction in major depressive disorder using structural MRI: a NeuroPharm study. NeuroImage Clin. 2022;36:103224.
Iosifescu DV, Greenwald S, Devlin P, Mischoulon D, Denninger JW, Alpert JE, et al. Frontal EEG predictors of treatment outcome in major depressive disorder. Eur Neuropsychopharmacol. 2009;19:772–7.
Leuchter AF, Cook IA, Marangell LB, Gilmer WS, Burgoyne KS, Howland RH, et al. Comparative effectiveness of biomarkers and clinical indicators for predicting outcomes of SSRI treatment in major depressive disorder: results of the BRITE-MD study. Psychiatry Res. 2009;169:124–31.
Godlewska BR, Harmer CJ. Cognitive neuropsychological theory of antidepressant action: a modern-day approach to depression and its treatment. Psychopharmacology. 2021;238:1265–78.
Spielmans GI, Berman MI, Usitalo AN. Psychotherapy versus second-generation antidepressants in the treatment of depression: a meta-analysis. J Nerv Ment Dis. 2011;199:142–9.
Cuijpers P, Sijbrandij M, Koole SL, Andersson G, Beekman AT, Reynolds CF. The efficacy of psychotherapy and pharmacotherapy in treating depressive and anxiety disorders: a meta-analysis of direct comparisons. World Psychiatry. 2013;12:137–48.
Amick HR, Gartlehner G, Gaynes BN, Forneris C, Asher GN, Morgan LC, et al. Comparative benefits and harms of second generation antidepressants and cognitive behavioral therapies in initial treatment of major depressive disorder: systematic review and meta-analysis. BMJ. 2015;351:h6019.
Cuijpers P, Karyotaki E, Weitz E, Andersson G, Hollon SD, van Straten A. The effects of psychotherapies for major depression in adults on remission, recovery and improvement: a meta-analysis. J Affect Disord. 2014;159:118–26.
Marwood L, Wise T, Perkins AM, Cleare AJ. Meta-analyses of the neural mechanisms and predictors of response to psychotherapy in depression and anxiety. Neurosci Biobehav Rev. 2018;95:61–72.
Yoshimura S, Okamoto Y, Onoda K, Matsunaga M, Okada G, Kunisato Y, et al. Cognitive behavioral therapy for depression changes medial prefrontal and ventral anterior cingulate cortex activity associated with self-referential processing. Soc Cogn Affect Neurosci. 2014;9:487–93.
Brown VM, Zhu L, Solway A, Wang JM, McCurry KL, King-Casas B, et al. Reinforcement learning disruptions in individuals with depression and sensitivity to symptom change following cognitive behavioral therapy. JAMA Psychiatry. 2021;78:1113.
Beck AT, Rush AJ. Cognitive therapy of depression. New York: Guilford Press; 1979.
Drevets WC, Raichle ME. Suppression of regional cerebral blood during emotional versus higher cognitive implications for interactions between emotion and cognition. Cognit Emot. 1998;12:353–85.
Phillips ML, Drevets WC, Rauch SL, Lane R. Neurobiology of emotion perception II: implications for major psychiatric disorders. Biol Psychiatry. 2003;54:515–28.
Etkin A, Egner T, Kalisch R. Emotional processing in anterior cingulate and medial prefrontal cortex. Trends Cogn Sci. 2011;15:85–93.
Pizzagalli DA. Frontocingulate dysfunction in depression: toward biomarkers of treatment response. Neuropsychopharmacology. 2011;36:183–206.
Davidson RJ. Affective neuroscience and psychophysiology: toward a synthesis. Psychophysiology. 2003;40:655–65.
Ray JP, Price JL. The organization of projections from the mediodorsal nucleus of the thalamus to orbital and medial prefrontal cortex in macaque monkeys. J Comp Neurol. 1993;337:1–31.
Stuhrmann A, Dohm K, Kugel H, Zwanzger P, Redlich R, Grotegerd D, et al. Mood-congruent amygdala responses to subliminally presented facial expressions in major depression: associations with anhedonia. J Psychiatry Neurosci. 2013;38:249–58.
Suslow T, Konrad C, Kugel H, Rumstadt D, Zwitserlood P, Schöning S, et al. Automatic mood-congruent amygdala responses to masked facial expressions in major depression. Biol Psychiatry. 2010;67:155–60.
Klug M, Enneking V, Borgers T, Jacobs CM, Dohm K, Kraus A, et al. Persistence of amygdala hyperactivity to subliminal negative emotion processing in the long-term course of depression. Mol Psychiatry. 2024. https://doi.org/10.1038/s41380-024-02429-4.
Dannlowski U, Ohrmann P, Bauer J, Kugel H, Arolt V, Heindel W, et al. Amygdala reactivity to masked negative faces is associated with automatic judgmental bias in major depression: a 3 T fMRI study. J Psychiatry Neurosci. 2007;32:423–9.
Fales CL, Barch DM, Rundle MM, Mintun MA, Snyder AZ, Cohen JD, et al. Altered emotional interference processing in affective and cognitive-control brain circuitry in major depression. Biol Psychiatry. 2008;63:377–84.
Siegle GJ, Thompson W, Carter CS, Steinhauer SR, Thase ME. Increased amygdala and decreased dorsolateral prefrontal BOLD responses in unipolar depression: related and independent features. Biol Psychiatry. 2007;61:198–209.
DeRubeis RJ, Siegle GJ, Hollon SD. Cognitive therapy versus medication for depression: treatment outcomes and neural mechanisms. Nat Rev Neurosci. 2008;9:788–96.
