Abstract
Anxiety disorders are common, complex psychiatric disorders with twin heritabilities of 30–60%. We conducted a genome-wide association study of Lifetime Anxiety Disorder (ncase = 25 453, ncontrol = 58 113) and an additional analysis of Current Anxiety Symptoms (ncase = 19 012, ncontrol = 58 113). The liability scale common variant heritability estimate for Lifetime Anxiety Disorder was 26%, and for Current Anxiety Symptoms was 31%. Five novel genome-wide significant loci were identified including an intergenic region on chromosome 9 that has previously been associated with neuroticism, and a locus overlapping the BDNF receptor gene, NTRK2. Anxiety showed significant positive genetic correlations with depression and insomnia as well as coronary artery disease, mirroring findings from epidemiological studies. We conclude that common genetic variation accounts for a substantive proportion of the genetic architecture underlying anxiety.
Similar content being viewed by others
Introduction
Anxiety disorders are amongst the most common classes of psychiatric disorders worldwide [1, 2] and have a global lifetime prevalence of ~16% [1]. They were responsible for an estimated 27 121 years lost due to disability globally in 2017 [3], have an early age of onset and significant impact on educational attainment and risk for subsequent disorders [1]. Anxiety disorders aggregate in families, and twin heritability estimates range from 20 to 60%, dependent upon the participant age and specific trait or disorder being studied [4, 5]. Although many candidate gene studies of anxiety disorders have been carried out, these associations have not proven robust [6]. As is the case with other psychiatric disorders, such as schizophrenia [7] and depression [8], it is likely that a multitude of common genetic variants with modest effects, in addition to environmental factors, underlie the risk for anxiety disorders [6, 9, 10].
Clinically, anxiety is divided into several sub-classifications, including generalised anxiety disorder, social anxiety disorder, panic disorder, agoraphobia, and specific phobias. However, there is evidence for commonalities across anxiety disorders at both the phenotypic [11] and genetic [6, 10, 12] level. The majority of the data indicating genetic overlap between different anxiety traits/disorders comes from the twin literature [12,13,14,15], with clear evidence that the shared genetic component between anxiety disorders is larger than the unique contributions to any one disorder.
Furthermore, the covariance between anxiety disorders and depression is best explained by a single genetic factor with some evidence for additional phobia specific genetic factors [13]. A recent review summarised a range of research strongly indicating that current diagnostic boundaries between anxiety disorders are unlikely to reflect biologically distinct disorders [6]. Similarly, several twin studies have found high genetic correlations between the personality trait of neuroticism and anxiety disorders (including generalised anxiety disorder [16,17,18], social and situational phobias and agoraphobia [18, 19]). As such, it is likely there is a single underlying liability distribution, with variation in a range of anxiety-related traits, including anxiety disorders, depression, and neuroticism, occurring to different degrees across the population, driven in part by common genetic variants [12, 14, 20, 21]. If this were the case, it is likely that anxiety disorders share a common polygenic influence, which may explain the shared phenotypic and genetic structure identified in the twin literature.
The detection of genetic variants and genes associated with anxiety disorders has made slow progress, limited by small sample sizes. There have been some suggestive hits including TMEM132D for panic disorders, but few have been replicated [6]. A meta-analysis of genome-wide association studies of several anxiety disorders (N = 18 186) identified one genome-wide significant locus associated with anxiety caseness (ncases = 3 695; ncontrols = 13 615) and a second with a quantitative factor score of broad anxiety [10]. Finally, a SNP within RBFOX1 was found to be significantly associated with anxiety sensitivity in a small cohort of twins [22]. The proportion of variation in generalised anxiety disorder symptoms explained by individual genetic variation (SNP heritability; h2SNP) was estimated at 7.2% in a modest sample (n = 12 282) of Hispanic/Latino adults [9], and 14% in the meta-analysis described above, which consisted largely of individuals with European ancestry [10].
To date, none of these findings have been replicated, although RBFOX1 was significantly associated with major depression in a recent genome-wide association analysis [8]. Analyses of significantly larger samples are required to further our understanding of the underlying genetic architecture of anxiety disorders and current anxiety symptoms at the population level. With this aim, we conducted genome-wide association studies (GWAS) of two anxiety phenotypes: Lifetime Anxiety Disorder, which combines self-reported lifetime diagnosis of an anxiety disorder by a clinician withprobable DSM-IV generalized anxiety disorder; and Current Anxiety Symptoms, here cases are anyone who reported at least moderate symptoms of generalised anxiety disorder in the two weeks preceding assessment. Data were drawn from the UK Biobank, a large community-based sample. Two cohorts containing anxiety disorder cases and controls were used for the purposes of replication analyses; the Anxiety NeuroGenetics Study (ANGST) [10], and a sample from the Danish iPSYCH study [23]. Further replication analyses were undertaken using summary statistics from two highly relevant phenotypes - neuroticism (in a distinct subset of the UK Biobank), and major depressive disorder (in a subset of the Psychiatric Genetics Consortium Major Depressive Disorder sample) [8].
Methods
Sample and phenotype definition
Samples were a subset of 126 443 (age 46–80) individuals of Western European ancestry who took part in the UK Biobank online mental health follow-up questionnaire that aimed to identify mental health disorders without the need for clinical ascertainment [24]. The UK Biobank is a large prospective cohort study of over 500 000 people in the United Kingdom.
