Abstract
Acute myeloid leukemia (AML) is considered a poor prognosis malignancy where patients exhibit altered glucose metabolism and stem cell signatures that contribute to AML growth and maintenance. Here, we report that the epigenetic factor, Ten-Eleven Translocation 3 (TET3) dioxygenase is overexpressed in AML patients and functionally validated human leukemic stem cells (LSCs), is required for leukemic growth by virtue of its regulation of glucose metabolism in AML cells. In human AML cells, TET3 maintains 5-hydroxymethylcytosine (5hmC) epigenetic marks and expression of early myeloid progenitor program, critical glucose metabolism and STAT5A signaling pathway genes, which also positively correlate with TET3 expression in AML patients. Consequently, TET3 depletion impedes hexokinase activity and L-Lactate production in AML cells. Conversely, overexpression of TET3 in healthy human hematopoietic stem progenitors (HSPCs) upregulates the expression of glucose metabolism, STAT5A signaling and AML associated genes, and impairs normal HSPC lineage differentiation in vitro. Finally, TET3 depletion renders AML cells highly sensitive to blockage of the TET3 downstream pathways glycolysis and STAT5 signaling via the combination of 2-Deoxy-D-glucose and STAT5 inhibitor which preferentially targets AML cells but spares healthy CD34+ HSPCs.
Similar content being viewed by others
Introduction
Nearly a century ago, Warburg et al. postulated aerobic glycolysis as a metabolic hallmark of cancer [1]. More recently, metabolic studies in acute myeloid leukemia (AML) confirmed that glucose metabolism is critical for leukemia initiation and progression, accompanied by altered glucose metabolism signatures and overexpression of several glycolytic enzymes [2, 3]. Furthermore, it has been demonstrated that AML bulk has higher glycolytic reserves than AML leukemic stem cells (LSCs), as the latter are mainly dependent on OXPHOS [4].
The TET family (TET1-3) of dioxygenase enzymes regulate active gene expression via establishment of 5-hydroxymethylcytosine (5hmC) epigenetic marks and are deeply embedded in the pathobiology of AML [5,6,7,8]. TET1 has been implicated as an oncogene in AML, while TET2 is frequently mutated and is characterized as a tumor suppressor of myeloid malignancies [6,7,8]. In contrast, recent studies demonstrate that TET3 is required for normal human erythropoiesis and plays a role in suppression of interferon signaling [9, 10]. However, TET3 is not mutated in AML patients and its functional role in human AML remains uninvestigated. 5hmC levels vary considerably in AML patients and high 5hmC levels correlate with inferior overall survival [11]. Whether the distribution of 5hmC marks in AML cells is TET3 dependent, the genomic location of TET3 dependent 5hmC marks and impact on gene expression is also unclear. It is however noteworthy that TET3 and 5hmC marks promote tumorigenesis in some human cancers [12, 13].
In this study, we uncover the growth-promoting role of TET3 in AML and describe a close link between TET3 induced 5hmC epigenetic marks, glycolysis, and the STAT5 pathway.
Methods and materials
Cell lines and transduction
All human leukemic cell lines used in this study were purchased from DSMZ. All cell lines except OCI-AML3 and Kasumi-1 were cultured in RPMI medium with 10% FBS and 1% Penicillin-streptomycin (P/S). OCI-AML3 and Kasumi-1 cells were cultured in RPMI medium supplemented with 20% FBS. SKNO-1 cells were cultured in RPMI medium with 10% FBS and 1% P/S, supplemented with 10 ng/ml Granulocyte macrophage colony stimulating factor.
Knockout (KO) of TET3 in AML cell lines was generated via transduction with lentiviral plasmids containing two different sgRNA and Cas9 (TET3 CRISPR Guide RNA 2 and 3, Genscript). sgRNA and Cas9 expressing cells were selected using puromycin and KO was confirmed in bulk cells by western blots. Please see detailed methods in the Supplementary Information section “Cell lines and transduction”.
Proliferation and colony forming cell assays
For normal CD34+ CB cells, FACS sorted CD34+ GFP+ cells were seeded into the complete culture medium with cytokines (1–2 × 104 cells cultured in serum free 20% BIT medium (Stemcell Technologies) containing IMDM media with 10−5M β-mercaptoethanol supplemented with cytokines (100 µg/ml SCF, 50 µg/ml FLT3-L, 10 µg/ml IL-3, IL-6, and TPO) and were counted after 7 and 14 days. The cells were subcultured in appropriate dilutions after first 7 days. The viable cells were counted using trypan blue exclusion. For CFC assays, 500 CD34+ CB cells were seeded into MethoCult H4434 methylcellulose medium. The cell colonies were counted after 14 days and scored as erythroid (BFU-E, CFU-E), granulocyte/macrophage (CFU-GM, CFU-M, CFU-G) and multipotential granulocyte-erythroid-macrophage-megakaryocyte (CFU-GEMM) colonies based on their morphology.
Transplantation
All experiments were performed in accordance to the guidelines of animal care committee of the Regierungspraesidium Tuebingen, Germany. Transplantation was performed on either C57Bl/6Ly-Peb3b × C3H/HeJ or NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ immunodeficient mice from the Jackson Laboratory and Charles Rivers, bred, and maintained in the animal facility, University of Ulm. For transplantation of murine MLL-AF9 (MA9) cells, 8–12 week-old Peb mice were lethally irradiated (1200 cGy) before i.v. injection. 5 × 104 MA9+ transduced cells were injected along with 5 × 105 helper cells (bulk BM cells from wild type (wt) mice).
For human AML cell lines, 12–15 week-old NSG mice were sub-lethally irradiated (325 cGy) and conditioned intra-peritoneally with 30 mg/kg human immunoglobulin IVIg (Privigen) 24 h before transplantation. For shRNA knockdown (KD) experiment, 5 × 105 or 1 × 106 transduced cells were transplanted to each mouse via tail vein. Mice were under careful observation for early signs of disease. Peripheral blood (PB) aspiration was performed after 4 weeks of transplantation and the presence of leukemic blasts in blood smears was examined. The mice were sacrificed and analyzed 5–6 weeks posttransplantation as the control mice showed signs of disease.