Canli T, Cooney RE, Goldin P, Shah M, Sivers H, Thomason ME, et al. Amygdala reactivity to emotional faces predicts improvement in major depression. NeuroReport. 2005;16:1267–70.
Williams LM, Korgaonkar MS, Song YC, Paton R, Eagles S, Goldstein-Piekarski A, et al. Amygdala reactivity to emotional faces in the prediction of general and medication-specific responses to antidepressant treatment in the randomized iSPOT-D trial. Neuropsychopharmacology. 2015;40:2398–408.
Siegle GJ, Carter CS, Thase ME. Use of fMRI to predict recovery from unipolar depression with cognitive behavior therapy. Am J Psychiatry. 2006;163:735–8.
Queirazza F, Fouragnan E, Steele JD, Cavanagh J, Philiastides MG. Neural correlates of weighted reward prediction error during reinforcement learning classify response to cognitive behavioral therapy in depression. Sci Adv. 2019;5:eaav4962.
Fu CHY, Williams SCR, Cleare AJ, Scott J, Mitterschiffthaler MT, Walsh ND, et al. Neural responses to sad facial expressions in major depression following cognitive behavioral therapy. Biol Psychiatry. 2008;64:505–12.
Costafreda SG, Khanna A, Mourao-Miranda J, Fu CHY. Neural correlates of sad faces predict clinical remission to cognitive behavioural therapy in depression. NeuroReport. 2009;20:637–41.
Ritchey M, Dolcos F, Eddington KM, Strauman TJ, Cabeza R. Neural correlates of emotional processing in depression: changes with cognitive behavioral therapy and predictors of treatment response. J Psychiatr Res. 2011;45:577–87.
Rubin-Falcone H, Weber J, Kishon R, Ochsner K, Delaparte L, Doré B, et al. Neural predictors and effects of cognitive behavioral therapy for depression: the role of emotional reactivity and regulation. Psychol Med. 2020;50:146–60.
Siegle GJ, Thompson WK, Collier A, Berman SR, Feldmiller J, Thase ME, et al. Toward clinically useful neuroimaging in depression treatment: prognostic utility of subgenual cingulate activity for determining depression outcome in cognitive therapy across studies, scanners, and patient characteristics. Arch Gen Psychiatry. 2012;69:913.
McGrath CL, Kelley ME, Dunlop BW, Holtzheimer PE, Craighead WE, Mayberg HS. Pretreatment brain states identify likely nonresponse to standard treatments for depression. Biol Psychiatry. 2014;76:527–35.
Messina I, Bianco F, Cusinato M, Calvo V, Sambin M. Abnormal default system functioning in depression: implications for emotion regulation. Front Psychol. 2016;7:202607.
Dunlop BW, Mayberg HS. Neuroimaging-based biomarkers for treatment selection in major depressive disorder. Dialogues Clin Neurosci. 2014;16:479–90.
Crowther A, Smoski MJ, Minkel J, Moore T, Gibbs D, Petty C, et al. Resting-state connectivity predictors of response to psychotherapy in major depressive disorder. Neuropsychopharmacol. 2015;40:1659–73.
Davis EG, Foland-Ross LC, Gotlib IH. Neural correlates of top-down regulation and generation of negative affect in major depressive disorder. Psychiatry Res Neuroimaging. 2018;276:1–8.
Straub J, Metzger CD, Plener PL, Koelch MG, Groen G, Abler B. Successful group psychotherapy of depression in adolescents alters fronto-limbic resting-state connectivity. J Affect Disord. 2017;209:135–9.
Hollon SD, Thase ME, Markowitz JC. Treatment and prevention of depression. Psychol Sci Public Interest. 2002;3:39–77.
Zhou D, Zhou X, Lin Q, Wang W, Lv Z, Chen X, et al. Nonpharmacological interventions for relapse prevention in unipolar depression: a network meta-analysis. J Affect Disord. 2021;282:1255–62.
Berlim MT, Turecki G. Definition, assessment, and staging of treatment—resistant refractory major depression: a review of current concepts and methods. Can J Psychiatry. 2007;52:46–54.
Nemeroff CB. Prevalence and management of treatment-resistant depression. J Clin Psychiatry. 2007;68:17–25.
Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163:28–40.
George MS, Wassermann EM, Williams WA, Callahan A, Ketter TA, Basser P, et al. Daily repetitive transcranial magnetic stimulation (rTMS) improves mood in depression. NeuroReport. 1995;6:1853–6.
Tik M, Hoffmann A, Sladky R, Tomova L, Hummer A, Navarro De Lara L, et al. Towards understanding rTMS mechanism of action: Stimulation of the DLPFC causes network-specific increase in functional connectivity. NeuroImage. 2017;162:289–96.
Cao X, Deng C, Su X, Guo Y. Response and remission rates following high-frequency vs. low-frequency Repetitive Transcranial Magnetic Stimulation (rTMS) over right DLPFC for treating Major Depressive Disorder (MDD): a meta-analysis of randomized, double-blind trials. Front Psychiatry. 2018;9:413.
Baeken C, Marinazzo D, Wu G-R, Van Schuerbeek P, De Mey J, Marchetti I, et al. Accelerated HF-rTMS in treatment-resistant unipolar depression: insights from subgenual anterior cingulate functional connectivity. World J Biol Psychiatry. 2014;15:286–97.
Liston C, Chen AC, Zebley BD, Drysdale AT, Gordon R, Leuchter B, et al. Default mode network mechanisms of transcranial magnetic stimulation in depression. Biol Psychiatry. 2014;76:517–26.
Fox MD, Buckner RL, White MP, Greicius MD, Pascual-Leone A. Efficacy of transcranial magnetic stimulation targets for depression is related to intrinsic functional connectivity with the subgenual cingulate. Biol Psychiatry. 2012;72:595–603.