(1) Self-report clinician provided or probable lifetime anxiety disorder (Lifetime Anxiety Disorder). This was our primary phenotype. Cases met one of two definitions. First was self-reporting a lifetime professional diagnosis of one of the core five anxiety disorders (generalised anxiety disorder, social phobia, panic disorder, agoraphobia or specific phobia; n = 21 108). Further cases were defined as meeting criteria for a likely lifetime diagnosis of DSM-IV generalised anxiety disorder based on anxiety questions from the Composite International Diagnostic Interview (CIDI) Short-form questionnaire [25] (n = 4 345). We note that 5 296 participants meet both criteria for case inclusion. We excluded individuals self-reporting a lifetime diagnosis of schizophrenia, bipolar disorder, autistic spectrum disorder, attention deficit hyperactivity disorder, or eating disorders (all of which have higher heritabilities than anxiety disorders) to enable assessment of shared genetic risk with psychiatric phenotypes that have a high rate of phenotypic co-occurrence in secondary analyses. Individuals meeting one or other of these criteria resulted in a total of 25 453 cases. Consistent with epidemiological findings [26] 66% of cases were female.
(2) Moderate/severe current generalised anxiety symptoms (Current Anxiety Symptoms). For this secondary phenotype, participants were defined as cases if they obtained a total score on a screening measure for anxiety symptoms over the past two weeks (GAD-7) of ≥10 out of a total score of 21. This is a standard threshold for recent moderate to severe generalised anxiety symptoms as measured by the GAD-7 [27] (n = 19 012).
(3) Healthy controls: A common control group was identified consisting of a set of screened healthy individuals who did not meet criteria for any mental health disorder or known substance abuse, were not prescribed medication for any psychiatric disorder and did not report having sought help for any mental health disorder (n = 58 113).
See Supplementary Information for more detailed description of each category.
Independent anxiety-related samples
Summary statistics from four other studies were used to replicate and extend our analyses. The first two of these represent lifetime anxiety diagnoses replications. These included GWAS summary statistics from the Anxiety NeuroGenetics Study [10] of individuals meeting DSM criteria for at least one of the five core anxiety disorders (generalized anxiety disorder, social phobia, panic disorder, agoraphobia or specific phobia; n = 3 695) during their lifetime; and controls (n = 13 615). In addition we include genetic correlations with summary statistics from the ANGST latent factor score derived from combining controls and individuals reporting full and sub-threshold symptoms of the five anxiety disorders (n = 18 186) [10]. Second, summary statistics from an unpublished subset of lifetime anxiety cases (n = 2 829) and controls (n = 10 386) from the iPSYCH cohort in Denmark [23, 28], defined using the same inclusion and exclusion criteria as for our primary phenotype, although diagnoses were made by psychiatrists rather than self-report of symptoms or clinical diagnosis. The latter two are phenotypes known to be closely related to anxiety. These included summary statistics from an analysis of total neuroticism score in all individuals who completed a baseline assessment in the UK Biobank (UKB Neuroticism), who were not included as either a case or control in the Lifetime Anxiety Disorder phenotype (n = 241 883). Finally we used summary statistics for major depressive disorder from the most recent Psychiatric Genomics Consortium depression analysis (PGC MDD) [8], excluding samples drawn from the UK Biobank or 23&Me (ncases = 45 591; ncontrols = 97 674).
In summary, anxiety-related phenotypes from four replication samples were available for comparison with the core UK Biobank Lifetime Anxiety Disorder phenotype; (1) ANGST Anxiety Disorder, (2) iPSYCH Anxiety Disorder, (3) UKB Neuroticism, and (4) PGC MDD. See Supplementary Table 1 for sample sizes for each phenotype.
Genotyping and quality control
Genotype data were collected and processed as part of the UK Biobank extraction and quality control pipeline [29]. SNPs imputed to the Haplotype Reference Consortium (HRC) reference panel or genotyped SNPs were used for these analyses (hg19 build). SNPs with a minor allele frequency >0.01 and INFO score >0.4 (indicating well imputed variants) were retained. Only data from unrelated individuals were included. For details see Supplementary Information.
Statistical analyses
Primary analyses
Genome-wide association analyses
Analyses were limited to individuals of European ancestry defined by 4-means clustering on the first two ancestry principal components. Covariates (age at time of assessment, sex, genotyping batch, assessment centre, and the first six genetic principal components) were regressed out of each phenotype using logistic regression performed using R v. 3.5.1 [30]. Resulting residuals were used as dependent variables in three linear genome-wide association analyses, using BGENIE v1.2 software [29]. Variants surpassing genome-wide significance (p < 5 × 10−8) were annotated using Region Annotator software to annotate known protein coding genes within the regions of significance (https://github.com/ivankosmos/RegionAnnotator).
SNP heritability
To estimate the proportion of variance explained by common genetic variants (h2SNP) for the two UK Biobank anxiety phenotypes we used variance component analyses conducted in BOLT-LMM [31] . Estimates (and standard errors) were converted to the liability scale assuming: (1) accurate sampling rate in the UK Biobank (sample prevalence = population prevalence), (2) sample prevalence < population prevalence; and (3) sample prevalence > population prevalence (Supplementary Table 2).
Genetic correlations
LD score regression [32] was used to estimate genetic correlations between (1) UK Biobank Lifetime Anxiety Disorder, Current Anxiety Symptoms and all replication samples, and (2) UK Biobank Lifetime Anxiety Disorder and other previously published phenotypes using a manually curated set of downloaded publicly available summary statistics. Correlations with 597 UK Biobank traits [33] available on LDhub [34] were not conducted as these “should not be regarded as a definitive set of curated and reviewed association statistics” [35]. Genetic correlations are considered significant at p < 0.0002, Bonferroni corrected for 251 independent tests. See Supplementary Information for more detail.