Analysis of experimental mice
The diseased mice were sacrificed by CO2 asphyxiation, isolated the total BM, splenic and PB cells and checked the engraftment of AML cell lines. The cells collected were resuspended in PBS and lysed RBCs using 1x RBC lysis buffer. For Peb mice, engraftment was analyzed using GFP positivity using flow cytometry. The blast percentage was analyzed using cytospins. For NSG mice, the cells were stained using a human-specific anti-CD45 antibody and determined the percentage of CD45-GFP double positive cells using flow cytometry. The body of experimental animals was preserved in 4% formalin solution for the histopathology. The animals with less than 1% leukemic cell engraftment in BM were excluded from the study.
Drug treatment of AML cells
AML cells were treated with STAT5i (Stem cell technologies, #73852) and 2-DG (Sigma #D8375) with the following concentrations STAT5i—5, 25, 50, 100 µM and 2-DG—100, 250, 500, 1000, 2000 nM, with the solvent DMSO as control. Cells were treated for 72 h and cell death was analyzed by flow cytometry using Annexin V and 7-AAD staining. IC50 was calculated using Prism software. For combination therapy, calculated IC50 values of 2-DG and STAT5i were used for treatment of AML cells and CD34+ enriched cord blood cells for 72 h.
The detailed methods are provided in the Supplementary Information.
Results
TET3 is overexpressed in the majority of AML patients and LSC populations
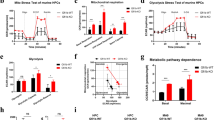
Overexpression of TET3 has been observed in solid cancers such as breast and pancreatic cancers, and is associated with oncogenicity [12, 13]. To determine the role of TET3 in AML, we first analyzed its mRNA expression in AML patients and AML stem cells harboring cell subfractions. Our initial analysis using published RNA-sequencing data revealed that TET3 is highest expressed in AML patients compared to the aforementioned cancers and other liquid and solid tumors (Fig. 1A) [14]. TET3 was also higher expressed in AML than its family member, TET1, a known oncogene in AML (Fig. S1A). qRT-PCR analysis revealed that albeit TET3 had a broad range of expression in AML patients and cell lines, it was overexpressed in the large majority of AML patients compared to healthy CD33+ myeloid cells and total bone marrow (BM) mononuclear cells (MNCs) (Fig. 1B). Among AML genotypes, TET3 was highest expressed in core binding factor and cytogenetically normal (CN)-AML patients and cell lines (Figs. 1C and S1B). TET3 expression did not differ significantly between TET2 or DNMT3A mutated vs. wt gene harboring CN-AML patients. However, NPM1 mutated and FLT3 wt patients exhibited significantly higher expression than NPM1 wt and FLT3 mutated patients (Figs. 1D, S1C, Supplementary Table 1). Furthermore, in our RNA-Seq data set of functionally validated primary human AML LSC subpopulations, TET3 exhibited significantly higher expression in AML LSCs harboring lymphoid-primed multipotential progenitors (LMPP) and granulocyte macrophage progenitors (GMPs) compared to their normal counterparts [15] (Fig. 1E). In normal hematopoietic subpopulations, TET3 expression was highest in healthy CD34+ BM derived HSPCS compared to BM myeloid, lymphoid and total MNCs (Fig. S1D). Among lineage positive cells TET3 was higher expressed in BM and PB derived myeloid cells compared to lymphoid cells (Fig. S1D–F).
A TET3 expression in RNA-Seq analysis of cancer patients (AML patients n = 173, where “n” = no. of biological replicates) [14]. B Quantitative real time PCR (qRT-PCR) expression of TET3 in healthy BM MNCs and BM CD33+ myeloid cells vs. AML patients divided into cytogenetic subtypes. Each dot represents an independent biological replicate indicated as “n”, horizontal bars represent median expression values and vertical error bars represent standard error of mean (SEM). C TET3 expression in cytogenetically normal (CN)-AML (n = 80), core binding factor (CBF) positive (inv(16)+ and AML1-ETO+ n = 18), MLL rearranged (n = 14) and PML-RAR+ AML patients (n = 16) from the TCGA data set. D TET3 expression among CN-AML patients harboring TET2 mutations (n = 23), TET2 wild type (n = 147), DNMT3A mutations (n = 42), DNMT3A wild type (n = 128), NPM1 mutation (n = 66), NPM1 wild type (n = 158), FLT3 mutations (n = 56), and FLT3 wild type (n = 167) from the Vizome RNA-Seq data set. E RNA-seq, mRNA counts of TET3 expression in functionally validated human AML-LSC subpopulations and lymphoid prime multipotent progenitor (LMPP) (n = 9) and granulocyte macrophage progenitor (GMP) (n = 9) and healthy BM LMPP (n = 3) and GMP (n = 3). Bars represent mean ± SEM and “n” indicates biological replicates. F Integrative genomics viewer (IGV) plot showing the TET3/Tet3 promoter and co-occupancy of RNA polymerase II, histone marks and TF binding in human AML (NB-4), murine c-Kit+ MLL-AF9 LSC harboring population and murine MLL-AF9 LSC vs. healthy HSC and GMP.
In line with its overexpression in AML cells, analysis of the TET3 promoter in the myeloid leukemia cell lines, murine MLL-AF9 c-Kit+ LSC harboring cell population, murine MLL-AF9 LSCs vs. healthy HSCs and GMPs from published data sets revealed an enrichment of euchromatic histone marks such as H3K4me3, H3K27Ac, H3K9Ac and the absence of the suppressive H3K27me3 mark (Figs. 1F and S1G, H). The euchromatic histone marks overlapped with RNA polymerase II and transcription factor (TF) binding sites, such as the TF MYC, its binding partner MAX among several others (Figs. 1F and S1G, H). Notably, MLL-AF9+ LSCs and AML patients exhibited higher enrichment of H3K4me3 marks on the TET3 promoter vs. healthy counterparts, suggesting that TET3 overexpression in AML is potentially epigenetically regulated (Figs. 1F and S1I).