Weigand A, Horn A, Caballero R, Cooke D, Stern AP, Taylor SF, et al. Prospective validation that subgenual connectivity predicts antidepressant efficacy of transcranial magnetic stimulation sites. Biol Psychiatry. 2018;84:28–37.
Cash RFH, Zalesky A, Thomson RH, Tian Y, Cocchi L, Fitzgerald PB. Subgenual functional connectivity predicts antidepressant treatment response to transcranial magnetic stimulation: independent validation and evaluation of personalization. Biol Psychiatry. 2019;86:e5–7.
Ge R, Downar J, Blumberger DM, Daskalakis ZJ, Vila-Rodriguez F. Functional connectivity of the anterior cingulate cortex predicts treatment outcome for rTMS in treatment-resistant depression at 3-month follow-up. Brain Stimulation. 2020;13:206–14.
Elbau IG, Lynch CJ, Downar J, Vila-Rodriguez F, Power JD, Solomonov N, et al. Functional connectivity mapping for rTMS target selection in depression. Am J Psychiatry. 2023;180:230–40.
Roalf DR, Figee M, Oathes DJ. Elevating the field for applying neuroimaging to individual patients in psychiatry. Transl Psychiatry. 2024;14:87.
Siddiqi SH, Khosravani S, Rolston JD, Fox MD. The future of brain circuit-targeted therapeutics. Neuropsychopharmacology. 2024;49:179–88.
Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, et al. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45:651–60.
Elias GJB, Germann J, Boutet A, Loh A, Li B, Pancholi A, et al. 3T MRI of rapid brain activity changes driven by subcallosal cingulate deep brain stimulation. Brain. 2022;145:2214–26.
Huebl J, Brücke C, Merkl A, Bajbouj M, Schneider G-H, Kühn AA. Processing of emotional stimuli is reflected by modulations of beta band activity in the subgenual anterior cingulate cortex in patients with treatment resistant depression. Soc Cogn Affect Neurosci. 2016;11:1290–8.
Riva-Posse P, Choi KS, Holtzheimer PE, Crowell AL, Garlow SJ, Rajendra JK, et al. A connectomic approach for subcallosal cingulate deep brain stimulation surgery: prospective targeting in treatment-resistant depression. Mol Psychiatry. 2018;23:843–9.
Lozano AM, Giacobbe P, Hamani C, Rizvi SJ, Kennedy SH, Kolivakis TT, et al. A multicenter pilot study of subcallosal cingulate area deep brain stimulation for treatment-resistant depression. J Neurosurg. 2012;116:315–22.
Redlich R, Opel N, Grotegerd D, Dohm K, Zaremba D, Bürger C, et al. Prediction of individual response to electroconvulsive therapy via machine learning on structural magnetic resonance imaging data. JAMA Psychiatry. 2016;73:557.
Argyelan M, Lencz T, Kaliora S, Sarpal DK, Weissman N, Kingsley PB. et al. Subgenual cingulate cortical activity predicts the efficacy of electroconvulsive therapy. Transl Psychiatry. 2016;6:e789
Cano M, Cardoner N, Urretavizcaya M, Martínez-Zalacaín I, Goldberg X, Via E, et al. Modulation of limbic and prefrontal connectivity by electroconvulsive therapy in treatment-resistant depression: a preliminary study. Brain Stimulation. 2016;9:65–71.
Leaver AM, Wade B, Vasavada M, Hellemann G, Joshi SH, Espinoza R, et al. Fronto-temporal connectivity predicts ECT outcome in major depression. Front Psychiatry. 2018;9:92.
Moreno-Ortega M, Prudic J, Rowny S, Patel GH, Kangarlu A, Lee S, et al. Resting state functional connectivity predictors of treatment response to electroconvulsive therapy in depression. Sci Rep. 2019;9:5071.
Sun H, Jiang R, Qi S, Narr KL, Wade BS, Upston J, et al. Preliminary prediction of individual response to electroconvulsive therapy using whole-brain functional magnetic resonance imaging data. NeuroImage Clin. 2020;26:102080.
Pang Y, Wei Q, Zhao S, Li N, Li Z, Lu F, et al. Enhanced default mode network functional connectivity links with electroconvulsive therapy response in major depressive disorder. J Affect Disord. 2022;306:47–54.
Van Waarde JA, Scholte HS, Van Oudheusden LJB, Verwey B, Denys D, Van Wingen GA. A functional MRI marker may predict the outcome of electroconvulsive therapy in severe and treatment-resistant depression. Mol Psychiatry. 2015;20:609–14.
Ge R, Downar J, Blumberger DM, Daskalakis ZJ, Lam RW, Vila-Rodriguez F. Structural network integrity of the central executive network is associated with the therapeutic effect of rTMS in treatment resistant depression. Prog Neuro-Psychopharmacol Biol Psychiatry. 2019;92:217–25.
Fan J, Tso IF, Maixner DF, Abagis T, Hernandez-Garcia L, Taylor SF. Segregation of salience network predicts treatment response of depression to repetitive transcranial magnetic stimulation. NeuroImage Clin. 2019;22:101719.
Cao B, Luo Q, Fu Y, Du L, Qiu T, Yang X, et al. Predicting individual responses to the electroconvulsive therapy with hippocampal subfield volumes in major depression disorder. Sci Rep. 2018;8:5434.
Wade BSC, Joshi SH, Njau S, Leaver AM, Vasavada M, Woods RP, et al. Effect of electroconvulsive therapy on striatal morphometry in major depressive disorder. Neuropsychopharmacol. 2016;41:2481–91.
Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2017;23:28–38.
Dunlop K, Grosenick L, Downar J, Vila-Rodriguez F, Gunning FM, Daskalakis ZJ, et al. Dimensional and categorical solutions to parsing depression heterogeneity in a large single-site sample. Biol Psychiatry. 2024:S0006322324000556.
Fox MD. Mapping symptoms to brain networks with the human connectome. N. Engl J Med. 2018;379:2237–45.
Thomas Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–65.
Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, et al. The human connectome project: a data acquisition perspective. Neuroimage. 2012;62:2222–31.
Siddiqi SH, Taylor SF, Cooke D, Pascual-Leone A, George MS, Fox MD. Distinct symptom-specific treatment targets for circuit-based neuromodulation. Am J Psychiatry. 2020;177:435–46.
Greenberg T, Fournier JC, Stiffler R, Chase HW, Almeida JR, Aslam H, et al. Reward related ventral striatal activity and differential response to sertraline versus placebo in depressed individuals. Mol Psychiatry. 2020;25:1526–36.
Chin Fatt CR, Cooper C, Jha MK, Aslan S, Grannemann B, Kurian B, et al. Dorsolateral prefrontal cortex and subcallosal cingulate connectivity show preferential antidepressant response in major depressive disorder. Biol Psychiatry Cognit Neurosci Neuroimaging. 2021;6:20–8.
Frodl T, Scheuerecker J, Schoepf V, Linn J, Koutsouleris N, Bokde ALW, et al. Different effects of mirtazapine and venlafaxine on brain activation: an open randomized controlled fMRI study. J Clin Psychiatry. 2011;72:448–57.
Little JT, Ketter TA, Kimbrell TA, Dunn RT, Benson BE, Willis MW, et al. Bupropion and venlafaxine responders differ in pretreatment regional cerebral metabolism in unipolar depression. Biol Psychiatry. 2005;57:220–8.
Perez-Caballero L, Torres-Sanchez S, Romero-López-Alberca C, González-Saiz F, Mico JA, Berrocoso E. Monoaminergic system and depression. Cell Tissue Res. 2019;377:107–13.
Cooper CM, Chin Fatt CR, Jha M, Fonzo GA, Grannemann BD, Carmody T, et al. Cerebral blood perfusion predicts response to sertraline versus placebo for major depressive disorder in the EMBARC trial. EClinicalMedicine. 2019;10:32–41.
Dang Y, Lu B, Vanderwal T, Castellanos FX, Yan C-G. Early-treatment cerebral blood flow change as a predictive biomarker of antidepressant treatment response: evidence from the EMBARC clinical trial. Psychol Med. 2024:1–10.
Poirot MG, Ruhe HG, Mutsaerts H-JMM, Maximov II, Groote IR, Bjørnerud A, et al. Treatment response prediction in major depressive disorder using multimodal MRI and clinical data: secondary analysis of a randomized clinical trial. Am J Psychiatry. 2024;181:223–33.
Zhao K, Xie H, Fonzo GA, Tong X, Carlisle N, Chidharom M, et al. Individualized fMRI connectivity defines signatures of antidepressant and placebo responses in major depression. Mol Psychiatry. 2023;28:2490–9.
Korb AS, Hunter AM, Cook IA, Leuchter AF. Rostral anterior cingulate cortex theta current density and response to antidepressants and placebo in major depression. Clin Neurophysiol. 2009;120:1313–9.
Widge AS, Bilge MT, Montana R, Chang W, Rodriguez CI, Deckersbach T, et al. Electroencephalographic biomarkers for treatment response prediction in major depressive illness: a meta-analysis. Am J Psychiatry. 2019;176:44–56.
Pizzagalli DA, Webb CA, Dillon DG, Tenke CE, Kayser J, Goer F, et al. Pretreatment rostral anterior cingulate cortex theta activity in relation to symptom improvement in depression: a randomized clinical trial. JAMA Psychiatry. 2018;75:547.
Tenke CE, Kayser J. Generator localization by current source density (CSD): implications of volume conduction and field closure at intracranial and scalp resolutions. Clin Neurophysiol. 2012;123:2328–45.
Müller K-R, Tangermann M, Dornhege G, Krauledat M, Curio G, Blankertz B. Machine learning for real-time single-trial EEG-analysis: From brain–computer interfacing to mental state monitoring. J Neurosci Methods. 2008;167:82–90.
Singh AK, Krishnan S. Trends in EEG signal feature extraction applications. Front Artif Intell. 2023;5:1072801.
Wu W, Zhang Y, Jiang J, Lucas MV, Fonzo GA, Rolle CE, et al. An electroencephalographic signature predicts antidepressant response in major depression. Nat Biotechnol. 2020;38:439–47.
Zhdanov A, Atluri S, Wong W, Vaghei Y, Daskalakis ZJ, Blumberger DM, et al. Use of machine learning for predicting escitalopram treatment outcome from electroencephalography recordings in adult patients with depression. JAMA Netw Open. 2020;3:e1918377.
ACNP 62nd Annual Meeting: Poster Abstracts P1 – P250. Neuropsychopharmacol. 2023;48:63–210.
Dunlop BW, Binder EB, Cubells JF, Goodman MM, Kelley ME, Kinkead B, et al. Predictors of remission in depression to individual and combined treatments (PReDICT): study protocol for a randomized controlled trial. Trials. 2012;13:106.
Dunlop BW, Rajendra JK, Craighead WE, Kelley ME, McGrath CL, Choi KS, et al. Functional connectivity of the subcallosal cingulate cortex and differential outcomes to treatment with cognitive-behavioral therapy or antidepressant medication for major depressive disorder. Am J Psychiatry. 2017;174:533–45.