Secondary analyses
Further analyses were carried out on the best powered core Lifetime Anxiety Disorder phenotype except where otherwise specified to investigate replication and dimensionality of anxiety symptoms.
Partitioned heritability
Stratified LD score regression [32] was used to partition SNP heritability by functional genomic categories to test for enrichment of heritability in biologically relevant genomic regions. See Supplementary Fig. 1 for results of this analysis.
Gene-wise analysis
Gene-wise associations were computed on GWA summary statistics using MAGMA [36] to test for the aggregated effect of multiple genetic markers simultaneously. This was done (a) to test which specific genes showed evidence for association with anxiety and (b) to use these gene-wide statistics to run biological pathway analyses. See Supplementary Information for additional details.
Polygenic prediction of anxiety
We examined the extent to which genome-wide polygenic signal from independent SNPs identified in the Lifetime Anxiety Disorder GWAS account for variance in liability to Lifetime Anxiety Disorder case status using a leave-one-out approach. All participants in the Lifetime Anxiety Disorder analysis were randomly divided into ten subgroups of approximately equal size with case:control ratio comparable to the primary analysis. Ten new GWAS were performed using PLINK 1.9 [37]. Each sub group was left out of one analysis and served as the target sample for polygenic scoring. The average R2 was derived across all ten iterations of the polygenic scoring at the p-threshold that was best predictive. For detail of polygenic score creation see Supplementary Information.
Testing the dimensionality of anxiety
To consider the degree to which differing levels of severity in anxiety traits reflect the same underlying genetic factors, three independent groups were identified using their current anxiety (GAD-7) symptom scores, including mild, (scoring 5–10, ncases = 49 144), moderate (10–15, ncases = 13 788) and severe (>15, ncases = 5 093). See Supplementary Table 3 for sample sizes for each analysis. Genetic correlations between these three groups were calculated using LD Score regression [32].
Replication of significant SNPs
All lead SNPs that surpassed genome-wide significance (5 × 10−8) in the core UK Biobank Lifetime Anxiety Disorder analysis were examined for direction and significance of effect in the four replication samples.
Meta-analysis
The three Lifetime Anxiety Disorder samples; UK Biobank Lifetime Anxiety Disorder, ANGST Anxiety Disorder and iPSYCH Anxiety Disorder were combined using inverse-variance weighted, fixed-effect meta-analyses in METAL [38]. Only SNPs shared across all three samples were included in this meta-analysis. Due to the risk of biased estimates of effect size when meta-analysing heterogeneous samples with small sample sizes [39] we consider these analyses as preliminary rather than primary.
Results
Genome-wide association
Results of the genome-wide association analyses for the two anxiety phenotypes (Lifetime Anxiety Disorder and Current Anxiety Symptoms) in UK Biobank are displayed in Fig. 1. Manhattan and Q-Q plots are shown for each phenotype. Supplementary Table 4 shows the results of region annotation for regions that surpassed genome-wide significance (P < 5 × 10−8).
Manhattan and Q-Q plots of p-values of single-nucleotide polymorphism (SNP) based association analyses of anxiety in the UK Biobank. In the Manhattan plots, the threshold for genome-wide significance (p < 5 x 10−8) is indicated by the red line, while the blue line indicates the threshold for suggestive significance (p < 1 x 10−5). The top panel shows results for our core lifetime anxiety disorder analysis. The bottom panel shows results for the current anxiety symptoms analysis
Lifetime anxiety disorder
Five regions were significant at the genome-wide threshold of 5 × 10−8 on chromosomes 9 (9p23 and 9q21.33), 7 (7q21.1), 5 (5q15) and 3 (3q11.2). The index SNP for the most significant region on 9p23 was rs10809485 (p = 1.6 × 10−12) which is in an intergenic region. The index SNP within 9q21.33 is rs1187280 (p = 5.2 × 10−8), which is in an intron for the protein coding gene Neurotrophic Receptor Tyrosine Kinase 2 (NTRK2). The index SNP of the chromosome 7 locus (rs3807866, p = 4.8 × 10−8) is within Transmembrane Protein 106B (TMEM106B). The chromosome 5 locus (rs2861139, p = 2.6 × 10−9) is in an intergenic region, and the chromosome 3 locus (rs4855559, p = 3.7 × 10−8) is in the intron for Myosin Heavy Chain 15 (MYH15). See Supplementary Figs. 2–6 for region plots.
Current anxiety symptoms
A single genome-wide significant locus was associated with our secondary phenotype in the intergenic region on chromosome 9p23, also associated with Lifetime Anxiety Disorder (LD r2 = 1) [40]. See Supplementary Fig. 7 for the region plot.
SNP heritability
Estimates of SNP heritability (h2SNP) converted to the liability threshold are 0.26 (SE = 0.011), and 0.31 (SE = 0.011), for Lifetime Anxiety Disorder and Current Anxiety Symptoms assuming sample prevalence of 0.20 and 0.18, respectively. We note that this is somewhat higher than epidemiological estimates of population prevalence of anxiety [1]. Supplementary Table 5 shows the estimates using different assumptions about population prevalence rates.
Genetic correlation between independent anxiety-related phenotypes
UK Biobank anxiety phenotypes have high genetic correlations with each other (rG = 0.93), and with the ANGST case-control phenotype (rG = 0.74 ± 0.01), UK Biobank neuroticism (rG = 0.73 ± 0.04) and PGC MDD (rG = 0.78 ± 0.10). UK biobank anxiety correlates moderately with the ANGST factor score (rG = 0.49 ± 0.03). Note that it was not possible to compare genetic correlations with the iPSYCH sample, as the estimate of h2SNP in that sample did not significantly differ from zero. See Supplementary Table 6 for genetic correlation and standard error between UK Biobank Lifetime Anxiety Disorder and all other anxiety-related phenotypes.
