Abstract
Extramedullary (EM) colonization is a rare complication of acute myeloid leukemia (AML), occurring in about 10% of patients, but the processes underlying tissue invasion are not entirely characterized. Through the application of RNAseq technology, we examined the transcriptome profile of 13 AMLs, 9 of whom presented an EM localization. Our analysis revealed significant deregulation within the extracellular matrix (ECM)-receptor interaction and focal-adhesion pathways, specifically in the EM sites. The transcription factor TWIST1, which is known to impact on cancer invasion by dysregulating epithelial–mesenchymal-transition (EMT) processes, was significantly upregulated in EM-AML. To test the functional impact of TWIST1 overexpression, we treated OCI-AML3s with TWIST1-siRNA or metformin, a drug known to inhibit tumor progression in cancer models. After 48 h, we showed downregulation of TWIST1, and of the EMT-related genes FN1 and SNAI2. This was associated with significant impairment of migration and invasion processes by Boyden chamber assays. Our study shed light on the molecular mechanisms associated with EM tissue invasion in AML, and on the ability of metformin to interfere with key players of this process. TWIST1 may configure as candidate marker of EM-AML progression, and inhibition of EMT-pathways may represent an innovative therapeutic intervention to prevent or treat this complication.

Similar content being viewed by others
Introduction
“Myeloid sarcomas” (MS) or otherwise defined extramedullary acute myeloid leukemias (EM-AML), indicate extramedullary masses of myeloid blasts, altering the tissue architecture [1,2,3].
MS may be present at the time of diagnosis or relapse in about 10–15% of AML, with or without simultaneous bone marrow (BM) involvement, which however inevitably occurs during the disease course in the majority of cases [4]. Recent data on large patient series have confirmed that MS is associated with dismal prognosis, with significantly lower probability of achieving complete remission, and reduced survival, as compared to the corresponding AML subtypes [3]. Most commonly affected tissues include soft/connective tissue (31–35%), skin/breast (11–46%), and gastrointestinal system (10–19%), with less frequent involvement of reproductive organs (1–10%), bone (5–16%), head and neck (6–14%), and central nervous system (CNS, 4–11%) [5,6,7,8]. The incidence of MS increases in the allogeneic stem cell transplantation (HSCT) setting, accounting for 10–25% of EM relapses, occurring at median intervals of 6–12 months after HSCT [9, 10].
Different attempts have been made to predict the risk of EM-AML localizations, but they have been unsuccessful to date. A monoblastic/myelomonocytic morphology is frequent in EM-AML, and the immunophenotype is usually positive for myeloid and monocytic markers, including CD11b, CD56, CD33, CD68, lysozyme, and the more immature CD117 and CD34, CD61, glycophorin and CD4 [11]. Clonal cytogenetic abnormalities have been observed in 54–70% of cases, but there is no clear correlation between the AML subtype and the development of EM disease [12]. The mutational topography also does not show a specific signature, predictive of synchronous or metachronous EM-AML, with frequencies of NPM1, NRAS, IDH2, DNMT3A and TET2 mutations overall similar to that of de novo AML [13,14,15].
In recent years, studies on EM-AML have shown deregulation in leukemic blasts of epithelial–mesenchymal transition (EMT) pathways, mirroring the metastasis formation process in solid tumors [16]. EMT is in turn controlled by several signal transduction pathways, leading to the activation of the master regulators TWIST1, SNAIL and ZEB [17]. These key EMT transcription factors (EMT-TF) may trigger cellular changes associated with invasiveness and cancer cell dissemination [18].
Given the paucity of data on the cell-specific features leading to EM localization of AML, and taking advantage of cases collected through a multi-institutional effort, we analyzed by RNA-Seq the expression profiles of EM localizations of AML as compared to that of paired BM blasts, isolated at the time of initial diagnosis or of relapse. The final goal was to identify predictive factors of EM-migration of AML cells, and possible druggable targets to be used to prevent or treat this prognostically adverse leukemic manifestation.
Materials And Methods
Study cohort
During the years 2019–2022, we collected samples from 16 AML patients, 12 of whom developed an extramedullary localization (7 at diagnosis and 5 at relapse). There were 6 males and 6 females, of a median age 63 years (range 40–79). Bone marrow mononuclear cells (BM-MNC) of 4 patients with AML, who did not develop an EM localization, and of 3 healthy donors (HDs) were included in the study as controls (Supplementary Fig. 1). According to the declaration of Helsinki, all patients gave informed consent for the study, that was approved by the Institutional Review Board of the Policlinico Tor Vergata of Rome, as part of the Mynerva program.
Clinical and biological features of the AML patient cohort are reported in Table 1. Skin was the secondary AML site in 6 cases (50%), while lymph nodes and bone localizations were present in 4 and 3 patients (33 and 25%, respectively). Paired samples of BM and sarcoma tissues at the time of diagnosis and/or relapse were available for 7 cases. The 4 AML samples used as controls had a monoblastic morphology, and did not develop a secondary localization at a median follow-up of 29.5 months from initial diagnosis (range 22–35 months).
