Abstract
Baseline cytogenetic studies at diagnosis remain the single most important determinant of outcome in patients with acute myeloid leukemia (AML). However, the prognostic role of the complete gamut of cytogenetic aberrations in AML patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT) is currently undefined. In addition, their significance in conjunction with FLT3-ITD status has not been addressed thus far. Using the ALWP/EBMT registry we conducted a retrospective analysis to determine the clinical outcomes of AML patients undergoing allo-HSCT with respect to specific recurring cytogenetic abnormalities complemented with FLT3-ITD status. We analyzed a cohort consisting of 8558 adult AML patients who underwent allo-HSCT from either a matched sibling or a matched unrelated donor. Patients with inv(3)(q21q26)/t(3;3)(q21;q26), del(5q), monosomy 7, chromosome 17p abnormalities, t(10;11)(p11-14;q13-23), t(6;11)(q27;q23), as well as those patients with a monosomal or complex karyotype experienced significantly inferior leukemia-free survival (LFS) compared to patients with a normal karyotype. Trisomy 14, del(9q), and loss of chromosome X were associated with improved LFS rates. A novel prognostic model delineating 5 distinct groups incorporating cytogenetic complexity and FLT3-ITD status was constructed with significant prognostic implications. Our analysis supports the added prognostic significance of FLT3-ITD to baseline cytogenetics in patients undergoing allo-HSCT.
Similar content being viewed by others
Introduction
In recent years the field of leukemia has witnessed a remarkable incremental gain in our knowledge of the intricate molecular underpinnings of acute myeloid leukemia (AML) which has allowed for a deeper understanding of the biology and disease process of AML [1,2,3,4,5,6]. Notwithstanding these remarkable scientific breakthroughs, the time-honored evaluation of cytogenetics at initial diagnosis still retains a crucial role in current practice and serves as the benchmark for predicting outcome and dictating the choice of post-remission therapy in AML patients as reflected in the recently published European LeukemiaNet (ELN) recommendations [7, 8]. Indeed, a fundamental premise of the risk stratification schemes employed in the past two decades has been that patients with specifically designated higher risk recurring cytogenetic abnormalities, namely those patients harboring a complex [9, 10] or monosomal [11,12,13] karyotype for example, are referred to transplant at first remission. Conversely, patients with better prognosis karyotypic lesions such as core binding factor (CBF), are typically consolidated with chemotherapy [14, 15]. Whereas the prognostic significance of these commonly occurring cytogenetic aberrations is clearly established, the clinical impact of the full gamut of less commonly recurring chromosomal changes was uncovered by the extensive analyses undertaken by the United Kingdom Medical Research Council (UKMRC) which diligently delineated the specific prognoses effected by the full spectrum of cytogenetic aberrations seen in AML [16]. Furthermore, the UKMRC investigators showed that increasing level of cytogenetic complexity was a robust predictor for worse overall outcome. However, the prognostic role this model assumes in the prognostication of AML patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT) has not been defined thus far. To address this question we paired cytogenetic analysis with comprehensive medical annotation using the large, registry-based cohort of the Acute Leukemia Working Party (ALWP) of the European Society for Blood and Marrow Transplantation (EBMT) multicenter research group. Disease and transplant-related outcomes of the patients were analyzed according to the specific cytogenetic aberration and degree of karyotype complexity coupled with molecular data which was available for a subset of patients.
Methods
Data source
Patient data were obtained from the EBMT multicenter registry. The EBMT is an international research collaborative group comprising over 650 transplant centers that contribute detailed data on consecutive allo-HSCTs to a statistical center at the EBMT study office at the Saint Antoine Hospital (Paris, France). Transplant centers in the EBMT submit patient outcomes data for transplants to the EBMT and undergo annual audits to ensure the integrity, quality, and completeness of the data. The study was approved by the ALWP review board and was performed in compliance with the Helsinki declaration and under guidance of the EBMT Institutional Review Board. All patients provided written informed consent authorizing the use of information for research purposes.
Study population
For this study, we selected adult AML patients undergoing a first allo-HSCT performed in first remission (CR1) and reported to the EBMT between 2006 and 2016. Our final study population consisted of 8558 patients transplanted from matched related donors and HLA matched unrelated donors with full karyotype available. Patients with t(15;17)(q24;q21) were excluded from the analysis. Data were contributed by 288 EBMT transplant centers. Karyotype analysis and assessment of FLT3-ITD status were performed at the time of initial AML diagnosis and were obtained from local labs at the treating medical center.