Genetic correlations with other traits
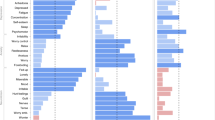
Significant genetic correlations between Lifetime Anxiety Disorder and a range of other previously published phenotypes are presented in Fig. 2. Genetic correlations with Current Anxiety Symptoms are in Supplementary Table 7.
The figure shows genetic correlations (rG) between UK Biobank “Lifetime Anxiety Disorder” and external phenotypes. Only traits where correlation exceeded the Bonferroni corrected threshold of p < 0.0002 are shown here. Depression and neuroticism were not included in this analysis as they have been included as replication samples in subsequent analyses. See Supplementary Table 6 for rG with neuroticism and MDD GWAS samples, including the UK Biobank. Asterisks indicate the source study sample wholly or partially included UK Biobank participants. Correlations are ordered first by rG within category. Bars show the standard error of the estimate
Gene-wise analysis
For gene-wise associations, significance was determined to be p < 2.80 × 10−6 (i.e. 0.05/17 831) to account for the number of comparisons. Supplementary Table 8 shows all gene-wise associations that are significantly associated with Lifetime Anxiety Disorder. Fig. 3 presents a Manhattan plot showing the top gene-wise associations with Lifetime Anxiety Disorder per chromosome. The most significantly associated gene in this analysis was the Transmembrane Protein 106B (TMEM106B; p = 6.28 × 10−10), while Glutamate Decarboxylase 2 (GAD2; p = 7.00 × 10−7) and Neurotrophic Receptor Tyrosine Kinase 2(NTRK2; p = 1.79 × 10−6) were also notably significant.
Manhattan plots of p-values of gene-wise association analyses of lifetime anxiety disorder in the UK Biobank. In the Manhattan plots, the threshold for gene-wide significance (p < 3 x 10−6) is indicated by the red line, while the blue line indicates the threshold for suggestive significance (p < 1 x 10−5)
Polygenic prediction of anxiety
Across all ten iterations, a higher polygenic score was significantly associated with increased likelihood of self-reporting Lifetime Anxiety Disorder, explaining on average 0.5% of the variance in outcome on the liability scale across all samples. The relatively low variance explained by the polygenic association with Lifetime Anxiety Disorder relative to our high estimates of h2SNP is consistent with what would be expected for a polygenic trait [41]. See Supplementary Tables 9–12 for detailed results.
Testing the dimensionality of anxiety
Severe current GAD symptoms showed genetic correlations of 0.76 (SE = 0.08) and 0.98 (SE = 0.08) with mild and moderate symptoms respectively. Genetic correlation between mild and moderate symptoms was 0.82 (SE = 0.05). See Supplementary Tables 13 and 14 for SNP heritability estimates of these phenotypes and genetic correlations with other anxiety phenotypes respectively.
Replication and meta-analyses
Replication of significant SNPs
Table 1 shows the p-value and direction of effect for all SNPs found to be genome-wide significant in our core Lifetime Anxiety Disorder analysis, and in the analysis from all four external samples. The locus at 9q21.33 (Index SNP rs10959883) is formally replicated (i.e. significant correcting for 20 independent tests, p < 5.25 × 10−3) in both the neuroticism and MDD samples and has an effect in the same direction in all four samples. The locus annotated to the NTRK2 (index SNP rs1187280) gene has the same direction of effect in iPSYCH Lifetime Anxiety Disorder and in both Neuroticism and PGC MDD. The locus annotated to the TMEM106B (index SNP rs3807866) formally replicates in Neuroticism, and the same direction of effect in iPSYCH Lifetime Anxiety Disorder, and Depression. The locus annotated to the MYH15 gene (index SNP rs4855559) shows the same direction of effect in the two Lifetime Anxiety Disorder samples, but not in the Neuroticism or Depression analyses.
Meta-analysis
Next, we meta-analysed the core UK biobank Lifetime Anxiety Disorder analysis with the two external Lifetime Anxiety Disorder samples (ANGST and iPSYCH), a combined sample of 114 019 (31 977 cases and 82 114 controls). See Fig. 4 for Manhattan and Q-Q plots for this analysis. Two loci were genome-wide significant. The region on chromosome 9q23 (rs10959577) is likely part of the same region associated with Lifetime Anxiety Disorder and Current Anxiety Symptoms in the UK Biobank alone (the lead SNPs are ~70 kb apart with an LD r2 = 0.27 [40]). The second region on chromosome 5 (rs7723509) falls in an intergenic region. See Supplementary Fig. 8 for Manhattan and Q-Q plots for leave-one-out meta-analyses of the UK Biobank, ANGST and iPSYCH samples.