McGrath CL, Kelley ME, Holtzheimer PE, Dunlop BW, Craighead WE, Franco AR, et al. Toward a neuroimaging treatment selection biomarker for major depressive disorder. JAMA Psychiatry. 2013;70:821–9.
Delaveau P, Jabourian M, Lemogne C, Guionnet S, Bergouignan L, Fossati P. Brain effects of antidepressants in major depression: a meta-analysis of emotional processing studies. J Affect Disord. 2011;130:66–74.
Fitzgerald PB, Laird AR, Maller J, Daskalakis ZJ. A meta‐analytic study of changes in brain activation in depression. Hum Brain Mapp. 2008;29:683–95.
Kelley ME, Choi KS, Rajendra JK, Craighead WE, Rakofsky JJ, Dunlop BW, et al. Establishing evidence for clinical utility of a neuroimaging biomarker in major depressive disorder: prospective testing and implementation challenges. Biol Psychiatry. 2021;90:236–42.
Rutherford BR, Sneed JR, Roose SP. Does study design influence outcome? The effects of placebo control and treatment duration in antidepressant trials. Psychother Psychosom. 2009;78:172-81.
Hannon K, Bijsterbosch J. Challenges in identifying individualized brain biomarkers of late life depression. Adv Geriatr Med Res. 2023;5:e230010.
Nichols TE, Das S, Eickhoff SB, Evans AC, Glatard T, Hanke M, et al. Best practices in data analysis and sharing in neuroimaging using MRI. Nat Neurosci. 2017;20:299–303.
Rutherford BR, Wall MM, Brown PJ, Choo T-H, Wager TD, Peterson BS, et al. Patient expectancy as a mediator of placebo effects in antidepressant clinical trials. Am J Psychiatry. 2017;174:135–42.
Gaudiano BA, Ellenberg SR, Schofield CA, Rifkin LS. A randomized survey of the public’s expectancies and willingness to participate in clinical trials of antidepressants versus psychotherapy for depression. Prim Care Companion CNS Disord. 2016;18 https://doi.org/10.4088/PCC.15m01879.
Fountoulakis KN, McIntyre RS, Carvalho AF. From randomized controlled trials of antidepressant drugs to the meta-analytic synthesis of evidence: methodological aspects lead to discrepant findings. Curr Neuropharmacol. 2015;13:605–15.
Fu CHY, Antoniades M, Erus G, Garcia JA, Fan Y, Arnone D, et al. Neuroanatomical dimensions in medication-free individuals with major depressive disorder and treatment response to SSRI antidepressant medications or placebo. Nat Ment Health. 2024;2:164–76.
Schmaal L, Pozzi E, C. Ho T, Van Velzen LS, Veer IM, Opel N, et al. ENIGMA MDD: seven years of global neuroimaging studies of major depression through worldwide data sharing. Transl Psychiatry. 2020;10:172.
Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, et al. The ENIGMA consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–82.
Fu CHY, Erus G, Fan Y, Antoniades M, Arnone D, Arnott SR, et al. AI-based dimensional neuroimaging system for characterizing heterogeneity in brain structure and function in major depressive disorder: COORDINATE-MDD consortium design and rationale. BMC Psychiatry. 2023;23:59.
Enck P, Bingel U, Schedlowski M, Rief W. The placebo response in medicine: minimize, maximize or personalize? Nat Rev Drug Discov. 2013;12:191–204.
Jones BDM, Razza LB, Weissman CR, Karbi J, Vine T, Mulsant LS, et al. Magnitude of the placebo response across treatment modalities used for treatment-resistant depression in adults: a systematic review and meta-analysis. JAMA Netw Open. 2021;4:e2125531.
Wager TD, Atlas LY. The neuroscience of placebo effects: connecting context, learning and health. Nat Rev Neurosci. 2015;16:403–18.
Dunlop BW, Kelley ME, Aponte-Rivera V, Mletzko-Crowe T, Kinkead B, Ritchie JC, et al. Effects of patient preferences on outcomes in the Predictors of Remission in Depression to Individual and Combined Treatments (PReDICT) study. Am J Psychiatry. 2017;174:546–56.
Wasmann KA, Wijsman P, van Dieren S, Bemelman W, Buskens C. Partially randomised patient preference trials as an alternative design to randomised controlled trials: systematic review and meta-analyses. BMJ Open. 2019;9:e031151.
Zimmerman M, Balling C, Chelminski I, Dalrymple K. Have treatment studies of depression become even less generalizable? Applying the inclusion and exclusion criteria in placebo-controlled antidepressant efficacy trials published over 20 years to a clinical sample. Psychother Psychosom. 2019;88:165–70.
Zimmerman M, Balling C, Chelminski I, Dalrymple K. Applying the inclusion/exclusion criteria in placebo-controlled studies to a clinical sample: a comparison of medications. J Affect Disord. 2020;260:483–8.
Kessler RC, Merikangas KR, Wang PS. Prevalence, comorbidity, and service utilization for mood disorders in the United States at the beginning of the twenty-first century. Annu Rev Clin Psychol. 2007;3:137–58.
Kaufman J, Charney D. Comorbidity of mood and anxiety disorders. Depress Anxiety. 2000;12:69–76.
Wiethoff K, Bauer M, Baghai TC, Möller H-J, Fisher R, Hollinde D, et al. Prevalence and treatment outcome in anxious versus nonanxious depression: results from the German Algorithm Project. J Clin Psychiatry. 2010;71:1047–54.
Richards D. Prevalence and clinical course of depression: a review. Clin Psychol Rev. 2011;31:1117–25.