Working definitions for chromosomal aberrations
Chromosomal abnormalities at diagnosis were assessed according to contemporary standard nomenclature (ISCN 2009, ISCN 2013, Shaffe, Karger publishing). To define the degree of cytogenetic complexity, we followed the methodology used previously by Grimwade et al. [16] whereby balanced translocations, chromosomal monosomies, and chromosomal trisomies were defined as a single abnormality. The gain of 2 chromosomes, unbalanced translocations, and the gain of a derivative chromosome were counted as two abnormalities. A monosomal karyotype was defined as by the presence of at least two autosomal monosomies or of one monosomy plus one or more structural aberrations (not including loss of a chromosome) [11]. A complex karyotype was defined by the presence of more than or equal to 4 chromosomal aberrations [17]. We only included cytogenetic aberrancies present in at least 20 patients.
Statistical analysis
The primary endpoint used for variable selection in this study was leukemia-free survival (LFS) which was defined as survival from the time of transplantation without evidence of disease relapse or progression. Secondary endpoints were relapse incidence (RI), non-relapse mortality (NRM), overall survival (OS), acute graft-versus-host disease (aGVHD) and chronic graft-versus-host-disease (cGVHD), GVHD-free/relapse-free survival (GRFS). Relapse was defined as the presence of 5% BM blasts and/or extramedullary disease. NRM was defined as death without evidence of relapse or progression. OS was defined as the time from allo-HSCT to death, regardless of the cause. GRFS was defined as events including grade 3–4 acute GVHD, extensive chronic GVHD, relapse, or death in the first post-HCT year. Cumulative incidence was used to estimate the endpoints of NRM, RI, acute and chronic to accommodate for competing risks. To study acute and chronic GVHD, we considered relapse and death to be competing events. Probabilities of OS, LFS, and GRFS were calculated using the Kaplan–Meier method. Univariate analyses were done using Gray’s test for cumulative incidence functions and the log-rank test for LFS, OS, and GRFS.
Multivariate analyses were performed using Cox proportional hazards regression for LFS. Stepwise selection methodology (backward and forward) was used for the selection of cytogenetics abnormalities (p ≤ 0.05) adjusted for all other well-known prognostic variables including patient age at transplant, gender, secondary disease, donor type, stem cell source (bone marrow versus peripheral blood), donor/patient cytomegalovirus matching, conditioning intensity, time from diagnosis to HSCT, gender matching (female to male vs other), and in vivo T-cell depletion.
Subsequently, we calculated LFS according to each selected cytogenetic abnormality and their association, and we were able to identify four risk groups according to favorable and adverse cytogenetic abnormalities significantly associated with LFS. Finally, we investigated the impact of fms-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) in the intermediate cytogenetic risk group and a prognostic model integrating cytogenetic aberrancy and FLT3-ITD status was constructed thereby identifying 5 distinct prognostic groups shown in the final EBMT classification. The performance of this classification was compared to the MRC classification in terms of Harrell C index using a non-parametric method developed by Kang et al. [18]. All tests were two-sided with the type I error rate fixed at 0.05 for the determination of factors associated with time-to-event outcomes. Statistical analyses were performed using the SPSS 24.0 (SPSS Inc, Chicago, IL, USA) and R 3.4.1 (R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/)) software packages.
Results
The spectrum of cytogenetic aberrancies seen in transplanted AML patients
Our final cohort consisted of 8558 adult patients with a median age of 52 years (range 18–77) who had received their first allo-HSCT between 2006 and 2016. Baseline clinical and demographic data are shown in Supplementary Table 1. In all, 4530 patients (52.6%) had a normal baseline karyotype. The most frequent abnormalities seen were trisomy 8 (8.8%), monosomy 7 (5.2%), and chromosome 5q abnormalities (4.6%). The complete range of the specific cytogenetic abnormalities of patients in this analysis is outlined in Table 1. A monosomal karyotype was seen in 533 patients (6.2%) whereas 507 patients (5.9%) were diagnosed with a complex karyotype. Of note, of the 76 patients with loss of chromosome X, 16 were male patients and 60 were female patients, and an association with t(8;21) was seen in 20 of these patients (3 males and 17 females). Supplementary Table 2 outlines the degree of chromosomal complexity seen for each given cytogenetic aberrancy.