High TET3 levels are required for AML growth and AML LSC, and its overexpression in healthy human HSPCs perturbs myeloid differentiation
To test whether high TET3 expression regulates AML growth, we performed KD or KO studies in AML cells and KD/KO efficiency was assessed using qRT-PCR and western blot (Fig. S2A, B). TET3 depletion significantly decreased the clonogenic and proliferative potential of AML cells (Figs. 2A, B and S2C, D). On the other hand, the T-ALL cell line Jurkat, used as an off-target control, did not show impairment of growth upon TET3 KD (Fig. S2C). Transplantation of TET3 KD and KO AML cell lines into xenografts resulted in severely reduced leukemic engraftment in BM of xenografts (Figs. 2C, D and S2E). TET3 KD in primary CN-AML patient cells inhibited their clonogenic potential in vitro (Fig. 2E). Similarly, stable KD of Tet3 in murine AML-LSC harboring c-Kit+ cell population from MLL-AF9 (MA9-LSCs) and AML1-ETO9A immortalized cells decreased their clonogenic potential in vitro and delayed the onset of MLL-AF9+ AML in vivo (Figs. 2F and S2F). The abrogation of TET3 expression induced loss of 5hmC marks in human AML cell lines, primary patient cells and murine LSCs, caused apoptosis, but did not significantly affect the cell cycle (Figs. 2G and S2G–L).
A Colony forming cell (CFC) assays with AML cell lines transduced with either scrambled control or shRNA against TET3. The colony numbers from initially seeded 500 cells were counted after 12–14 days. Bars represent mean ± SEM. B CFC assays with AML cell lines transduced with either Cas9 and sgScr or sgTET3. Bars represent mean ± SEM. C Upper panel, the AML cell lines engraftment measured as the percentage of human CD45+ GFP+ cells in BM of mice, measured by flow cytometry after 5–6 weeks posttransplantation of NB-4 and OCI-AML3 cells transduced with shTET3-A or scrambled control. Each mouse was transplanted with either 1 × 106 cells (Scrambled vs. shB) or with 0.5 × 106 (Scrambled vs. shA). Each dot represents an individual mouse and horizontal bar represents mean ± SD. Lower panel—histological analysis of diseased mice. D The leukemic cell engraftment measured as the percentage of CD45+ GFP+ AML cells in BM of mice, measured by flow cytometry after 5–6 weeks posttransplantation of OCI-AML3 cells transduced with sgScrambled control or sgTET3. E CFC assays with primary AML cells from patients transduced with either scrambled control or shRNA against TET3 (n = 4). The colony numbers from initially seeded 1 × 105 cells were counted after 12–14 days. F Kaplan–Meyer plot of mice transplanted with c-Kit+ murine AML-LSCs transduced with either scrambled control or shRNA against Tet3. G Left panel: intracellular staining followed by flow cytometry for 5hmC marks in OCI-AML3 and SKNO-1 transduced with sgScrambled control and sgTET3. Right panel: representative histogram shows global 5hmC marks in OCI-AML3 cell line.
Interestingly, stable overexpression of TET3FL (TET3 full length) and TET3ΔCXXC (lacking CXXC domain) in AML cell lines increased the corresponding mRNA levels and protein levels in vitro (Fig. S3A–C). The overexpression significantly augmented cell growth in vitro and enhanced global 5hmC levels (Figs. 3A, B and S3D). However, overexpression of full length TET1 in our hands (data not shown) and TET2 in a published study [16] suppressed human AML growth, suggesting that unlike other TET members, TET3 is a growth-promoting factor in AML.
A Cell growth analysis of human AML cell lines transduced with TET3 catalytic domain (TET3ΔCXXC) or TET3 full length (TET3FL) and empty vector control, cells were counted after 48 h time intervals (n = 3). B Left panel: representative figure of intracellular staining followed by confocal microscopy performed on transduced OCI-AML3 cells using DAPI to visualize nucleus and antibody against 5hmC detecting global 5hmC marks. Scale bar shown indicates 50 µm. Right panel: representative histogram of intracellular staining followed by flow cytometry showing intracellular levels of 5hmC (n = 3). C Erythroid and myeloid colony morphology of TET3FL and TET3ΔCXXC transduced HSPCs under light microscope. D FACS analysis of cells derived from CD34+ transduced HSPCs in the CFC assay (after 14 days) using an antibody against (left panel) mature myeloid markers CD11b and CD15 (control and TET3ΔCXXC; n = 5, TET3FL; n = 4), and (right panel) mature erythroid marker GLYA (control and TET3ΔCXXC; n = 7, TET3FL; n = 5). E Absolute number of mature myeloid cells (CD11b+, CD15+ and CD11b+ CD15+ cells) in liquid culture assay on day 7 (n = 2).
Next, we aimed to test whether aberrant expression of TET3 perturbs normal function of healthy HSPCs. Therefore, we stably overexpressed TET3FL and TET3ΔCXXC in CD34+ CB HSPCs. The overexpression of TET3FL and TET3ΔCXXC increased global 5hmC marks in healthy human HSPCs and skewed normal differentiation program in vitro, with reduction of terminally differentiated mature myeloid CD11B+ and CD15+ colonies in CFCs and reduction in absolute cell number of mature myeloid cells in liquid culture assays (Figs. 3C–E and S3E, F). We also observed an increased number of GLYA+ erythroid cells in the CFC assays, which goes in line with a previous study which reports that TET3 depletion in healthy human CD34+ HSPCs impairs erythroid differentiation (Figs. 3C, D and S3E, F) [9]. These data indicate that balanced expression of TET3 is important for the normal erythroid-myeloid lineage commitment. In sum, our data demonstrate that high TET3 levels promote AML growth and its high expression in normal HSPCs impairs normal myeloid differentiation.