Manhattan and Q-Q plots of p-values of single nucleotide polymorphism (SNP) based association analyses of combined sample meta-analysis across three lifetime anxiety disorder cohorts (UK Biobank, ANGST and iPSYCH). In the Manhattan plots, the threshold for genome-wide significance (p < 5 x 10−8) is indicated by the red line, while the blue line indicates the threshold for suggestive significance (p < 1 x 10−5)
Discussion
To our knowledge this is the largest GWAS on anxiety conducted, and our core analysis identified five genome-wide significant loci. The first locus is an intergenic region on chromosome 9 previously associated with neuroticism [42, 43] and depression [8]. The second novel locus on chromosome 9 was in NTRK2, a Brain Derived Neurotrophic Factor (BDNF) receptor. Together NTRK2 and BDNF regulate both short-term synaptic functions and long-term potentiation of brain synapses (OMIM *600456; https://www.omim.org/entry/600456). Given this key role in brain function, NTRK2 has been widely investigated in a range of neuropsychiatric traits and disorders [44,45,46,47,48,49,50,51,52]. The third locus, on chromosome 7, is in Transmembrane Protein 106B, a gene associated with lysosomal enlargement and cell toxicity implicated in depression [8, 53] and coronary artery disease [54]. A further locus on chromosome 5 is in an intergenic region. Finally, a locus on chromosome 3 located in Myosin Heavy Chain 15, a gene coding for a class of motor proteins that is highly expressed in the brain in humans. Our GWAS is considerably better powered than previous anxiety GWAS, including 25 453 cases compared to 3 695 cases [10], in addition to having a homogenous study design for all UK Biobank analyses. While we did not see evidence for replication in two smaller anxiety cohorts, two of the five loci we identify had significant evidence for formal replication in larger independent samples for the related phenotypes of trait neuroticism and major depressive disorder. Phenotypic and genetic correlations of these phenotypes with anxiety are substantial, thus while these cannot be called pure replications of anxiety, the replication of significant loci in these highly related and well powered samples suggests that these are robust associations.
Gene level analyses of Lifetime Anxiety Disorder confirmed the association with Neurotrophic Receptor Tyrosine Kinase 2 (NTRK2; p = 1.79 × 10−6) and Transmembrane Protein 106B (TMEM106B; p = 6.28 × 10−10) and enabled the identification of several additional protein coding genes associated with anxiety disorder. Specifically Phospholipase C Gamma 1 (PLCG1; p = 2.96 × 10−7), Ring Finger and WD Repeat Domain 2 (RFWD2, p = 4.60 × 10−7), Zinc Fingers and Homeoboxes 3 (ZHX3 p = 5.21 × 10−7), Glutamate Decarboxylase 2 (GAD2; p = 7.00 × 10−7), and Lipin 3 (LPIN3; p = 2.54 × 10−6). NTRK2 has been implicated in several neuropsychiatric traits and disorders including emotional arousal [44], autism [45], suicide [46,47,48], Alzheimer’s disease [49], alcohol dependence [52], and treatment response [50, 51], although many of these are candidate gene studies and are thus less likely to represent robust findings. TMEM106B was recently found to be associated with both a broad depression phenotype and probable major depression in the UK Biobank [53], and with major depression in the Psychiatric Genomic Consortium meta-analysis [8]. This is unsurprising given the phenotypic and genetic overlap between anxiety and depression [12]. TMEM106B is widely expressed throughout normal human cell types and tissue, including the fetal brain, and adult frontal cortex. A recent investigation demonstrated that a noncoding variant on chromosome 7 (rs1990620) interacts with TMEM106B via long-range chromatin looping interactions, a physical folding of the chromosome that brings distal regions of the genome into close proximity, mediating increased TMEM106B expression. This is strongly associated with lysosome enlargement and cell toxicity [55]. Conditional analyses indicate that the locus associated with Lifetime Anxiety Disorder in the UK Biobank is the same locus which results in increased expression of TMEM106B, suggesting a plausible role for lysosome enlargement and cell toxicity in anxiety (see Supplementary Figs. 9–10). This is in line with research showing a role for neurotoxicity in fear learning in mice [56].
Glutamate decarboxylase 2 (GAD2) encodes for an enzyme that synthesises gamma-aminobutyric acid (GABA) from L-glutamic acid. GABA is the principal inhibitory neurotransmitter in the mammalian central nervous system and has well-established associations anxiety. Specifically, variation in glutamate decarboxylase genes have been associated with internalising disorders [57]. Furthermore, GABAergic deficits are observed in patients with, and animal models of, anxiety and depression [58]. Finally, benzodiazepines, a class of pharmacological agent with strong anxiolytic effects, act through binding to a specific GABA receptor, facilitating GABAergic inhibitory effects [59].
Several findings relating to the polygenicity of anxiety are notable. First, SNP-heritability (h2SNP) estimates for Lifetime Anxiety Disorder (h2SNP = 25.7%) and Current Anxiety Symptoms (h2SNP = 30.8%) in UK Biobank were especially large. Typically h2SNP estimates are less than half twin h2 (40–60%) [4, 5]. Heritability estimates from twin studies take into account genetic influence of both common and rare genetic variants, whereas h2SNP only takes into account the additive effect of common variants. Our results suggest that a large proportion of heritable variance in anxiety is attributable to common genetic variants, with the effects spread over many hundreds or thousands of loci [60]. Our h2SNP estimates are also much higher than those derived from previous studies of anxiety [9, 10], which may reflect the homogeneity of the UK Biobank sample in terms of assessment, sampling, and genotyping.
We also explored genetic overlap between anxiety phenotypes. These suggest that common genetic effects on anxiety are shared across different levels of severity, as well as across varying samples and definitions, and with both neuroticism and depression. We note that the phenotypic overlap between anxiety and depression is also high, with 53% of our sample reporting a comorbid diagnosis of depression. This is in line with epidemiological observations in primary care settings [61]. See Supplementary Table 15 for genetic correlations between lifetime anxiety with and without self-reported or probable depression cases and our primary lifetime anxiety phenotype (rG = 0.98, SE = 0.01 and rG = 0.92, SE = 0.04, respectively). The high shared genetic risk and phenotypic overlap with depression suggests substantial genetic pleiotropy that may make it challenging to identify anxiety-specific risk variants.