Association of specific cytogenetic aberrancies and post-transplant outcomes
Next, a multivariate analysis was performed to evaluate the impact of specific karyotypic abnormalities on the rate of LFS. As shown in Table 2, a wide array of cytogenetic abnormalities were associated with adverse outcome with the notable exception of del(9q), trisomy 14, and loss of chromosome X, all of which were significantly associated with better LFS rates. The 2-year outcomes of patients with specific cytogenetic aberrancies are outlined in Supplementary Table 3. We then evaluated whether increasing cytogenetic complexity impacted significantly on disease and transplant-related outcomes. The analysis outlined in Supplementary Table 4, revealed that an increasing number of cytogenetic abnormalities was tightly correlated with inferior rates of relapse incidence, leukemia-free survival, and overall survival. Additionally, patients with monosomal or complex karyotypes also experienced significantly worse disease related outcomes. Of note, patients with inv(16)(p13q22)/t(16;16)(p13;q22), experienced favorable LFS irrespective of the number of concomitant chromosomal abnormalities.
Classification of cytogenetic risk groups in transplanted patients
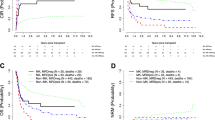
The initial classification of patients into specific prognostic groups was initially carried out by allocating patients into four prognostic groups based on a hazard ratio of less or more than 1 after stepwise selection of the rate of LFS. Patients were thus classified either as favorable risk or poor risk, respectively, while patients with a karyotype consisting of both complex cytogenetics and a monosomal karyotype were classified as very poor (Fig. 1). Thus, the favorable risk group consisted of patients with the following cytogenetic features: deletion of 9q, including add(9q) irrespective of additional cytogenetic abnormalities, trisomy 14 in the absence of a complex karyotype, inv(16)(p13q22)/t(16;16)(p13;q22) irrespective of additional cytogenetic abnormalities, and patients with loss of chromosome X in the absence of a complex karyotype. Patients in the poor risk group were those with the following high risk features: inv(3)(q21q26)/t(3;3)(q21;q26), del(5q), monosomy 7, t(10;11)(p11-14;q13-23), t(6;11)(q27;q23), abnormality of 17p, trisomy 19, and the presence of either a monosomal or complex karyotype.
Patients classified as favorable risk experienced 2 year RI and LFS rates of 17.8 and 72.2%, respectively, sharply contrasting with the very poor outcomes seen in patients with both complex cytogenetics and a monosomal karyotype where the 2 year RI and LFS rates were 66.5 and 18.9%, respectively. The outcome of patients according to the MRC classification and the EBMT classification are outlined in Supplementary Figure 1. Harrell’s C index was used to measure how well the prognostic model discriminated patients with or without the event of interest. The calculated C-indexes were 0.576 [95% CI: 0.564–0.588] and 0.609 [95% CI: 0.597–0.621] for the MRC and the EBMT classifications, respectively. In Supplementary Table 5, a comparison between specific cytogenetic abnormalities and their respective risk designation in the MRC and our classification is provided.
Incorporation of FLT3-ITD mutation status into the prognostic model
Since FLT3-ITD is a pivotal determinant of patient outcome in AML, an additional classification model including FLT3-ITD mutational status was evaluated for the subset of patients with available molecular data (n = 2473). In all, 51% of evaluable patients were FLT3-ITD mutated and as shown in Supplementary Table 2, patients with trisomy 4, t(6;9)(p23;q34), and t(8;21)(q22;q22) were with the highest rate of FLT3-ITD positivity. In this model, five distinct groups in terms of prognosis could be identified (Table 3, Fig. 2). Notably, we found that the presence of FLT3-ITD was significantly associated with outcome only in the intermediate cytogenetic risk group, thus in the expanded 5 group model, we were able to include also the patients from the favorable risk, high risk and very high-risk groups in the expanded classification, resulting in a total number of 5115 patients. The best outcome with regard to RI, LFS, and OS was seen for those patients with either favorable risk cytogenetic abnormalities (n = 489) or intermediate risk cytogenetics and not FLT3-ITD mutated (n = 1749) (2-year LFS rates of 72 and 66%, respectively). Patients mutated for FLT3-ITD with an intermediate-risk karyotype (n = 1476) had poorer outcomes compared to patients with intermediate risk cytogenetics and not harboring the FLT3-ITD mutation (2-year LFS rate of 57%). Finally, patients with poor-risk cytogenetics (n = 1116) experienced significantly worse outcomes (2 year LFS rate of 41%) whereas the least favorable outcomes were seen in the group of patients (n = 285) with the composite of a monosomal and complex karyotype (2 year LFS rate of 19%).