TET3 downregulates expression of genes involved in glucose metabolism and LSC function in AML cells
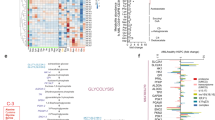
Next, we sought to examine the impact of high TET3 levels on the gene expression patterns in AML cells. RNA-Seq analysis of TET3 KD NB-4 cells revealed 1409 differentially expressed genes, out of which 60% were significantly downregulated (Supplementary Table 2). Gene Set Enrichment Analysis (GSEA) revealed the downregulation of genes associated with LSC function (LSC signature), STAT5 signaling and glucose metabolism pathways upon TET3 depletion (Fig. 4A). In particular, factors central to glucose metabolism and glucose transport were downregulated, such as enzymes HK1, HK2, ENO2, PC, ALDH2, ALDOC, PKFP, PGAM1, PDK1, PDK3, PDP1, the lncRNA H19, TF ZEB-1, its target SLC2A3 and several HNF4A targets were also downregulated (Figs. 4B and S4A and Supplementary Table 2). To ensure that the differential expression of glucose metabolism genes was specific to the KD of TET3 and not a general phenomenon the KD of TET family members or other oncogenes, we conducted a supervised analysis of glucose metabolism and transport genes in the context of TET1-3 depletion, and KD of the AML proto-oncogene CDX2. The analysis using our own RNA-Seq data set of TET3, TET1 and CDX2 KD in AML cell lines and published RNA-Seq of TET2 KO in the AML cell line THP-1 revealed that gross downregulation of glucose metabolism genes was exclusively induced by TET3 depletion in AML cells (Figs. 4B and S4B–D) [17]. In line with its effect on LSC genes and its high expression in leukemic LMPP and GMP, CellRadar analysis illustrated that higher levels of TET3 maintained the expression of early myeloid progenitor associated gene signature (CMP and GMP) and suppressed the expression of mature myeloid lineage (monocytes) associated gene signature in NB-4 cell line (Fig. 4C). We observed a decreased expression of TET1 in TET3 depleted NB-4 cells, but TET1 has been shown to be dispensable for NB-4 cell growth (Supplementary Table 2) [18].
A Gene Set enrichment Analysis (GSEA) performed for TET3 DEGs (n = 3, p value < 0.05, FDR < 0.05). B Heatmap of 116 genes associated with glucose metabolism in NB-4 cells transduced with Scrambled control vs. shTET3-A. C CellRadar plot of DEGs showing downregulation of CMP/GMP/Myelocytic signature and upregulation of Monocytic lineage signature in TET3 KD NB-4 cells vs. scrambled (https://karlssong.github.io/cellradar/). D Heatmap representing the 1530 DEGs upon TET3-CD overexpression compared to vector control in healthy CD34+ cord blood HSPCs after 48 post sorting (n = 3, p value <0.05, FDR < 0.05). E GSEA analysis performed for TET3 DEGs recorded in healthy CD34+ HSPCs transduced with TET3ΔCXXC and vector control (n = 3, p value < 0.05, FDR < 0.05). F CellRadar analysis of DEGs upon TET3 overexpression and TET3 KD in CD34+ cord blood cells and TET3 KD in NB-4 cells, respectively.
Overexpression of TET3 in healthy human HSPCs induces genes involved in glucose metabolism and AML associated pathways
Next, we tested whether aberrant overexpression of TET3 in normal CD34+ CB HSPCs induced expression of glucose metabolism, STAT5 and AML specific and myeloid progenitor associated gene signatures. Due to the better transduction efficiency, higher cell number, resultant better RNA quality and similar phenotype of TET3 ΔCXXC vs. TET3FL transduced HSPCs, we chose to perform our experiments using TET3ΔCXXC. GSEA analysis showed that overexpression of TET3ΔCXXC in HSPCs upregulated STAT5A, the glucose metabolism associated HNF4A pathway and the AML LSC, progenitor and erythroid development pathways (Fig. 4D, E). CellRadar analysis illustrated that TET3 overexpression in normal HSPCs prompted suppression of the mature myeloid cell associated gene signature (monocytic), which was found upregulated in TET3 KD NB-4 cells (Fig. 4F). Genes associated with myeloid differentiation (such as CEBPA and TET2) were downregulated in TET3ΔCXXC overexpressing HSPCs. However, TET3ΔCXXC triggered expression of erythroid lineage gene signature (MEP). These data are in line with our functional data and published KD data that demonstrate reduced number of mature myeloid cells and perturbation in erythroid lineage cells, respectively [9]. Notably, 190 genes that were upregulated in TET3ΔCXXC overexpressing HSPCs were downregulated in TET3 KD NB-4 cells and were deemed as putative TET3 targets, such as STAT5A and glucose metabolism associated factors HK1, ENO2, and SLC2A3.
TET3 regulates expression of genes involved in glucose metabolism and LSC function via 5hmC marks
Next, we analyzed the impact of TET3 depletion on 5hmC marks genome wide by hydroxymethylated DNA immunoprecipitation-sequencing (hMeDIP-Seq) and assessed its relationship with deregulated gene expression. hMeDIP-Seq analysis confirmed that TET3 depletion in NB-4 cells resulted in a global reduction of 5hmC marks compared to scrambled control (Fig. 5A and Supplementary Table 3). Strikingly, 5hmC marks were lost at promoters or gene body (GB) regions of genes that are associated with early myeloid progenitor (CMP and GMP) gene signature (Fig. S5A). The comparison of the hMeDIP-Seq and RNA-Seq data from TET3 KD NB-4 revealed that more than 44% (372/845) of downregulated genes exhibited loss of 5hmC marks on promoters or GB. The majority of these genes belonged to AML associated, glucose metabolism pathways and were associated with early myeloid progenitor gene signature (Figs. 5B and S5A and Supplementary Table 2). Moreover, glucose metabolism genes and STAT5A were among the 94 genes that experienced loss of 5hmC marks, downregulation of expression upon TET3 KD in NB-4 cells and upregulation in TET3ΔCXXC overexpressing HSPCs (Fig. 5C and Supplementary Table 4).