Routine morphologic, immunophenotypic and genetic analysis were carried out in all cases at presentation or relapse. Conventional karyotyping was performed on BM aspirates after short-term culture and analyzed after G-banding. For molecular analysis, MNCs were isolated from patients BM by Ficoll-Hypaque gradient centrifugation using Lympholyte®- H (Cederlane, ON, Canada) and DNA was extracted using the column-based QIAamp DNA Mini Kit (Qiagen), following the manufacturer’s instructions.
The mutational landscape was studied by targeted next-generation sequencing (NGS) of 30 myeloid genes, using the Myeloid-Solution-Panel by SOPHiA GENETICS (SOPHiA GENETICS, Saint Sulpice, Swiss) and the sequencing platform MiniSeq® Illumina (Illumina, San Diego, CA, USA). FASTQ sequencing files were uploaded to the SOPHiA DDM® platform (coverage > 1000X), that enables the detection, annotation and classification of genetic alterations (SNVs, Single Nucleotide Variants and insertions/deletions) through artificial intelligence. Only variants with a variant allele frequency (VAF) greater than 1% and alterations identified as highly or probably pathogenic were considered, and analysis was performed according to standard procedures.
Diagnosis of EM-AML was confirmed by immunohistochemistry (IHC) and histopathological analysis of biopsy specimens. Immunohistochemical staining was performed using an automated slide preparation system (Benchmark XT, Ventana, Tuscon, AZ), a Ventana Enhanced DAB Detection Kit (Ventana, Tucson, AZ), and commercially available prediluted monoclonal antibodies: CD163 (NeoMarkers), CD4 (Biocare Medical), myeloperoxidase, lysozyme, CD3, CD4, CD8, CD15, CD20, CD34, CD43, CD56, CD68, CD79a, CD117, Factor VIII (FVIII), and glycophorin A (all Ventana, Tucson, AZ) (Supplementary Fig. 2 shows a myeloid sarcoma immunohistochemistry, derived from UPN1).
RNA isolation and RNA-Seq
Total RNA was extracted using TRIzolTM Reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) from BM-MNC samples of AML patients, and from the OCI-AML3 cell line. RNA isolation from paraffin-embedded tissue was performed using the RecoverAllTM kit (RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE; Thermo Fisher Scientific, Waltham, MA, USA). At least 50 ng of RNA was reverse transcribed with the SuperScript™ IV VILO™ Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). The reaction mix was incubated in a thermal cycler at the following conditions: 25 °C for 10 min, 50 °C for 10 min, 85 °C for 5 min and holding at 4 °C. cDNA corresponding to 50 ng RNA was used for qRT-PCR.
For RNA-Seq, RNA was quantified by spectrophotometry and evaluated for integrity using the Agilent 4200 TapeStation system. In addition to the RNA Integrity Number (RIN) value, the Distribution Value 200 (DV200), corresponding to the percentage of fragments with dimensions >200 bp, was also evaluated for RNA isolated from FFPE (formalin-fixed, paraffin-embedded) tissues. RNA sequences with RIN >2 and DV200 >25% were selected for transcriptomic analysis.
The library preparation was carried out using the Takara Bio SMARTer Stranded Total RNA-Seq Kit v2-Pico Input Mammalian, with an input of 8–10 ng RNA isolated from fresh blood samples or of 20–50 ng RNA isolated from FFPE samples. RNA libraries were quantified by fluorimetry (Qubit™ dsDNA HS Assay Kit), and analyzed for quality check and final quantification by the Agilent TapeStation 4200 with the High Sensitivity D1000 ScreenTape Assay kit system. The overall quality and the presence of adapter sequences was checked by the FastQC program (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All paired reads were then aligned to the reference human genome (GRCh38/hg38) using the aligner splice-aware STAR v2.5.0c (26), sorted and indexed by Samtools (27).
The reads for each gene were calculated using the quantMode geneCounts option and then elaborated by the Bioconductor DESeq2 v1.30 (28).
Differential gene expression and functional enrichment analysis
Expression data derived from DESeq2 package was used to perform differential gene expression analysis. Statistical significance threshold for differentially expressed genes (DEGs) was set using a Benjamini–Hochberg adjusted P-value < 0.1 and |log2(Fold Change)| ≥ 2 to identify upregulated or downregulated genes. DEGs were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (v. 2021, https://david.ncifcrf.gov) [19, 20], using the Kyoto Encyclopedia of Gene and Genome pathways database (KEGG pathway enrichment analysis). In detail, a Gene Functional Annotation Chart option was applied, which collects all significant terms of the list as inputs to visualize KEGG pathways. Statistically significant pathways were selected using a Benjamini–Hochberg adjusted P-value < 0.01 as cut-off criteria.
A second computational analysis was carried out using gene set enrichment analysis (GSEA), using the GSEA software v.3.0, and a gene-based permutation test with 1000 replications. Significantly enriched pathways were defined according to P-value of < 0.01 and false discovery rate (FDR) < 0.25.