Lueken U, Zierhut KC, Hahn T, Straube B, Kircher T, Reif A, et al. Neurobiological markers predicting treatment response in anxiety disorders: a systematic review and implications for clinical application. Neurosci Biobehav Rev. 2016;66:143–62.
Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62.
Beck AT, Steer RA, Brown G. Manual for the Beck depression inventory-II. San Antonio, TX: Psychological Corporation; 1996.
Bagby RM, Ryder AG, Schuller DR, Marshall MB. The hamilton depression rating scale: has the gold standard become a lead weight? Am J Psychiatry. 2004;161:2163–77.
Williams LM. Defining biotypes for depression and anxiety based on large-scale circuit dysfunction: a theoretical review of the evidence and future directions for clinical translation. Depress Anxiety. 2017;34:9–24.
Grunebaum MF, Keilp JG, Ellis SP, Sudol K, Bauer N, Burke AK, et al. SSRI versus bupropion effects on symptom clusters in suicidal depression: post hoc analysis of a randomized clinical trial. J Clin Psychiatry. 2013;74:872–9.
Stewart JG, Harkness KL. Symptom specificity in the acute treatment of major depressive disorder: a re-analysis of the treatment of depression collaborative research program. J Affect Disord. 2012;137:87–97.
Uher R, Maier W, Hauser J, Marušič A, Schmael C, Mors O, et al. Differential efficacy of escitalopram and nortriptyline on dimensional measures of depression. Br J Psychiatry. 2009;194:252–9.
Dunlop BW, Cole SP, Nemeroff CB, Mayberg HS, Craighead WE. Differential change on depressive symptom factors with antidepressant medication and cognitive behavior therapy for major depressive disorder. J Affect Disord. 2018;229:111–9.
Ballard ED, Yarrington JS, Farmer CA, Lener MS, Kadriu B, Lally N, et al. Parsing the heterogeneity of depression: an exploratory factor analysis across commonly used depression rating scales. J Affect Disord. 2018;231:51–57.
Fried EI, Nesse RM. Depression sum-scores don’t add up: why analyzing specific depression symptoms is essential. BMC Med. 2015;13:72.
Sheehan DV, Harnett-Sheehan K, Raj BA. The measurement of disability. Int Clin Psychopharmacol. 1996;11:89–95.
Soares CN, Endicott J, Boucher M, Fayyad RS, Guico-Pabia CJ. Predictors of functional response and remission with desvenlafaxine 50 mg/d in patients with major depressive disorder. CNS Spectr. 2014;19:519–27.
Soares CN, Zhang M, Boucher M. Categorical improvement in functional impairment in depressed patients treated with desvenlafaxine. CNS Spectr. 2019;24:322–32.
Fu CHY, Costafreda SG, Sankar A, Adams TM, Rasenick MM, Liu P, et al. Multimodal functional and structural neuroimaging investigation of major depressive disorder following treatment with duloxetine. BMC Psychiatry. 2015;15:82.
Zhou R, Chen J, Zhao G, Wang Z, Peng D, Xia W, et al. Neural biomarker of functional disability in major depressive disorder: a structural neuroimaging study. Prog Neuropsychopharmacol Biol Psychiatry. 2021;111:110337.
Solomonov N, Lee J, Banerjee S, Flückiger C, Kanellopoulos D, Gunning FM, et al. Modifiable predictors of nonresponse to psychotherapies for late-life depression with executive dysfunction: a machine learning approach. Mol Psychiatry. 2021;26:5190–8.
Lewis CC, Simons AD, Kim HK. The role of early symptom trajectories and pretreatment variables in predicting treatment response to cognitive behavioral therapy. J Consult Clin Psychol. 2012;80:525–34.
Kelley ME, Dunlop BW, Nemeroff CB, Lori A, Carrillo-Roa T, Binder EB, et al. Response rate profiles for major depressive disorder: Characterizing early response and longitudinal nonresponse. Depress Anxiety. 2018;35:992–1000.
Frässle S, Marquand AF, Schmaal L, Dinga R, Veltman DJ, van der Wee NJA, et al. Predicting individual clinical trajectories of depression with generative embedding. Neuroimage Clin. 2020;26:102213.
Schmaal L, Marquand AF, Rhebergen D, van Tol M-J, Ruhé HG, van der Wee NJA, et al. Predicting the naturalistic course of major depressive disorder using clinical and multimodal neuroimaging information: a multivariate pattern recognition study. Biol Psychiatry. 2015;78:278–86.
Gonzalez-Castillo J, Kam JWY, Hoy CW, Bandettini PA. How to interpret resting-state fmri: ask your participants. J Neurosci. 2021;41:1130–41.
Jiang R, Zuo N, Ford JM, Qi S, Zhi D, Zhuo C, et al. Task-induced brain connectivity promotes the detection of individual differences in brain-behavior relationships. Neuroimage. 2020;207:116370.
Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen X, Constable RT, et al. A neuromarker of sustained attention from whole-brain functional connectivity. Nat Neurosci. 2016;19:165–71.
Zhao W, Makowski C, Hagler DJ, Garavan HP, Thompson WK, Greene DJ, et al. Task fMRI paradigms may capture more behaviorally relevant information than resting-state functional connectivity. NeuroImage. 2023;270:119946.
Greene AS, Gao S, Scheinost D, Constable RT. Task-induced brain state manipulation improves prediction of individual traits. Nat Commun. 2018;9:2807.
Elliott ML, Knodt AR, Cooke M, Kim MJ, Melzer TR, Keenan R, et al. General functional connectivity: Shared features of resting-state and task fMRI drive reliable and heritable individual differences in functional brain networks. Neuroimage. 2019;189:516–32.