Discussion
Notwithstanding the incorporation of novel innovative molecular risk stratification models into modern prognostication schemas of AML patients [8, 19, 20], baseline cytogenetic studies still serve as an invaluable prognostic tool for prediction of outcome at key clinical time points during the therapeutic sequence of AML patients [7]. Indeed, cytogenetic abnormalities have been previously established as robust predictors of the risk of primary induction resistance [21], risk of relapse [22, 23], and outcome following transplant for primary refractory disease [24]. Yet, to what extent these chromosomal abnormalities impact on patients undergoing allo-HSCT has not been fully addressed hitherto. Herein, we outline the specific clinical outcomes experienced for the full range of the cytogenetic spectrum, and we establish a novel prognostic model incorporating specific cytogenetic aberrations with cytogenetic complexity and FLT3-ITD status.
While in general, our results were consistent with what Grimwade et al. had previously published with the MRC analysis there were several notable differences. Unlike the MRC publication, our analysis indicates that t(3;5), t(11;19), and t(9;22) were not significant in terms of LFS. There are several possible explanations to the different outcome experienced by these patient segments which first and foremost most likely result from the fact that our analysis consisted only of patients which underwent transplant whereas the MRC data analyzed all comers. Furthermore, patients with t(11;19) were more significantly represented in our cohort compared to the MRC analysis; 93 patients with t(11;19) in our registry compared to 30 in the MRC dataset. It is also worth noting that while t(11;19) is an established poor prognostic factor [25, 26], data from the MD Anderson Cancer Center suggests that transplantation had a significant benefit for this patient segment resulting in superior overall survival and relapse-free survival [27]. It is not completely clear to us why patients with t(9;22) and t(3;5), two uncommon cytogenetic presentations of AML [28], did better in our analysis, however it may be possible that transplantation or the increased use of tyrosine kinase inhibitors improved the suboptimal prognosis of these patients. Additionally, we note that a smaller study of 7 patients with t(3;5) suggested these patients had intermediate prognosis [29]. Interestingly and diverging from the MRC data, our findings reveal that trisomy of chromosome 19 is associated with worse outcomes. Isolated trisomy 19 is a rare abnormality that has been reported previously with some cases evolving from antecedent myelodysplasia with no clear prognostic influence [30]. Conversely, in our analysis loss of chromosome X, chromosome 9q deletion and trisomy 14 were found to be correlated with significantly improved clinical outcome in our patient cohort. Deletion of the long arm of chromosome 9 can present as an isolated abnormality with no measurable impact prognosis [31] or in combination with other cytogenetic aberrations such as t(8;21) where according to data from the French CBF-AML intergroup it does negatively impact on overall survival and leukemia-free survival [32]. Loss of chromosome X is a frequent finding in patients with t(8;21) [33] but has not been found to impact on patient outcome in AML. Trisomy 14 was not seen in the MRC analysis and was diagnosed in 53 patients in our cohort, this rare chromosomal abnormality has been reported to occur mostly in elderly male AML patients with a significant component of dysplasia [34]. In accordance with previous literature, we found that outcomes for CBF patients diverged whereby patients with inv(16)(p13q22) experienced superior outcomes compared to patients with t(8;21)(q22;q22). Notably, similar observations were made previously by the Japan Society for Hematopoietic Cell Transplantation group [35], The German-Austrian AML Study Group [6], and more recently by Halaburda and colleagues from the EBMT in CBF patients undergoing transplant in CR2 [36]. Finally, whereas in the 2017 ELN classification t(6;9)(p23;q34.1) is considered an adverse prognostic cytogenetic lesion, our data, supported by the MRC analysis as well as by transplant data from Japan [37], indicate that t(6;9)(p23;q34.1) is prognostically associated with intermediate-risk disease.