To control for the different RNA sources [formalin-fixed, paraffin-embedded (FFPE) EM localization vs BM-MNC], non-differentially expressed genes were analyzed by DESeq2, and by Pearson correlation. The top 10% most expressed genes were then used as input for pathway enrichment analysis. Pathways with Benjamini-Hochberg-corrected P-value < 0.1 were considered significantly enriched.
Cell cultures and RNA interference
The human leukemia cell line OCI-AML3 was obtained by the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ, Braunschweig, Germany) and cultured in RPMI 1640 Medium (Gibco Roswell Park Memorial Institute, Sigma-Aldrich, St. Louis, MO, USA), supplemented with 20% FBS (fetal bovine serum), 1% l-glutamine and 1% penicillin/streptomycin (Lonza Sales, Basel, Switzerland). Cells were then plated in sterile flasks and maintained in culture in a thermostat incubator, at 37 °C under 5% CO2 atmosphere.
A siRNA oligonucleotide targeting the human TWIST1 cDNA sequence was purchased from Invitrogen (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA, siRNA s14524 sense 5′−3′: GCACCAUCCUCACACCUCUtt, siRNA s14524 antisense 5′−3′: AGAGGUGUGAGGAUGGUGCcg). A siRNA sequence not matching any mRNA present within the cells was used as negative control. Forty-eight hours after transfection, downregulation of RNA expression was confirmed by qRT-PCR.
Metformin treatment
OCI-AML3 cells were seeded into 6-well plates at a density of 1.5 × 106 cells per well in 3 mL complete medium, and metformin (Selleck Chemicals, Houston, TX, USA), dissolved in H2O, was added at increasing concentrations (10, 20 and 40 mM) for 24 and 48 h. After metformin treatment, cell viability was studied by Trypan-Blue dye exclusion assay (0.5% w/v) (Sigma-Aldrich, St. Louis, MO, USA) by a phase contrast microscope. Cell suspension was then collected for RNA extraction, using TRIzolTM Reagent (Invitrogen, Waltham, MA, USA). qRT-PCR was performed using SYBR Green chemistry (iTaq™ Universal SYBR® Green Supermix, BIO-RAD, Hercules, California, USA). Gene expression was calculated using the 2−ΔΔCt method, and normalized to B2M levels. Primers’ sequences and qRT-PCR conditions are reported in Supplementary Table 1.
Migration and invasion assays
A 24-well plate, 8-μm pore size Costar transwell plate (Corning, Cambridge, MA, USA) was used to test the migration and invasion ability of OCI-AML3. For the invasion assay, the chamber was coated with GeltrexTM matrix (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), to mimic the extracellular matrix. A total of 2 × 105 cells pre-treated with siRNA or metformin and the corresponding controls were resuspended in 200 μL of RPMI 1640 medium, and seeded in the upper chamber of the transwell, while the lower well was filled with 600 μL RPMI 1640 medium containing 20% FBS. After incubation for 4 h at 37 °C, transmigrated cells were counted by flow cytometry for 5 min, after labeling with an anti-CD45 human monoclonal antibody.
Statistical analysis
Statistical analysis was carried out using the GraphPad Prism software v8.0 (GraphPad Software, San Diego, CA). Specifically, the non-parametric Kruskal–Wallis test was applied to compare gene expression levels by RT-PCR in BM and EM samples. Wilcoxon matched-pairs test was used for the evaluation of expression levels of genes of interest after treatment with siRNA or metformin. Data are presented as the median with range of the experiments.
Results
Mutational landscape of EM-AML
The mutational landscape of AMLs with EM localizations, at diagnosis or relapse, was analyzed by targeted-NGS on DNA extracted from BM-MNC samples (n = 12) (Supplementary Fig. 1). We identified the presence of a median of 4 mutations per patient (range 0–7) (Fig. 1), with no detectable mutations in the BM samples of 2 patients who had less than 5% BM blasts. As shown in Fig. 1, the most commonly mutated genes were SRSF2 (41% of cases), RUNX1 and NPM1 (33% of cases each).
Transcriptome profile of EM-AML reveals dysregulation of EMT-related pathways
The expression profile of BM and EM-AML localizations was then characterized by RNA sequencing of samples isolated from 13 patients, 9 of whom were complicated by an EM localization. BM-MNC of 4 AML patients who did not develop extramedullary disease served as AML controls (Supplementary Fig. 1).
The trascriptomic profile of diagnostic BM-MNCs of patients who developed a myeloid sarcoma at the time of diagnosis (n = 4) or relapse (n = 4) was compared to that of BM-MNCs of 4 AML Ctrl. Comparing these BM samples, no evident cluster separation could be identified by Principal Component Analysis (PCA) (Fig. 2A).
A PCA of control AML patients (AML Ctrl, n = 4) versus BM at diagnosis of patients who presented EM-AML at diagnosis (n = 4) or at relapse (n = 4). B PCA of expression profiles of AML Ctrl and BM-MNC of patients who experienced both BM and EM relapse (n = 5). C PCA of paired samples of extramedullary localizations as compared to BM (n = 7). The blue dots show the AML Ctrl patients, the red dots show the patients who developed the extramedullary localization. The graphic legends are Created with BioRender.com.