Dunlop K, Rizvi SJ, Kennedy SH, Hassel S, Strother SC, Harris JK, et al. Clinical, behavioral, and neural measures of reward processing correlate with escitalopram response in depression: a Canadian Biomarker Integration Network in Depression (CAN-BIND-1) report. Neuropsychopharmacology. 2020;45:1390–7.
Admon R, Kaiser RH, Dillon DG, Beltzer M, Goer F, Olson DP, et al. Dopaminergic enhancement of striatal response to reward in major depression. Am J Psychiatry. 2017;174:378–86.
Kraus C, Klöbl M, Tik M, Auer B, Vanicek T, Geissberger N, et al. The pulvinar nucleus and antidepressant treatment: dynamic modeling of antidepressant response and remission with ultra-high field functional MRI. Mol Psychiatry. 2019;24:746–56.
Walsh EC, Eisenlohr-Moul TA, Minkel J, Bizzell J, Petty C, Crowther A, et al. Pretreatment brain connectivity during positive emotion upregulation predicts decreased anhedonia following behavioral activation therapy for depression. J Affect Disord. 2019;243:188–92.
Karim HT, Wang M, Andreescu C, Tudorascu D, Butters MA, Karp JF, et al. Acute trajectories of neural activation predict remission to pharmacotherapy in late-life depression. Neuroimage Clin. 2018;19:831–9.
Crane NA, Jenkins LM, Bhaumik R, Dion C, Gowins JR, Mickey BJ, et al. Multidimensional prediction of treatment response to antidepressants with cognitive control and functional MRI. Brain. 2017;140:472–86.
Miller S, McTeague LM, Gyurak A, Patenaude B, Williams LM, Grieve SM, et al. Cognition-childhood maltreatment interactions in the prediction of antidepressant outcomes in major depressive disorder patients: results from the iSPOT-D trial. Depress Anxiety. 2015;32:594–604.
Hack LM, Tozzi L, Zenteno S, Olmsted AM, Hilton R, Jubeir J, et al. A cognitive biotype of depression and symptoms, behavior measures, neural circuits, and differential treatment outcomes: a prespecified secondary analysis of a randomized clinical trial. JAMA Netw Open. 2023;6:e2318411.
Finn ES, Bandettini PA. Movie-watching outperforms rest for functional connectivity-based prediction of behavior. Neuroimage. 2021;235:117963.
Welvaert M, Rosseel Y. On the definition of signal-to-noise ratio and contrast-to-noise ratio for fMRI data. PLoS one. 2013;8:e77089.
Kundu P, Inati SJ, Evans JW, Luh W-M, Bandettini PA. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage. 2012;60:1759–70.
Kundu P, Brenowitz ND, Voon V, Worbe Y, Vértes PE, Inati SJ, et al. Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proc Natl Acad Sci USA. 2013;110:16187–92.
Kundu P, Voon V, Balchandani P, Lombardo MV, Poser BA, Bandettini PA. Multi-echo fMRI: a review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage. 2017;154:59–80.
DuPre E, Salo T, Ahmed Z, Bandettini P, Bottenhorn K, Caballero-Gaudes C, et al. TE-dependent analysis of multi-echo fMRI with tedana. J Open Sour Softw. 2021;6:3669.
Steel A, Garcia BD, Silson EH, Robertson CE. Evaluating the efficacy of multi-echo ICA denoising on model-based fMRI. Neuroimage. 2022;264:119723.
Cohen AD, Chang C, Wang Y. Using multiband multi-echo imaging to improve the robustness and repeatability of co-activation pattern analysis for dynamic functional connectivity. Neuroimage. 2021;243:118555.
Power JD, Plitt M, Gotts SJ, Kundu P, Voon V, Bandettini PA, et al. Ridding fMRI data of motion-related influences: removal of signals with distinct spatial and physical bases in multiecho data. Proc Natl Acad Sci USA. 2018;115:E2105–14.
Gonzalez-Castillo J, Panwar P, Buchanan LC, Caballero-Gaudes C, Handwerker DA, Jangraw DC, et al. Evaluation of multi-echo ICA denoising for task based fMRI studies: block designs, rapid event-related designs, and cardiac-gated fMRI. Neuroimage. 2016;141:452–68.
Gilmore AW, Agron AM, González-Araya EI, Gotts SJ, Martin A. A comparison of single- and multi-echo processing of functional MRI data during overt autobiographical recall. Front Neurosci. 2022;16:854387.
Lynch CJ, Power JD, Scult MA, Dubin M, Gunning FM, Liston C. Rapid precision functional mapping of individuals using multi-echo fMRI. Cell Rep. 2020;33:108540.
Lynch CJ, Elbau I, Liston C. Improving precision functional mapping routines with multi-echo fMRI. Curr Opin Behav Sci. 2021;40:113–9.
Kong R, Li J, Orban C, Sabuncu MR, Liu H, Schaefer A, et al. Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cereb Cortex. 2019;29:2533–51.
Seitzman BA, Gratton C, Laumann TO, Gordon EM, Adeyemo B, Dworetsky A, et al. Trait-like variants in human functional brain networks. Proc Natl Acad Sci USA. 2019;116:22851–61.
Wang D, Li M, Wang M, Schoeppe F, Ren J, Chen H, et al. Individual-specific functional connectivity markers track dimensional and categorical features of psychotic illness. Mol Psychiatry. 2020;25:2119–29.
Greene AS, Shen X, Noble S, Horien C, Hahn CA, Arora J, et al. Brain-phenotype models fail for individuals who defy sample stereotypes. Nature. 2022;609:109–18.