As molecular data was available only in a small patient subset of the MRC cohort (n = 215), an additional aim of our analysis was to incorporate FLT3-ITD into our working model to further stratify patient outcome in the intermediate cytogenetic risk group. Our final model which was performed on over 2400 AML patients with available FLT3-ITD data did delineate quite different outcomes for transplanted patients in terms of LFS and RI. Indeed, FLT3-ITD patients experienced a 2-year relapse rate of nearly 30% compared with 19% of patient with FLT3-WT. Thus, and not surprisingly, FLT3-ITD adds an additional important prognostic facet to cytogenetics in the prediction of clinical outcome of transplanted AML patients which is consistent with previous publications in the field [38,39,40].
An extensive body of literature published over the past two decades firmly demonstrates that AML patients with either a monosomal karyotype or a complex karyotype experience significantly worse outcomes compared to patients absent of these cytogenetic features. Furthermore, Haferlach and colleagues suggest that the combination of a monosomal karyotype and complex cytogenetics confers even worse prognosis [41]. Our analysis reaffirms the poor prognosis seen in monosomal karyotype patients previously published by the Center for International Blood and Marrow Transplant Research (CIBMTR) [42] and others [12, 43,44,45,46]. Our data clearly show that an increasing degree of cytogenetic complexity directly translates into an increased risk of relapse and significantly worse leukemia-free survival and overall survival. Indeed, patients with a single karyotypic abnormality had a 2-year relapse rate of 28% compared to the rate of 56% seen in patients with 5 or more chromosomal abnormalities. These findings are in line with the MRC experience as well as with those of the Study Alliance Leukemia who also found that patients with 4 or more cytogenetic abnormalities to have worse prognosis compared to those with only 3 cytogenetic abnormalities [17]. Notably, in an analysis of over 1500 patients enrolled on the ECOG-ACRIN trials, having a monosomal karyotype had no incremental impact on survival of patients harboring a complex karyotype of 5 or more chromosomal abnormalities, thus leading the authors of the study to suggest that the prognostic impact of a monosomal karyotype being limited to patients with less complex karyotypes [47]. Our data also concur with a CIBMTR analysis conducted in 821 adults transplanted AML patients showing that complex karyotype (≥ 4 abnormalities) was still an adverse prognostic factor following transplantation [48].
There are several noteworthy limitations to our study. First, this being a large registry study did not allow for centralized review of cytogenetic studies which may have provided further information such as the distinction between KMT2A-MLLT1 (MLL-ENL) and KMT2A-ELL (MLL-ELL) fusion genes in t(11;19)(q23;q13). In addition, further aspects relating to cytogenetics were not captured by our registry such as clonal heterogeneity which has been shown previously to significantly impact on patient outcome [49].
In conclusion, in this analysis, we define the cytogenetic horizon of AML patients who underwent allo-HSCT during the past decade and delineate the clinical outcome of patients according to baseline cytogenetics studies coupled with FLT-ITD mutational status. We believe the EBMT score can complement the ELN 2017 recommendations for those patients referred to transplant (based on ELN 2017) and further aid in prediction of post-transplant outcomes by providing clinicians a general sense of outcomes. Furthermore, the risk of relapse as predicted by the EBMT score, may possibly influence clinical decision-making regarding the intensity of post-transplant maintenance strategies. This study further underscores the notion that despite the paramount anti-leukemia effect exerted by allogeneic transplantation, baseline cytogenetic studies retain pivotal and lasting prognostic consequences on patient outcome. Ongoing and future trials with FLT3 inhibitors and other targeted agents following transplantation will hopefully mitigate some of the detrimental consequences associated with a baseline adverse risk karyotype and FLT3-ITD.
References
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10.
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150: 264–78.
Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33.
Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–76.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–21.
Cornelissen JJ, Blaise D. Hematopoietic stem cell transplantation for patients with AML in first complete remission. Blood. 2016;127:62–70.
Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47.
Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–83.
Ciurea SO, Labopin M, Socie G, Volin L, Passweg J, Chevallier P, et al. Relapse and survival after transplantation for complex karyotype acute myeloid leukemia: A report from the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation and the University of Texas MD Anderson Cancer Center. Cancer. 2018;124; 2134–41.