We then compared the expression profiles of AML Ctrl to that of BM-MNC of patients who experienced both BM and EM relapse (n = 5) (Fig. 2B). Differential gene expression analysis revealed deregulation of three pathways involved in blast cells adhesion to endothelial surfaces, including cytokine-cytokine receptor interaction, toll-like receptor and chemokine signaling (Supplementary Fig. 3). In particular, 22 DEGs linked to cytokine-cytokine receptor interaction, were upregulated in AML Ctrl compared to EM-AMLs. These data were validated by qRT-PCR, confirming the upregulation of 4 out of 22 selected genes (CCL3, CCL4, CCR1 and CCR2). (Supplementary Fig. 4).
We then investigated the expression profiles of paired samples from 7 patients, consisting of BM-MNCs and EM localizations at the time of diagnosis (n = 4) or relapse (n = 3). As a control for the different RNA sources, we scored non-differentially expressed genes in FFPE EM localizations versus BM-MNCs, corresponding to 9783 of 31,835 genes. The two datasets were strongly correlated (Pearson’s r = 0.751, Supplementary Fig. 5), indicating a negligible effect of FFPE preservation on global gene expression. The transcriptional regulation of granulopoiesis pathway was significantly enriched (top 10 percentiles), as expected in AML (data not shown).
Using PCA, a clear separation of the BM-MNC profiles versus secondary localizations became evident (Fig. 2C), with deep differences in the transcriptome pattern between BM-MNC and extramedullary site. In particular, we identified 3572 DEGs, of which were 2889 upregulated, and 683 downregulated in EM localizations, as showed by the volcano plot (Supplementary Fig. 6).
The functional enrichment analysis of DEGs using DAVID, showed involvement of 25 KEGG pathways for upregulated DEGs, and 11 for downregulated genes (P-value < 0.01 and FDR < 0.01) (Supplementary Fig. 7A, B). The same analysis was then repeated using the GSEA software, and resulted in abnormal regulation of 40 pathways, with enrichment of 24 pathways in EM localizations compared to BM-MNC (P-value < 0.01 and FDR < 0.25, Supplementary Fig. 7C, D). Upregulated pathways identified by DAVID and GSEA were mainly associated with those with biological activity in cancer stem cells from several types of tumors [21, 22]. On the contrary, downregulated pathways were related to impaired antitumor properties [23].
Importantly, combination of the two analysis converged on enrichment of gene networks (n = 13) involved in ECM-receptor interaction and focal adhesion, which may exert a functional impact on the migration and invasive ability of the leukemic cells (Fig. 3A). Specifically, intersection of DEGs related to ECM signaling and focal adhesion of the corresponding leading-edge gene-sets, allowed us to identify 32 common genes (Fig. 3B) upregulated in patients who experienced an EM localization (Fig. 3C), that were likely critical for the deregulation of EMT-associated pathways.
A Venn plot of the deregulated pathways identified by DAVID and GSEA in extramedullary localizations samples as compared to BM. The pathways are sorted alphabetically by pathway name. B Venn plot of the upregulated genes, linked to ECM-receptor interaction (in yellow) and Focal adhesion (in purple), in common between DAVID and GSEA. C Heatmap showing the differential gene expression of the 32 DEGs upregulated in the extramedullary localizations as compared to the respective bone marrow samples. Stronger relationships between samples are given by shorter distances in the hierarchical clustering and the upregulation by the darker green color in the heatmap. BM bone marrow, EM extramedullary.
We then studied by qRT-PCR the expression of 22 of the 32 genes upregulated in the EM localizations. Significant overexpression of 19 of 22 genes (86%) was confirmed in three paired EM and BM-MNC samples from the retrospective study cohort (UPN4, 5 and 7, P-value < 0.0001), and in an independent paired prospective study cohort (n = 3) (Fig. 4A). Median gene upregulation was 743 folds (range 48–15,489) and 59 folds (range 0–10,474), in the retrospective and prospective patient cohorts, respectively.
A Differential gene expression of EM-AML patients as compared to paired BM samples in a retrospective (n = 3) and prospective (n = 3) study cohorts. B Comparison of BM from patients with myeloid sarcoma (n = 3) and healthy donors (n = 3) for selected genes. Fold Change = 2−ΔΔCt; 2−ΔΔCt = 2−(ΔCt EM − ΔCt BM); Fold Change = 2−ΔΔCt; 2−ΔΔCt = 2−(ΔCt BM EM-AMLS − ΔCt BM HDs); ΔCt = Ct test gene – Ct housekeeping gene; housekeeping gene = B2M; The significant FC cut-off is shown with dotted gray line on the y-axis. BM bone marrow, EM extramedullary localization, HDs healthy donors. Data are presented as the median with range. The graphic legends are Created with BioRender.com.
The analysis of the same set of 19 genes in BM-MNCs of patients who had EM-AMLs versus healthy donors, did not show any evidence of differential expression, suggesting that these genes are selectively modulated only in the extramedullary context (Fig. 4B).