A hMeDIP-Seq analysis; promoter regions were defined as TSS of ±5 kb. Losses and gains 5hmC marks across different genomic regions in TET3 depleted NB-4 cells vs. scrambled control. Heatmap shows centered 5hmC peaks, marks in the TSS region. The upper panel represents loci enriched with 5hmC marks in the scrambled arm but reduced or absent in the shRNA arm, while the lower panel shows regions enriched in the shRNA arm but reduced in the scrambled arm. B Top 10 pathways associated with both downregulated genes and 5hmC loss (KEGG pathway analysis, p value < 0.05, FDR < 0.05). C Venn diagram showing overlap of TET3 dependent 5hmC promoters and gene bodies (T3-5hmC) with genes significantly lost expression upon TET3 KD in NB-4 cells and gained the expression upon TET3ΔCXXC overexpression in CD34+ healthy HSPCs cells. D IGV plot of glucose metabolism associated genes showing distribution of 5hmC marks at promoter and/or gene body in scrambled controls, shTET3 transduced NB-4 and shTET1 transduced Kasumi-1 cells (unpublished data) along with co-occupancy of TET3 (hg19). E qRT-PCR analysis of glucose metabolism genes in TET3 depleted AML cell line. F Western blot of HK2 and STAT5A in scrambled and TET3 depleted NB-4 and OCI-AML3 AML cell lines. Beta-Actin represents endogenous control (n = 2).
5hmC sequencing data, from our own and published data sets, of AML cell lines and primary patients representing different AML genotypes revealed that glucose metabolism genes are enriched with 5hmC marks and hyperhydroxymethylated in AML cells compared with corresponding loci of glucose metabolism genes in healthy CD34+ HSPCs, suggesting that their mRNA expression is in-part regulated by TET proteins across AML subtypes (Fig. S5B, C). Furthermore, glucose metabolism genes also exhibited binding sites for TET3 which overlapped with sites of 5hmC enrichment (Figs. 5D and S5B) [19]. Notably, while TET3 KD lead to the loss of 5hmC marks on glucose metabolism genes, neither TET1 depleted Kasumi-1 cells nor TET2 mutation harboring AML patients reflected loss of 5hmC marks on the corresponding loci when compared to scrambled control and TET2 wt harboring AML patients, respectively (Figs. 5D and S5B) [20]. Therefore, in sum, the presence of 5hmC marks across AML subtypes, overlap with known TET3 binding sites, the exclusive loss of 5hmC marks and concomitant downregulation of mRNA expression upon TET3 depletion together indicated that factors critical for glucose metabolism are targets of TET3 and 5hmC mediated gene regulation, not involving TET2 and TET1. However, our study does not rule out that TET1-2 could regulate the expression of the aforementioned genes in other cancers or cell types.
qRT-PCR and western blots further confirmed the decreased expression of TET3 targets, Hexokinase 2 (HK2) and STAT5A, in KD and KO experiments in other AML cell lines, such as SKNO-1 and OCI-AML3 (Fig. 5E, F). The majority of the above-mentioned genes positively correlated with TET3 expression in AML patients, i.e., patients that exhibited higher expression of TET3 also exhibited higher expression of STAT5A, HK1, HK2, ENO2, etc. (Fig. S5D). Concordant with TET3 expression, these genes were also highly expressed in functionally validated LSCs compared to healthy counterparts (Fig. S5E, F). Taken together, our and published RNA-Seq and 5hmC sequencing data suggest that in the context of AML cells, TET3 regulates the expression of glucose metabolism and STAT5A via 5hmC marks.
TET3 depleted AML cells are vulnerable to glucose deprivation or chemical inhibitors targeting TET3 downstream genes HK2 and STAT5A
Next, we tested whether TET3 could alter the ability of AML cells to metabolize glucose. Time course measurements of glucose uptake using 2-NBDG revealed that TET3 depleted NB-4 cells exhibited a trend toward increased glucose uptake and L-Lactate production analysis revealed a clear decrease in lactate production in TET3 depleted NB-4 cells (Fig. 6A, B). These data were further supported by, colorimetric enzyme assays for hexokinase activity which demonstrated that TET3 depleted AML cells exhibited decreased hexokinase activity and showed reduced ability to convert glucose to glucose-6-phosphate (Fig. 6C). Conversely, TET3FL overexpressing AML cells exhibited enhanced hexokinase activity (Fig. 6D). In sum, these data demonstrated that TET3 has a direct impact on the ability of AML cells to metabolize glucose. The association of TET3 with glucose metabolism was further supported by the fact that short term glucose deprivation (72 h) lead to the upregulation of TET3 expression and its downstream targets HK2 and STAT5A in AML cell lines, while the expression of TET1-2 remained unchanged (Figs. 6E and S6A, B). This suggests that TET3 likely shares a feedback link with glucose levels and maintains glucose metabolism via regulation of vital genes in AML cells. Consistent with this hypothesis, TET3 depleted AML cells were unable to survive 48 h of glucose deprivation while TET1 depleted AML cells were unaffected by glucose deprivation even up to 72 h, demonstrating that TET3 depletion renders AML cells sensitive to glucose deprivation (Figs. 6F and S6C).
A Glucose uptake assay using 2-NBDG, analyzed by flow cytometry in TET3 depleted NB-4 cells vs. scrambled. B Lactate production in TET3 depleted cells vs. scrambled. C Hexokinase activity levels in TET3 depleted AML cells. D Hexokinase activity levels in TET3FL overexpressing AML cells compared to empty vector control. E Gene expression analyzed via qRT-PCR of TET1-3 in glucose deprived AML cell lines (10% of normal glucose levels in RPMI medium) compared to AML cells grown in regular RPMI growth medium. F Percentage of live cells in TET3 depleted and scrambled control transduced AML cells, grown under glucose deprivation vs. normal medium conditions.