Arslan S, Ktena SI, Makropoulos A, Robinson EC, Rueckert D, Parisot S. Human brain mapping: a systematic comparison of parcellation methods for the human cerebral cortex. NeuroImage. 2018;170:5–30.
Marek S, Tervo-Clemmens B, Calabro FJ, Montez DF, Kay BP, Hatoum AS, et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603:654–60.
Spisak T, Bingel U, Wager TD. Multivariate BWAS can be replicable with moderate sample sizes. Nature. 2023;615:E4–7.
Sun X, Sun J, Lu X, Dong Q, Zhang L, Wang W, et al. Mapping neurophysiological subtypes of major depressive disorder using normative models of the functional connectome. Biol Psychiatry. 2023;94:936–47.
Buch AM, Vértes PE, Seidlitz J, Kim SH, Grosenick L, Liston C. Molecular and network-level mechanisms explaining individual differences in autism spectrum disorder. Nat Neurosci. 2023;26:650–63.
Jha MK, Chin Fatt C, Minhajuddin A, Mayes TL, Trivedi MH. Accelerated brain aging in adults with major depressive disorder predicts poorer outcome with sertraline: findings from the EMBARC study. Biol Psychiatry Cogn Neurosci Neuroimaging. 2023;8:462–70.
Fusar-Poli P, Manchia M, Koutsouleris N, Leslie D, Woopen C, Calkins ME, et al. Ethical considerations for precision psychiatry: a roadmap for research and clinical practice. Eur Neuropsychopharmacol. 2022;63:17–34.
Stein LA. Making the best use of radiological resources in Canada. Healthpapers. 2005;6:18–23.
Lacson R, Pianykh O, Hartmann S, Johnston H, Daye D, Flores E, et al. Factors associated with timeliness and equity of access to outpatient MRI examinations. J Am Coll Radio. 2024;S1546-1440:00001–2.
Siegel JS, Pearson C, Lenze EJ. Better biomarkers, faster drugs, stronger models: progress towards precision psychiatry. Mo Med. 2023;120:292–8.
ClinicalTrials.gov Identifier: NCT05017311: Optimized Predictive Treatment In Medications for Unipolar Major Depression (OPTIMUM-D) (CAN-BIND-17). https://clinicaltrials.gov/study/NCT05017311.
ClinicalTrials.gov Identifier: NCT04041479: Biomarker-guided rTMS for Treatment Resistant Depression (BioTMS). https://clinicaltrials.gov/study/NCT04041479.
Ochsner KN, Knierim K, Ludlow DH, Hanelin J, Ramachandran T, Glover G, et al. Reflecting upon feelings: an fMRI study of neural systems supporting the attribution of emotion to self and other. J Cogn Neurosci. 2004;16:1746–72.
Scharnowski F, Nicholson AA, Pichon S, Rosa MJ, Rey G, Eickhoff SB, et al. The role of the subgenual anterior cingulate cortex in dorsomedial prefrontal-amygdala neural circuitry during positive-social emotion regulation. Hum Brain Mapp. 2020;41:3100–18.
Niendam TA, Laird AR, Ray KL, Dean YM, Glahn DC, Carter CS. Meta-analytic evidence for a superordinate cognitive control network subserving diverse executive functions. Cogn Affect Behav Neurosci. 2012;12:241–68.
Dosenbach NUF, Fair DA, Cohen AL, Schlaggar BL, Petersen SE. A dual-networks architecture of top-down control. Trends Cogn Sci. 2008;12:99–105.
Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–56.
Mayberg HS, Liotti M, Brannan SK, McGinnis S, Mahurin RK, Jerabek PA, et al. Reciprocal limbic-cortical function and negative mood: converging PET findings in depression and normal sadness. Am J Psychiatry. 1999;156:675–82.
RajMohan V, Mohandas E. The limbic system. Indian J Psychiatry. 2007;49:132.
Sergerie K, Chochol C, Armony JL. The role of the amygdala in emotional processing: a quantitative meta-analysis of functional neuroimaging studies. Neurosci Biobehav Rev. 2008;32:811–30.
Rolls ET. Limbic systems for emotion and for memory, but no single limbic system. Cortex. 2015;62:119–57.
Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–70.
Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex. 2012;22:158–65.
Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL. Functional-anatomic fractionation of the brain’s default network. Neuron. 2010;65:550–62.
Smallwood J, Bernhardt BC, Leech R, Bzdok D, Jefferies E, Margulies DS. The default mode network in cognition: a topographical perspective. Nat Rev Neurosci. 2021;22:503–13.
Tozzi L, Zhang X, Chesnut M, Holt-Gosselin B, Ramirez CA, Williams LM. Reduced functional connectivity of default mode network subsystems in depression: meta-analytic evidence and relationship with trait rumination. NeuroImage Clin. 2021;30:102570.
Sridharan D, Levitin DJ, Menon V. A critical role for the right fronto-insular cortex in switching between central-executive and default-mode networks. Proc Natl Acad Sci USA. 2008;105:12569–74.
Northoff G. Psychopathology and pathophysiology of the self in depression — Neuropsychiatric hypothesis. J Affect Disord. 2007;104:1–14.
Funding
KD is supported by a University of Toronto Department of Psychiatry Academic Scholars Award.
Author information
Authors and Affiliations
Contributions
Authors KD and SP conceived, wrote, and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Prompiengchai, S., Dunlop, K. Breakthroughs and challenges for generating brain network-based biomarkers of treatment response in depression. Neuropsychopharmacol. (2024). https://doi.org/10.1038/s41386-024-01907-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41386-024-01907-1
- Springer Nature Switzerland AG