TWIST1 silencing affects FN1 expression, but not the migratory capacity of OCI-AML3 cells
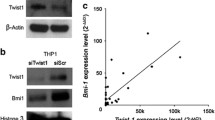
TWIST1, a transcription factor involved in EMT signaling, was among the genes significantly upregulated in EM-AML samples (Fig. 5A). To explore the functional effects of its upregulation, we silenced TWIST1 using a siRNA directed against a specific portion of the 3’ untranslated region (UTR). As reported in Fig. 5B, silencing of TWIST1 induced a moderate time-dependent reduction of cell proliferation, as compared to control. After 48 h of transfection, TWIST1 mRNA expression decreased by 76.8% [corresponding to median Fold Change (FC) of −4.32], as compared to control siRNA. Similarly, we observed a statistically significant decrease in FN1 expression (corresponding to median FC of −2.17, P-value < 0.05), a TWIST1 target involved in extracellular matrix remodeling and cancer cells invasion (Fig. 5C). However, the Boyden chamber assay, performed to test the impact of TWIST1 silencing on migration capability of OCI-AML3 cells, showed no significant differences in the migration rate between siRNA-treated and control cells, and there was only a slight reduction (20.28%) of the capability of invasion of treated cells through the GeltrexTM matrix (Supplementary Fig. 8).
A Bar chart displaying the Fold Change of TWIST1 expressed as 2−ΔΔCt of EM-AML samples in both retrospective (n = 3) and prospective study cohort (n = 3). Data are presented as the median with range. The significant FC cut-off is shown with dotted gray line on the y-axis. Fold Change = 2−ΔΔCt; 2−ΔΔCt = 2−(ΔCt EM − ΔCt BM); ΔCt = Ct test gene – Ct housekeeping gene; housekeeping gene = B2M. B Silencing of TWIST1 by siRNA inhibits cell proliferation. C Effects of TWIST1 and its target (FN1) silencing using 10 nM siRNA, as compared to negative control siRNA. Error bars indicate the median with range of three experiments. Fold Change = 2−ΔCt; ΔCt = Ct test gene – Ct housekeeping gene; housekeeping gene = B2M. The significant FC cut-off is shown with dotted gray line on the y-axis. *P < 0.05.
Metformin treatment results in downregulation of EMT-related genes and interfere with metastatic behavior of OCI-AML3 cells
Since metformin has been shown to interfere with the metastatic behavior of cancer cells, we tested the cytotoxicity of this drug on OCI-AML3 cells, cultured with 10 and 20 mM metformin for 24 and 48 h. The treatment induced a significant decrease of cell number at 48 h (Fig. 6A), which was associated with a significant decrease of mRNA expression of TWIST1 and of its target FN1, in a dose and time-dependent fashion (P-value < 0.05), whereas a trend for SNAI2 was observed at 20 mM metformin (P-value = 0.06) (Fig. 6B). In the same line, metformin also impaired cell migration and invasion by 66.3 and 50%, respectively (P-value = 0.0313) (Fig. 7).
A Effects of metformin treatment on OCI-AML3 cell proliferation. B mRNA levels of TWIST1, FN1 and SNAI2 in OCI-AML3 cells treated with metformin. The black bar represents the negative control, while the bars with increasing intensity of blue represent the scaling up doses of metformin (10 and 20 mM). The significant FC cut-off is shown with dotted gray line on the y-axis. The results are presented as the median with range of three experiments. *P < 0.05. NS not statistically significant.
Flow cytometry analysis of migration (A, B) and invasion (C, D) capacities of OCI-AML3 cells treated with metformin. In (A) and (C) panels are shown the results of one experiment representative of those that gave similar results. In (B) and (D) panels are reported the median number of events as a result of three independent experiments. *P < 0.05.
Discussion
Extramedullary soft tissue localizations are rare in myeloid neoplasms. The most common manifestation is myeloid sarcoma, onsetting at AML diagnosis or relapse. The precise mechanism of EM involvement is not fully understood [3, 4]. In this study, we demonstrate that the transcriptome profile of extramedullary localizations in AML is significantly different from the corresponding BM blasts. On the contrary, genetic profiles of AML patients developing MS were similar to that of de novo AML, as previously reported [24], with SRSF2, RUNX1 and NPM1 as the most frequently mutated genes. Mutational profile was not studied on DNA extracted from the EM-AML samples in our patients, due to the poor quality of available DNA. No differences in the type and frequency of mutated genes in EM vs BM samples has been reported in the literature [14, 25].
Of the highest importance, we studied the gene expression profiles of BM vs EM samples to try to dissect the mechanisms that allow leukemic blasts to invade and colonize extramedullary sites. Among the top deregulated pathways, we identified those involved in the migration and invasive ability of tumor cells, and genes with major functions in the microenvironment, promoting cancer progression [26]. Among these, were specific cytokines and molecules increasing the cellular ability of invasion and metastatization. Proteins making up the extracellular matrix participate in the abovementioned processes, in particular genes encoding for collagen, laminins, integrins and fibronectins [27, 28]. ECM proteins have been suggested to constitute a niche for the cancer stem cells (CSCs) by giving structural and molecular support for their proliferation, self-sustain and differentiation [29]. In our patients who developed an EM localization, we observed upregulation of genes encoding for six collagen isoforms (COL), glycoproteins, fibronectin (FN1), tenascin (TNC), thrombospondin (THBS2), and laminins (LAMA4/LAMB1), which are part of the structure of the basal membranes, support self-renewal of tumor cells, increase the EMT process and lead to drug resistance.