In AML cells, skewed metabolic pathways are pivotal for proliferation but also serve as therapeutic vulnerabilities. In this regard, altered glucose metabolism and overexpression of several glycolytic enzymes in AML cells can be targeted via the competitive chemical inhibitor of HK, 2-deoxy-D-glucose (2-DG). As HK was a downstream target of TET3, we utilized the 2-DG to antagonize AML cells. In addition, we targeted another TET3 downstream gene, STAT5A (STAT5i), previously shown to induce death of AML cells [2, 18, 21]. We tested the impact of 2-DG and STAT5i on MA9-LSC harboring cells and observed an IC50 of 500 nM and 28 µM, respectively (Fig. S7A, B). Importantly, 2-DG and STAT5i exhibited 2.5 times lower IC50 levels on Tet3 depleted MA9-LSC harboring cells compared to scrambled (Fig. 7A, B). Combination therapy with STAT5i and 2-DG on MA9-LSC harboring cells also resulted in a 2.5-fold reduction in IC50 concentrations of both drugs compared to monotherapy (Fig. 7C). Comparable effects were observed on human AML cell lines and primary AML patients with regard to cell viability and AML cell lines with regard to clonogenicity (Figs. 7D–F and S7C). Contrary to the impact on AML cells, 2-DG and STAT5i, neither mono nor combination therapy induced any significant impact on cell viability of healthy CD34+ HSPCs and only the combination therapy induced a minor impact on colony formation (Fig. S7D, E) compared to AML cells. Importantly, combination therapy reduced colony formation by an average of 96% in AML cells compared to only 28% in healthy HSPCs. Taken together, our data supported the use of inhibitors against TET3 downstream pathways, glucose metabolism, and STAT5 signaling, to preferentially target AML bulk and stem cell population over healthy stem cells.
A IC50 of 2-DG in c-Kit+ leukemic stem cell (LSC) harboring MLL-AF9 cells, transduced with scrambled or shRNA against Tet3. Concentrations tested- 2-DG: 125, 250, 500, 1000 nM for 72 h. B IC50 of STAT5i in c-Kit+ LSC harboring MLL-AF9 cells, transduced with scrambled or shRNA against Tet3. Concentrations tested- STAT5i: 5, 10, 50, 100 µM for 72 h. C IC50 of STAT5 inhibitor (STAT5i) as a single treatment and in combination with 250 nM of 2-DG in c-Kit+ LSC harboring MLL-AF9 cells. Concentrations tested for STAT5i- 5, 12.5, 25, 50 µM for 72 h. D Cell viability of primary AML patient cells treated with mono or combination therapy of IC50 concentrations of STAT5i (50 µM) and 2-DG (2 µM) vs. DMSO. CFC assay of E NB-4 and F OCI-AML3 cells treated with DMSO control, 2-DG (2 µM), STAT5i (50 µM) or combination of 2-DG and STAT5i, with an initial input of 500 cells.
Discussion
TET family of DNA demethylases was subjected to number of studies on malignant hematopoiesis in the last decade, yet the role of TET3 in human AML has remained obscure. In this study, we demonstrate that TET3 is aberrantly overexpressed in the human AML bulk and functionally validated AML-LSC populations and its overexpression is potentially regulated by euchromatic histone marks in AML cells. This study shows that higher TET3 level is required for the growth of AML bulk, LSCs and the maintenance of 5hmC marks as its deficiency in human AML cells and murine AML LSCs impairs their growth in vitro, in vivo, induces apoptosis and the loss of 5hmC marks. Furthermore, in contrast to wt TET1 and TET2, overexpression of wt TET3 in AML cell lines further enhances cell growth demonstrating that unlike other TET members, wt TET3 is a growth-promoting factor in AML. In line with our data, published studies in solid cancers such as esophageal squamous cell carcinoma and breast cancer demonstrate that the overexpression of TET3 is associated with cancer growth and cancer stem cell maintenance [22, 23]. Interestingly, the DNA binding domain lacking TET3ΔCXXC could also augment AML growth, which is not surprising as TET3 isoform lacking the CXXC domain can also bind DNA, presumably through its interacting partners, and is able to alter gene expression and 5hmC marks [24]. Our study indicates that the balanced expression of TET3 is important for the normal human HSPC function, as its forced expression in normal HSPCs impairs myeloid and erythroid differentiation in vitro. These data are consistent with a previous study which reports that TET3 depletion in healthy human HSPCs impairs erythroid differentiation [9].
Reports in the murine Tet2 KO model suggests that Tet3 could be a tumor suppressor in murine AML, especially in the context of Tet2 deletion [25, 26]. However, in human AML, the lack of spontaneous TET3 mutations, in the context of TET2 mutation or otherwise, indicates that TET3 mutations do not confer a selection advantage in human AML as they do in the murine model. On the contrary, our data suggest that human AML cells harboring TET2 mutations would be more dependent on TET3 enzymatic activity and thereby vulnerable to TET3 depletion. This notion is supported by a recent study where TET3 depletion in TET2 mutated AML cells caused a dramatic impact on 5hmC marks, apoptosis and AML growth [27]. In sum, its overexpression in the majority of AML patients and our functional data in human AML strongly suggest that TET3 function varies according to the cellular and species context, and is a dependency factor in human AML.