Polygenic scores derived from primary phenotype predicted ~0.5% of the variance in Lifetime Anxiety Disorder case control status. We note that the moderate to high h2SNP for the UK Biobank anxiety phenotypes suggests a PRS for anxiety will improve as larger discovery GWAS samples are used for the creation of PRS, increasing the signal to noise ratio in the construction of polygenic scores.
Genetic overlap was detected between anxiety and a broad range of traits. Significant positive genetic correlations were detected between Lifetime Anxiety Disorder and several psychiatric phenotypes (schizophrenia, bipolar disorder, insomnia, ADHD and cross-disorder analyses), indicating a degree of shared genetic risk between psychiatric traits more generally. Interestingly, given some epidemiological evidence for an association between these phenotypes [62], a significant genetic correlation was observed between anxiety and coronary artery disease. This may reflect the previously known epidemiological [63], and genomic overlap between major depressive disorder and coronary artery disease [8]. We interpret these findings in the context of several limitations. There is substantial sample overlap within the two UK Biobank anxiety phenotypes and between UK Biobank anxiety and several of the other phenotypes which also use the UK Biobank sample. However, LD score regression is robust to sample overlap [64], so genetic correlations are unlikely to be biased by this.
Although the intergenic genome-wide significant loci on chromosome 9 previously associated with neuroticism [42, 43] and depression [8] replicates in the combined Lifetime Anxiety Disorder meta-analysis, statistical significance is attenuated. Nonetheless, this provides good evidence for a role of this region in lifetime anxiety. Not all genome-wide significant SNPs observed in the individual cohorts survive the combined sample Lifetime Anxiety Disorder meta-analysis. This lack of replication may be due to a winner’s curse effect [65], the relatively modest power gain from the additional samples, or phenotypic and genetic heterogeneity across the studies. As noted above, the UK Biobank is a highly homogenous group of individuals, all based within the UK with the same sampling and phenotyping instrument used. The iPSYCH sample is also highly homogenous group of individuals, all based within Denmark. Conversely, the ANGST study is comprised of many smaller samples, using different GWAS arrays and varied phenotyping from across multiple continents. Thus, it is possible that sample heterogeneity contributes to the non-replication of some of our GWAS significant loci. Given that our analyses suggest anxiety is a highly polygenic trait, driven by small effects across many SNPs, it is likely that it remains somewhat underpowered, despite the sample being substantially larger than any prior genome-wide study of anxiety to date.
Although homogeneity within UK Biobank may be a significant factor in the success of our analyses and the high heritabilities found, this sample homogeneity is itself a limitation, as our analyses were thus limited across all three samples to individuals of European ancestry [66]. Furthermore, the UK Biobank is not representative for either age or socioeconomic status [67]. Another limitation is that the Lifetime Anxiety Disorder phenotype relies both on retrospective recall by the subject and accurate diagnosis by an unknown clinician, each of which are likely to contain error. However, there are high genetic correlations between this category and every other anxiety phenotype assessed, suggesting the category of Lifetime Anxiety Disorder has utility. Finally, we have not excluded cases who may have comorbid PTSD, which might be viewed as a limitation.
This study used the largest currently available dataset to investigate the role of common genetic variation in anxiety. Although no sufficiently powered studies are available to enable strict statistical replication [68], the study provides the first well powered characterisation of the common genetic architecture of anxiety. Analyses demonstrate that a large proportion of the broad heritability of severe and pathological anxiety is attributable to common genetic variants, and that this is shared between anxiety phenotypes and other closely related psychiatric disorders. Several genomic regions, which include loci with known associations with internalising disorders were found to be significantly associated with anxiety in these analyses. These findings are likely to be of interest for targeted characterisation of the underlying biology of anxiety.
References
Kessler RC, Aguilar-Gaxiola S, Alonso J, Chatterji S, Lee S, Ormel J, et al. The global burden of mental disorders: an update from the WHO World Mental Health (WMH) surveys. Epidemiol Psichiatr Soc. 2009;18:23–33.
Wittchen H-U, Jacobi F. Size and burden of mental disorders in Europe–a critical review and appraisal of 27 studies. Eur Neuropsychopharmacol. 2005;15:357–76.
GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–858.
Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–9.
Craske MG, Stein MB, Eley TC, Milad MR, Holmes A, Rapee RM, et al. Anxiety disorders. Nat Rev Dis Prim. 2017;3:17024.
Smoller JW. The genetics of stress-related disorders: PTSD, depression, and anxiety disorders. Neuropsychopharmacology. 2016;41:297–319.
Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–9.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
Dunn EC, Sofer T, Gallo LC, Gogarten SM, Kerr KF, Chen C-Y, et al. Genome-wide association study of generalized anxiety symptoms in the Hispanic Community Health Study/Study of Latinos. Am J Med Genet B, Neuropsychiatr Genet. 2017;174:132–43.
Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG, et al. Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry. 2016;21:1391–9.
Craske MG, Rauch SL, Ursano R, Prenoveau J, Pine DS, Zinbarg RE. What is an anxiety disorder? Depress Anxiety. 2009;26:1066–85.
Waszczuk MA, Zavos HMS, Gregory AM, Eley TC. The phenotypic and genetic structure of depression and anxiety disorder symptoms in childhood, adolescence, and young adulthood. JAMA Psychiatry. 2014;71:905–16.