Specifically, comparing BM samples of AML Ctrl to those of patients at the time of the EM relapse, we observed deregulation of the cytokine-cytokine interaction pathway, which has been previously reported to play a role in tumor development and progression [30, 31]. This suggests impairment of the ability of leukemic blasts to remain anchored to the hematopoietic niche in patients who develop a myeloid sarcoma, together with the acquisition of the ability to escape from the BM microenvironment, and marginalize at other sites [28, 32]. In this line, leukemic cells disrupt the BM stroma, which is a hub for normal hematopoietic cells’ support, creating a tumor microenvironment and altering the interactions between stromal molecules and their receptors [33, 34]. The prominent role of adhesion molecules in the homing and proliferation of progenitor cells is confirmed by the use in clinical practice of Plerixafor, a CXCR4 inhibitor that enhances progenitor cell mobilization for transplantation purposes [35,36,37].
Aberrant gene expression in EMT-related pathways, such as ECM-receptor interaction and focal adhesion, is one of the hallmarks of cancer invasion and metastasis. Genes involved in this reprogramming process inducing a mesenchymal phenotype include SNAI1 (SNAIL), SNAI2 (SLUG), ZEB, and TWIST1 [38,39,40]. The latter is part of a family of transcription factors characterized by a basic-helix-loop-helix (bHLH) domain, and is responsible for both primary tumor formation and metastasis [41, 42]. In hematologic malignancies, TWIST1 interaction with several molecules and proteins (e.g., adhesion molecules) is essential for the regulation of apoptosis and EMT [43,44,45]. In our patient cohort, we show that TWIST1 and its interactor FN1 [46] were significantly upregulated in EM-AML localizations, and that siRNA-mediated suppression of TWIST1 expression leads to FN1 downregulation. Similarly, Yang et al. and Kwok et al., showed that TWIST1 siRNA block metastasis formation and induce apoptosis of breast and prostate cancer cells [47, 48].
Among drugs able to interfere with the EMT process, metformin has been shown to specifically inhibit cancer invasion [49,50,51,52]. In particular this drug blocks the activation of numerous pathways, as AMPK and PI3K/Akt/mTOR signaling, and downregulates directly TWIST1 expression [43, 45]. Interestingly, in the present study, we found that metformin decreased AML cell growth in a dose and time-dependent manner. This was associated with downregulation of TWIST1 expression, and with decreased expression of its targets FN1 and SNAI2. However, silencing of TWIST1 alone was not sufficient to affect the migration potential of OCI-AML3 cells, indicating that the EMT process involves different pathways and numerous other factors. On the contrary, in cells silenced for TWIST1, there was a trend towards inhibition of the invasion process. We thus speculate that TWIST1 inhibition may block several proteins, such as metalloproteases, which promote cell invasion by degrading the extracellular matrix.
On the contrary, metformin treatment significantly reduced both the invasion and migration capabilities of the cells. This relates in part to the activity of metformin against TWIST1, but also to the inhibition of its target SNAI2, as previously reported [53].
In conclusion, taken together, our data show that deregulation of the ECM-receptor interaction and focal-adhesion pathways may play an important role in the functional alterations of leukemic cells, favouring their migration from the bone marrow to extramedullary sites. Moreover, since there are no specific approaches to prevent or treat extramedullary localizations, which are often resistant to standard AML treatment, we provide evidence that metformin may be relevant for the development of a targeted combined treatment approach.
Data availability
The datasets generated during the current study are available in the SRA repository, https://www.ncbi.nlm.nih.gov/sra/PRJNA1003124, BioProject PRJNA1003124. Request for additional information should be addressed to the corresponding author.
References
Kahali B. Myeloid sarcoma: the other side of acute leukemia. In: Hematology - latest research and clinical advances. 2018. https://doi.org/10.5772/INTECHOPEN.74931.
Mohammadiasl J, Khosravi A, Shahjahani M, Azizidoost S, Saki N. Molecular and cellular aspects of extramedullary manifestations of acute myeloid leukemia Quick Response Code. J Cancer Metastasis Treat. 2016. https://doi.org/10.4103/2394-4722.167230.
Shallis RM, Gale RP, Lazarus HM, Roberts KB, Xu ML, Seropian SE, et al. Myeloid sarcoma, chloroma, or extramedullary acute myeloid leukemia tumor: a tale of misnomers, controversy and the unresolved. Blood Rev. 2021;47:100773.
Loscocco GG, Vannucchi AM. Myeloid sarcoma: more and less than a distinct entity. Ann Hematol. 2023;102. https://doi.org/10.1007/S00277-023-05288-1.