Our RNA-Seq and hMeDIP-Seq data illustrate that TET3 supports AML growth via the epigenetic regulation of proliferation, stem cell and cell survival associated pathways. Higher TET3 levels positively regulate gene expression by maintaining 5hmC marks on the promoters of genes associated with early myeloid progenitor signature, glycolysis and STAT5A signaling. Our CellRadar analysis illustrates that TET3 maintains the expression of early myeloid progenitor associated gene signature (CMP and GMP), which is in line with its high expression in human leukemic LMPP and GMP. Our and published data collectively demonstrates that the aforementioned pathways, especially glycolytic pathway genes, are direct targets of TET3 and not TET1 or 2 in AML cells. Among the direct targets, the glucose metabolism pathway genes are of great interest due to their established role in AML metabolism, cell survival and their potential as drug targets. Enzymes, HK1, HK2, ENO2 and ALDH2 and lncRNA H19, are all critical players in glycolysis, while HK2 is also an anti-apoptotic factor [2, 3, 28, 29]. We demonstrate that TET3 not only regulates their expression but also in-turn regulates the glucose metabolism rate of AML cells. Furthermore, our data also hint at the existence of a feedback loop between serum glucose levels and TET3 expression and involvement of TET3 in glucose homeostasis in AML cells, as glucose deprived AML cells upregulate TET3. This is consistent with a study in hepatic cells where TET3 is acutely induced upon fasting and plays a critical role in hepatic glucose production to maintain systemic glucose homeostasis [30]. Furthermore, acute and chronic hypoxia is associated with upregulation of glycolysis. Hypoxia also triggers TET3 expression in cancer and healthy erythroid cells [31, 32]. It is likely that TET3 is a mediator of hypoxia and glucose deprivation stress associated upregulation of glycolytic enzymes, but this hypothesis requires detailed investigation.
Our RNA-Seq analysis also shows that higher TET3 levels not only in AML cells but also in healthy human HSPCs induces the expression of genes associated with STAT5A signaling and glucose metabolism associated HNF4A pathway. Interestingly, contrary to expectation, TET3 overexpression led to a greater downregulation of gene expression in healthy HSPCs, especially of myeloid differentiation genes possibly through a repressive function. In support of this, CellRadar analysis showed that mature myeloid lineage (monocytes) specific genes expression signature was suppressed in TET3 overexpressing healthy HSPCs and upregulated in TET3 KD AML cells. Since TET3 contains a highly conserved Sin3 interaction domain and is known to associate with gene expression suppressing factors SIN3A and histone deacetylases (HDACs), it is possible that TET3 function in AML cells extends beyond 5hmC mark mediated positive gene regulation of growth-promoting factors to suppression of myeloid differentiation genes via association with repressive complexes [10]. This hypothesis is backed by evidence that TET3, independent of its catalytic function, inhibits interferon production by recruiting HDAC1 to promoters of interferon genes [10, 19].
Lastly, our study makes a case for the combination therapy of AML cells with inhibitors of TET3 downstream targets, glucose metabolism, and STAT5A signaling. Activation of STAT5 signaling is required to maintain AML growth and LSC function, at least in-part, relies on the STAT5 pathway [33, 34]. Quiescent LSCs preferentially depend on OXPHOS [35]. However, it is likely that non-quiescent, cycling AML LSCs, as in the case of MLL leukemia [36], depend on glucose metabolism and TET3 expression. Our data demonstrate that MLL-AF9 c-kit+ LSC population harboring cells are dependent on TET3 expression. The close association of glucose metabolism and STAT5A signaling with AML growth and the minimal impact of their inhibitors on healthy HSPCs in our data provides a good rationale for targeting the pathways in AML patients [2, 18]. The collective inhibition of glycolysis and STAT5A signaling pathways attenuates growth of AML bulk and AML LSC harboring cells in vitro and induces their near complete cell death at IC50 concentrations within 48 h and reduces IC50 of both inhibitors by 2.5-fold. In sum, our study extends the knowledge about TET proteins in AML beyond TET1 and TET2 and links TET3 to the epigenetic regulation of metabolic and growth associated pathways in AML.
References
Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–30.
Chen WL, Wang JH, Zhao AH, Xu X, Wang YH, Chen TL, et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–54.
Chen WL, Wang YY, Zhao A, Xia L, Xie G, Su M, et al. Enhanced fructose utilization mediated by SLC2A5 is a unique metabolic feature of acute myeloid leukemia with therapeutic potential. Cancer Cell. 2016;30:779–91.
Jones CL, Stevens BM, D’Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell. 2018;34:724–40.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5.
Huang H, Jiang X, Li Z, Li Y, Song CX, He C, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110:11994–9.
An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun. 2015;6:10071.
Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170:1079–95.
Yan H, Wang Y, Qu X, Li J, Hale J, Huang Y, et al. Distinct roles for TET family proteins in regulating human erythropoiesis. Blood. 2017;129:2002–12.
Xue S, Liu C, Sun X, Li W, Zhang C, Zhou X, et al. TET3 inhibits type I IFN production independent of DNA demethylation. Cell Rep. 2016;16:1096–105.
Kroeze LI, Aslanyan MG, van Rooij A, Koorenhof-Scheele TN, Massop M, Carell T, et al. Characterization of acute myeloid leukemia based on levels of global hydroxymethylation. Blood. 2014;124:1110–8.
Wu MZ, Chen SF, Nieh S, Benner C, Ger LP, Jan CI, et al. Hypoxia drives breast tumor malignancy through a TET-TNFalpha-p38-MAPK signaling axis. Cancer Res. 2015;75:3912–24.
Bhattacharyya S, Pradhan K, Campbell N, Mazdo J, Vasantkumar A, Maqbool S, et al. Altered hydroxymethylation is seen at regulatory regions in pancreatic cancer and regulates oncogenic pathways. Genome Res. 2017;27:1830–42.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Rawat VPS, Götze M, Rasalkar A, Vegi NM, Ihme S, Thoene S, et al. The microRNA miR-196b acts as a tumor suppressor in Cdx2-driven acute myeloid leukemia. Haematologica. 2020;105:e285–9.
Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015;57:662–73.