Roberson-Nay R, Eaves LJ, Hettema JM, Kendler KS, Silberg JL. Childhood separation anxiety disorder and adult onset panic attacks share a common genetic diathesis. Depress Anxiety. 2012;29:320–7.
Hettema JM, Prescott CA, Myers JM, Neale MC, Kendler KS. The structure of genetic and environmental risk factors for anxiety disorders in men and women. Arch Gen Psychiatry. 2005;62:182–9.
Tambs K, Czajkowsky N, Røysamb E, Neale MC, Reichborn-Kjennerud T, Aggen SH, et al. Structure of genetic and environmental risk factors for dimensional representations of DSM-IV anxiety disorders. Br J Psychiatry. 2009;195:301–7.
Mackintosh M-A, Gatz M, Wetherell JL, Pedersen NL. A twin study of lifetime Generalized Anxiety Disorder (GAD) in older adults: genetic and environmental influences shared by neuroticism and GAD. Twin Res Hum Genet. 2006;9:30–7.
Hettema JM, Prescott CA, Kendler KS. Genetic and environmental sources of covariation between generalized anxiety disorder and neuroticism. Am J Psychiatry. 2004;161:1581–7.
Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006;163:857–64.
Bienvenu OJ, Hettema JM, Neale MC, Prescott CA, Kendler KS. Low extraversion and high neuroticism as indices of genetic and environmental risk for social phobia, agoraphobia, and animal phobia. Am J Psychiatry. 2007;164:1714–21.
Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–8.
McGrath LM, Weill S, Robinson EB, Macrae R, Smoller JW. Bringing a developmental perspective to anxiety genetics. Dev Psychopathol. 2012;24:1179–93.
Davies MN, Verdi S, Burri A, Trzaskowski M, Lee M, Hettema JM, et al. Generalised anxiety disorder—a twin study of genetic architecture, genome-wide association and differential gene expression. PLoS One. 2015;10:e0134865.
Pedersen CB, Bybjerg-Grauholm J, Pedersen MG, Grove J, Agerbo E, Bækvad-Hansen M, et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry. 2018;23:6–14.
Davis KAS, Coleman JRI, Adams M, Allen N, Breen G, Cullen B, et al. Mental health in UK Biobank: development, implementation and results from an online questionnaire completed by 157 366 participants. BJPsych Open. 2018;4:83–90.
Kessler RC, Andrews G, Mroczek D, Ustun B, Wittchen H-U. The World Health Organization Composite International Diagnostic Interview short-form (CIDI-SF). Int J Methods Psychiatr Res. 1998;7:171–85.
McLean CP, Asnaani A, Litz BT, Hofmann SG. Gender differences in anxiety disorders: prevalence, course of illness, comorbidity and burden of illness. J Psychiatr Res. 2011;45:1027–35.
Spitzer RL, Kroenke K, Williams JBW, Löwe B. A brief measure for assessing generalized anxiety disorder: the GAD-7. Arch Intern Med. 2006;166:1092–7.
Meier S, Trontti K, Als TD, Laine M, Pedersen MG, Bybjerg-Grauholm J et al. Genome-wide Association Study of Anxiety and Stress-related Disorders in the iPSYCH Cohort. BioRxiv 2018. https://doi.org/10.1101/263855.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K et al. Genome-wide genetic data on ~500,000 UK Biobank participants. BioRxiv 2017. https://doi.org/10.1101/166298.
R Foundation for Statistical Computing RCT. R: A Language and Environment for Statistical Computing. Vienna; 2017.
Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–90.
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
Rapid GWAS of thousands of phenotypes for 337,000 samples in the UK Biobank—Neale lab. http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank. Accessed 18 Jul 2019.
Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–9.
Details and considerations of the UK Biobank GWAS — Neale lab. http://www.nealelab.is/blog/2017/9/11/details-and-considerations-of-the-uk-biobank-gwas. Accessed 18 Jul 2019.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1.
Evangelou E, Ioannidis JPA. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013;14:379–89.
Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–7.
International Schizophrenia Consortium, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52.
Okbay A, Baselmans BML, De Neve J-E, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–33.
Smith DJ, Escott-Price V, Davies G, Bailey MES, Colodro-Conde L, Ward J, et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatry. 2016;21:749–57.
Spalek K, Coynel D, Freytag V, Hartmann F, Heck A, Milnik A, et al. A common NTRK2 variant is associated with emotional arousal and brain white-matter integrity in healthy young subjects. Transl Psychiatry. 2016;6:e758.
Correia CT, Coutinho AM, Sequeira AF, Sousa IG, Lourenço Venda L, Almeida JP, et al. Increased BDNF levels and NTRK2 gene association suggest a disruption of BDNF/TrkB signaling in autism. Genes Brain Behav. 2010;9:841–8.
Kohli MA, Salyakina D, Pfennig A, Lucae S, Horstmann S, Menke A, et al. Association of genetic variants in the neurotrophic receptor-encoding gene NTRK2 and a lifetime history of suicide attempts in depressed patients. Arch Gen Psychiatry. 2010;67:348–59.
Murphy TM, Ryan M, Foster T, Kelly C, McClelland R, O’Grady J, et al. Risk and protective genetic variants in suicidal behaviour: association with SLC1A2, SLC1A3, 5-HTR1B &NTRK2 polymorphisms. Behav Brain Funct. 2011;7:22.
Torres CM, Siebert M, Bock H, Mota SM, Castan JU, Scornavacca F, et al. Tyrosine receptor kinase B gene variants (NTRK2 variants) are associated with depressive disorders in temporal lobe epilepsy. Epilepsy Behav. 2017;71:65–72.