Eckardt JN, Stölzel F, Kunadt D, Röllig C, Stasik S, Wagenführ L, et al. Molecular profiling and clinical implications of patients with acute myeloid leukemia and extramedullary manifestations. J Hematol Oncol. 2022;15. https://doi.org/10.1186/S13045-022-01267-7.
Ganzel C, Manola J, Douer D, Rowe JM, Fernandez HF, Paietta EM, et al. Extramedullary disease in adult acute myeloid leukemia is common but lacks independent significance: analysis of patients in ECOG-ACRIN Cancer Research Group Trials, 1980-2008. J Clin Oncol. 2016;34:3544–53.
Goyal G, Bartley AC, Patnaik MM, Litzow MR, Al-Kali A, Go RS. Clinical features and outcomes of extramedullary myeloid sarcoma in the United States: analysis using a national data set. Blood Cancer J. 2017;7:e592.
Movassaghian M, Brunner AM, Blonquist TM, Sadrzadeh H, Bhatia A, Perry AM, et al. Presentation and outcomes among patients with isolated myeloid sarcoma: a Surveillance, Epidemiology, and End Results database analysis. Leuk Lymphoma. 2015;56:1698–703.
Solh M, DeFor TE, Weisdorf DJ, Kaufman DS. Extramedullary relapse of acute myelogenous leukemia after allogeneic hematopoietic stem cell transplantation: better prognosis than systemic relapse. Biol Blood Marrow Transplant. 2012;18:106–12.
Yoshihara S, Ando T, Ogawa H. Extramedullary relapse of acute myeloid leukemia after allogeneic hematopoietic stem cell transplantation: an easily overlooked but significant pattern of relapse. Biol Blood Marrow Transplant. 2012;18:1800–7.
Juncà J, Garcia-Caro M, Granada I, Rodríguez-Hernández I, Torrent A, Morgades M, et al. Correlation of CD11b and CD56 expression in adult acute myeloid leukemia with cytogenetic risk groups and prognosis. Ann Hematol. 2014;93:1483–9.
Ball S, Knepper TC, Deutsch YE, Samra W, Watts JM, Bradley TJ, et al. Molecular annotation of extramedullary acute myeloid leukemia identifies high prevalence of targetable mutations. Cancer. 2022;128:3880–7.
Werstein B, Dunlap J, Cascio MJ, Ohgami RS, Fan G, Press R, et al. Molecular discordance between myeloid sarcomas and concurrent bone marrows occurs in actionable genes and is associated with worse overall survival. J Mol Diagn. 2020;22:338–45.
Greenland NY, Van Ziffle JA, Liu YC, Qi Z, Prakash S, Wang L. Genomic analysis in myeloid sarcoma and comparison with paired acute myeloid leukemia. Hum Pathol. 2021;108:76–83.
Engel NW, Reinert J, Borchert NM, Panagiota V, Gabdoulline R, Thol F, et al. Newly diagnosed isolated myeloid sarcoma-paired NGS panel analysis of extramedullary tumor and bone marrow. Ann Hematol. 2021;100:499–503.
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Investig. 2009;119:1420–8.
Their JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54.
Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–21.
Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49.
Golubovskaya VM, Kwen FA, Cance WG. Focal adhesion kinase and cancer. Histol Histopathol. 2009;24:503–10.
Pankaew S, Potier D, Grosjean C, Nozais M, Quessada J, Loosveld M, et al. Calcium signaling is impaired in PTEN-Deficient T cell acute lymphoblastic leukemia. Front Immunol. 2022;13. https://doi.org/10.3389/FIMMU.2022.797244.
Network TCGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059.
Solh M, Solomon S, Morris L, Holland K, Bashey A. Extramedullary acute myelogenous leukemia. Blood Rev. 2016;30:333–9.
Xu S, Xu H, Wang W, Li S, Li H, Li T, et al. The role of collagen in cancer: from bench to bedside. J Transl Med. 2019;17:309.
Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol. 2010;341:126–40.
Graf F, Horn P, Ho AD, Boutros M, Maercker C. The extracellular matrix proteins type I collagen, type III collagen, fibronectin, and laminin 421 stimulate migration of cancer cells. FASEB J. 2021;35. https://doi.org/10.1096/FJ.202002558RR.
Huang J, Zhang L, Wan D, Zhou L, Zheng S, Lin S, et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct Target Ther. 2021;6:1–24.
Gruszka AM, Valli D, Restelli C, Alcalay M. Adhesion deregulation in acute myeloid leukaemia. Cells. 2019;8:66.
Luciano M, Krenn PW, Horejs-Hoeck J. The cytokine network in acute myeloid leukemia. Front Immunol. 2022;13:1000996.
Faaij CMJM, Willemze AJ, Révész T, Balzarolo M, Tensen CP, Hoogeboom M, et al. Chemokine/chemokine receptor interactions in extramedullary leukaemia of the skin in childhood AML: differential roles for CCR2, CCR5, CXCR4 and CXCR7. Pediatr Blood Cancer. 2010;55:344–8.