Morinishi L, Kochanowski K, Levine RL, Wu LF, Altschuler SJ. Loss of TET2 affects proliferation and drug sensitivity through altered dynamics of cell-state transitions. Cell Syst. 2020;11:86–94.
Jiang X, Hu C, Ferchen K, Nie J, Cui X, Chen CH, et al. Targeted inhibition of STAT/TET1 axis as a therapeutic strategy for acute myeloid leukemia. Nat Commun. 2017;8:2099.
Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–55.
Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–55.
Muller J, Sperl B, Reindl W, Kiessling A, Berg T. Discovery of chromone-based inhibitors of the transcription factor STAT5. Chembiochem. 2008;9:723–7.
Darvin P, Sasidharan Nair V, Elkord E. PD-L1 expression in human breast cancer stem cells is epigenetically regulated through posttranslational histone modifications. J Oncol. 2019;2019:3958908.
Xu F, Liu Z, Liu R, Lu C, Wang L, Mao W, et al. Epigenetic induction of tumor stemness via the lipopolysaccharide-TET3-HOXB2 signaling axis in esophageal squamous cell carcinoma. Cell Commun Signal. 2020;18:17.
Perera A, Eisen D, Wagner M, Laube SK, Kunzel AF, Koch S, et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11:283–94.
Yue X, Lio CJ, Samaniego-Castruita D, Li X, Rao A. Loss of TET2 and TET3 in regulatory T cells unleashes effector function. Nat Commun. 2019;10:2011.
Shrestha R, Sakata-Yanagimoto M, Maie K, Oshima M, Ishihara M, Suehara Y, et al. Molecular pathogenesis of progression to myeloid leukemia from TET-insufficient status. Blood Adv. 2020;4:845–54.
Guan Y, Tiwari AD, Phillips JG, Hasipek M, Grabowski DR, Pagliuca S, et al. A therapeutic strategy for preferential targeting of TET2 mutant and TET-dioxygenase deficient cells in myeloid neoplasms. Blood Cancer Disco. 2021;2:146–61.
Venton G, Perez-Alea M, Baier C, Fournet G, Quash G, Labiad Y, et al. Aldehyde dehydrogenases inhibition eradicates leukemia stem cells while sparing normal progenitors. Blood Cancer J. 2016;6:e469.
Zhang TJ, Zhou JD, Zhang W, Lin J, Ma JC, Wen XM, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47.
Da L, Cao T, Sun X, Jin S, Di X, Huang X, et al. Hepatic TET3 contributes to type-2 diabetes by inducing the HNF4alpha fetal isoform. Nat Commun. 2020;11:342.
Lin G, Sun W, Yang Z, Guo J, Liu H, Liang J. Hypoxia induces the expression of TET enzymes in HepG2 cells. Oncol Lett. 2017;14:6457–62.
Cao JZ, Liu H, Wickrema A, Godley LA. HIF-1 directly induces TET3 expression to enhance 5-hmC density and induce erythroid gene expression in hypoxia. Blood Adv. 2020;4:3053–62.
Polak A, Bialopiotrowicz E, Krzymieniewska B, Wozniak J, Stojak M, Cybulska M, et al. SYK inhibition targets acute myeloid leukemia stem cells by blocking their oxidative metabolism. Cell Death Dis. 2020;11:956.
Wingelhofer B, Maurer B, Heyes EC, Cumaraswamy AA, Berger-Becvar A, de Araujo ED, et al. Pharmacologic inhibition of STAT5 in acute myeloid leukemia. Leukemia. 2018;32:1135–46.
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–41.
Iwasaki M, Liedtke M, Gentles AJ, Cleary ML. CD93 marks a non-quiescent human leukemia stem cell population and is required for development of MLL-rearranged acute myeloid leukemia. Cell Stem Cell. 2015;17:412–21.
Acknowledgements
The authors would like to thank all members of the animal facility, genomics and flow cytometry core facilities of the University of Ulm, Germany. The work was supported by a grant received by VPSR from the Ministry of Science, Research and the Art (MWK), Baden-Württemberg, Germany (Junior-professor Program). CB was funded by a grant from the DFG (SFB 1074 project A4 to CB). We thank Prof. Konstanze Döhner and Prof. Hartmut Döhner (Department of Internal Medicine III, University Hospital Ulm) for providing patient samples. We thank Marc Young (Institute of Immunology, University Hospital Ulm) for his assistance with generating Fig. 1A.
Author information
Authors and Affiliations
Contributions
VPSR designed the project, CB discussed and edited the design of the project. AJP, SB, TA and FM performed experiments. AJP, KF and TM performed transplantations. AJP, SB, CB, and VPSR analyzed the data. SB and AJP performed the RNA-seq, ChIP-seq data analysis in collaboration with AS. UK and LQM performed histopathology. NMV contributed research material. AJP, SB, VPSR, and CB contributed to interpretation of patient data. SB, AJP, CB, and VPSR wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pulikkottil, A.J., Bamezai, S., Ammer, T. et al. TET3 promotes AML growth and epigenetically regulates glucose metabolism and leukemic stem cell associated pathways. Leukemia 36, 416–425 (2022). https://doi.org/10.1038/s41375-021-01390-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01390-3
- Springer Nature Limited
This article is cited by
-
Emerging DNA Methylome Targets in FLT3-ITD-Positive Acute Myeloid Leukemia: Combination Therapy with Clinically Approved FLT3 Inhibitors
Current Treatment Options in Oncology (2024)
-
Metabolic dependencies of acute myeloid leukemia stem cells
International Journal of Hematology (2024)
-
Direct inhibition of dioxygenases TET1 by the rheumatoid arthritis drug auranofin selectively induces cancer cell death in T-ALL
Journal of Hematology & Oncology (2023)
-
miR-488-5p mitigates hepatic stellate cell activation and hepatic fibrosis via suppressing TET3 expression
Hepatology International (2023)
-
Role of TET dioxygenases in the regulation of both normal and pathological hematopoiesis
Journal of Experimental & Clinical Cancer Research (2022)