Chen Z-Y, Jing D, Bath KG, Ieraci A, Khan T, Siao C-J, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–3.
Dong C, Wong ML, Licinio J. Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans. Mol Psychiatry. 2009;14:1105–18.
Wang Z, Fan J, Gao K, Li Z, Yi Z, Wang L, et al. Neurotrophic tyrosine kinase receptor type 2 (NTRK2) gene associated with treatment response to mood stabilizers in patients with bipolar I disorder. J Mol Neurosci. 2013;50:305–10.
Xu K, Anderson TR, Neyer KM, Lamparella N, Jenkins G, Zhou Z, et al. Nucleotide sequence variation within the human tyrosine kinase B neurotrophin receptor gene: association with antisocial alcohol dependence. Pharmacogenomics J. 2007;7:368–79.
Howard DM, Adams MJ, Shirali M, Clarke T-K, Marioni RE, Davies G et al. Genome-wide association study of depression phenotypes in UK Biobank (n = 322,580) identifies the enrichment of variants in excitatory synaptic pathways. BioRxiv 2017. https://doi.org/10.1101/168732.
van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–43.
Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald AL, et al. A dementia-associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am J Hum Genet. 2017;101:643–63.
Jing H, Hao Y, Bi Q, Zhang J, Yang P. Intra-amygdala microinjection of TNF-α impairs the auditory fear conditioning of rats via glutamate toxicity. Neurosci Res. 2015;91:34–40.
Hettema JM, An SS, Neale MC, Bukszar J, van den OordEJCG, Kendler KS, et al. Association between glutamic acid decarboxylase genes and anxiety disorders, major depression, and neuroticism. Mol Psychiatry. 2006;11:752–62.
Möhler H. The GABA system in anxiety and depression and its therapeutic potential. Neuropharmacology. 2012;62:42–53.
Farach FJ, Pruitt LD, Jun JJ, Jerud AB, Zoellner LA, Roy-Byrne PP. Pharmacological treatment of anxiety disorders: current treatments and future directions. J Anxiety Disord. 2012;26:833–43.
Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, et al. Psychiatric genomics: an update and an agenda. Am J Psychiatry. 2018;175:15–27.
Hirschfeld RMA. The comorbidity of major depression and anxiety disorders: recognition and management in primary care. Prim Care Companion J Clin Psychiatry. 2001;3:244–54.
Cohen BE, Edmondson D, Kronish IM. State of the art review: depression, stress, anxiety, and cardiovascular disease. Am J Hypertens. 2015;28:1295–302.
Carney RM, Freedland KE. Depression and coronary heart disease. Nat Rev Cardiol. 2017;14:145–55.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Kraft P. Curses–winner’s and otherwise–in genetic epidemiology. Epidemiology. 2008;19:649–51. discussion 657.
Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–49.
Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–34.
Huffman JE. Examining the current standards for genetic discovery and replication in the era of mega-biobanks. Nat Commun. 2018;9:5054.
Acknowledgements
This research has been conducted using the UK Biobank Resource, under application 16577. This study represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. High performance computing facilities were funded with capital equipment grants from the GSTT Charity (TR130505) and Maudsley Charity (980). The research reported in this publication was supported by the National Institute of Mental Health of the US National Institutes of Health under Award Number U01 MH109514. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. CK currently receives salary support from National Institute for Health Research (NIHR) and has previously received salary support from the Novo Nordisk UK Research Foundation, NIHR Biomedical Research Centre for Mental Health at South London and from the Maudsley National Health Service (NHS) Foundation Trust in the past. CR is supported by a grant from Fondation Peters to TE and GB. JH is supported by the National Institutes of Health grant R01 MH113665. The iPSYCH team acknowledges funding from the Lundbeck Foundation (grant no R102-A9118 and R155-2014-1724), the Novo Nordisk Foundation for supporting the Danish National Biobank resource, and grants from Aarhus and Copenhagen Universities and University Hospitals, including support to the iSEQ Center, the GenomeDK HPC facility, and the CIRRAU Center. K.L.P acknowledges funding from the Alexander von Humboldt Foundation and the Medical Research Council UK.
Author information
Authors and Affiliations
Contributions
KP, TE, MH, KKN and GB conceived the study. KP, JC, SMM, CR, HG and SWC performed statistical analyses. CH, CK, HG, JC and KP performed phenotype and data QC for the UKBB samples. MM supervised the pre and post GWAS analysis pipeline for the iPSYCH sample. OM, MN, MBH, JBG, PBM, TW, DMH and ADB provided and processed samples for the iPSYCH sample. KP, JC, TE, GB wrote the paper. MH, KD, JH, JD, AM, MM gave advice and feedback at several stages of data generation and paper writing. All authors reviewed the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Purves, K.L., Coleman, J.R.I., Meier, S.M. et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry 25, 3292–3303 (2020). https://doi.org/10.1038/s41380-019-0559-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0559-1
- Springer Nature Limited
This article is cited by
-
The genetic architecture of youth anxiety: a study protocol
BMC Psychiatry (2024)
-
The structure of psychiatric comorbidity without selection and assortative mating
Translational Psychiatry (2024)
-
Estimating the impact of transmitted and non-transmitted psychiatric and neurodevelopmental polygenic scores on youth emotional problems
Molecular Psychiatry (2024)
-
Examining intergenerational risk factors for conduct problems using polygenic scores in the Norwegian Mother, Father and Child Cohort Study
Molecular Psychiatry (2024)
-
Gene × environment effects and mediation involving adverse childhood events, mood and anxiety disorders, and substance dependence
Nature Human Behaviour (2024)