Bhagat TD, Chen S, Bartenstein M, Barlowe AT, Von Ahrens D, Choudhary GS, et al. Epigenetically aberrant stroma in MDS propagates disease via Wnt/β-catenin activation. Cancer Res. 2017;77:4846–57.
Konopleva MY, Jordan CT. Leukemia stem cells and microenvironment: biology and therapeutic targeting. J Clin Oncol. 2011;29:591–9.
Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM, et al. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 2012;119:3917.
Andreeff M, Zeng Z, Kelly MA, Wang R, McQueen T, Duvvuri S, et al. Mobilization and elimination of FLT3-ITD+ acute myelogenous leukemia (AML) stem/progenitor cells by Plerixafor/G-CSF/Sorafenib: results from a phase I trial in relapsed/refractory AML patients. Blood. 2012;120:142.
Burger JA, Bürkle A. The CXCR4 chemokine receptor in acute and chronic leukaemia: a marrow homing receptor and potential therapeutic target. Br J Haematol. 2007;137:288–96.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178.
Savagner P, Yamada KM, Thiery JP. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–19.
Cobaleda Ć, Ṕerez-Caro M, Vicente-Duẽnas C, Śanchez-García I. Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu Rev Genet. 2007;41:41–61.
Zhu QQ, Ma C, Wang Q, Song Y, Lv T. The role of TWIST1 in epithelial-mesenchymal transition and cancers. Tumour Biol. 2016;37:185–97.
Kang Y, Massagué J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–9.
Chen SC, Liao TT, Yang MH. Emerging roles of epithelial-mesenchymal transition in hematological malignancies. J Biomed Sci. 2018;25:1–8.
Li X, Marcondes AM, Gooley TA, Deeg HJ. The helix-loop-helix transcription factor TWIST is dysregulated in myelodysplastic syndromes. Blood. 2010;116:2304–14.
Li X, Xu F, Chang C, Byon J, Papayannopoulou T, Deeg HJ, et al. Transcriptional regulation of miR-10a/b by TWIST-1 in myelodysplastic syndromes. Haematologica. 2013;98:414.
Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Investig. 2009;119:1429–37.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39.
Kwok WK, Ling MT, Lee TW, Lau TCM, Zhou C, Zhang X, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–62.
Chen YC, Li H, Wang J. Mechanisms of metformin inhibiting cancer invasion and migration. Am J Transl Res. 2020;12:4885.
Liu Q, Tong D, Liu G, Xu J, Do K, Geary K, et al. Metformin reverses prostate cancer resistance to enzalutamide by targeting TGF-β1/STAT3 axis-regulated EMT. Cell Death Dis. 2017;8:e3007.
Park JH, Kim YH, Park EH, Lee SJ, Kim H, Kim A, et al. Effects of metformin and phenformin on apoptosis and epithelial-mesenchymal transition in chemoresistant rectal cancer. Cancer Sci. 2019;110:2834–45.
Xu J, Zhang W, Yan XJ, Lin XQ, Li W, Mi JQ, et al. DNMT3A mutation leads to leukemic extramedullary infiltration mediated by TWIST1. J Hematol Oncol. 2016;9:1–12.
Zhang N, Ng AS, Cai S, Li Q, Yang L, Kerr D. Novel therapeutic strategies: targeting epithelial–mesenchymal transition in colorectal cancer. Lancet Oncol. 2021;22:e358–68.
Acknowledgements
This work was supported by: AIRC 5 × 1000 call “Metastatic disease: the key unmet need in oncology” to MYNERVA project, #21267 (Myeloid Neoplasms Research Venture AIRC. A detailed description of the MYNERVA project is available at http://www.progettoagimm.it); MUR-PNRR M4C2I1.3 PE6 project PE00000019 Heal Italia; PRIN grant No. 2017WXR7ZT; Ministero della Salute, Rome, Italy (Finalizzata 2018, NET-2018-12365935, Personalized medicine program on myeloid neoplasms: characterization of the patient’s genome for clinical decision making and systematic collection of real-world data to improve quality of health care) to MTV. CG was supported by a grant from the Edward P. Evans Foundation. We thank Anna Paglia (Department of Anatomical Pathology, F. Spaziani Hospital, Frosinone, Italy) for her contributions to FFPE samples collection.
Author information
Authors and Affiliations
Contributions
TO, GS and MTV designed the study, interpreted the data, wrote the manuscript; RP helped in RNA-Seq data analysis and performed statistical analysis; ST helped in data interpretation, and gave helpful intellectual insights during the study; CG took part in data collection; FM, LG, SI, RA, AB, LF and MN participated in samples and data collection; AMN, EF, EA, MD, PR and MAIC performed the experiments, AM, AV, LA provided clinical data; MTV took responsibility for the integrity and the accuracy of the data presented; and all authors reviewed and approved the final version of this manuscript.
Corresponding author
Ethics declarations
competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ottone, T., Silvestrini, G., Piazza, R. et al. Expression profiling of extramedullary acute myeloid leukemia suggests involvement of epithelial–mesenchymal transition pathways. Leukemia 37, 2383–2394 (2023). https://doi.org/10.1038/s41375-023-02054-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-02054-0
- Springer Nature Limited