Breems DA, Van Putten WL, De Greef GE, Van Zelderen-Bhola SL, Gerssen-Schoorl KB, Mellink CH, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26:4791–7.
Cornelissen JJ, Breems D, van Putten WL, Gratwohl AA, Passweg JR, Pabst T, et al. Comparative analysis of the value of allogeneic hematopoietic stem-cell transplantation in acute myeloid leukemia with monosomal karyotype versus other cytogenetic risk categories. J Clin Oncol. 2012;30:2140–6.
Brands-Nijenhuis AV, Labopin M, Schouten HC, Volin L, Socie G, Cornelissen JJ, et al. Monosomal karyotype as an adverse prognostic factor in patients with acute myeloid leukemia treated with allogeneic hematopoietic stem-cell transplantation in first complete remission: a retrospective survey on behalf of the ALWP of the EBMT. Haematologica. 2016;101:248–55.
Kurosawa S, Miyawaki S, Yamaguchi T, Kanamori H, Sakura T, Moriuchi Y, et al. Prognosis of patients with core binding factor acute myeloid leukemia after first relapse. Haematologica. 2013;98:1525–31.
Hospital MA, Prebet T, Bertoli S, Thomas X, Tavernier E, Braun T, et al. Core-binding factor acute myeloid leukemia in first relapse: a retrospective study from the French AML Intergroup. Blood. 2014;124:1312–9.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65.
Stolzel F, Mohr B, Kramer M, Oelschlagel U, Bochtler T, Berdel WE, et al. Karyotype complexity and prognosis in acute myeloid leukemia. Blood Cancer J. 2016;6:e386.
Kang L, Chen W, Petrick NA, Gallas BD. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med. 2015;34:685–703.
Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016;22:792–9.
Wang M, Lindberg J, Klevebring D, Nilsson C, Mer AS, Rantalainen M, et al. Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling. Leukemia. 2017;31:2029–36.
Herold T, Jurinovic V, Batcha AMN, Bamopoulos SA, Rothenberg-Thurley M, Ksienzyk B, et al. A 29-gene and cytogenetic score for the prediction of resistance to induction treatment in acute myeloid leukemia. Haematologica. 2018;103:456–65.
Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100:4325–36.
Hackl H, Astanina K, Wieser R. Molecular and genetic alterations associated with therapy resistance and relapse of acute myeloid leukemia. J Hematol Oncol. 2017;10:51.
Brissot E, Labopin M, Stelljes M, Ehninger G, Schwerdtfeger R, Finke J, et al. Comparison of matched sibling donors versus unrelated donors in allogeneic stem cell transplantation for primary refractory acute myeloid leukemia: a study on behalf of the Acute Leukemia Working Party of the EBMT. J Hematol Oncol. 2017;10:130.
Krauter J, Wagner K, Schafer I, Marschalek R, Meyer C, Heil G, et al. Prognostic factors in adult patients up to 60 years old with acute myeloid leukemia and translocations of chromosome band 11q23: individual patient data-based meta-analysis of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2009;27:3000–6.
Bhatnagar B, Blachly JS, Kohlschmidt J, Eisfeld AK, Volinia S, Nicolet D, et al. Clinical features and gene- and microRNA-expression patterns in adult acute leukemia patients with t(11;19)(q23; p13.1) and t(11;19)(q23;p13.3). Leukemia. 2016;30:1586–9.
Chen Y, Kantarjian H, Pierce S, Faderl S, O’Brien S, Qiao W, et al. Prognostic significance of 11q23 aberrations in adult acute myeloid leukemia and the role of allogeneic stem cell transplantation. Leukemia. 2013;27:836–42.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Dumezy F, Renneville A, Mayeur-Rousse C, Nibourel O, Labis E, Preudhomme C. Acute myeloid leukemia with translocation t(3;5): new molecular insights. Haematologica. 2013;98:e52–4.
Johansson B, Billstrom R, Mauritzson N, Mitelman F. Trisomy 19 as the sole chromosomal anomaly in hematologic neoplasms. Cancer Genet Cytogenet. 1994;74:62–5.
Peniket AJ. Del(9q) acute myeloid leukaemia: clinical and cytological characteristics and prognostic implications. Br J Haematol. 2005;130:969. author reply
Prebet T, Boissel N, Reutenauer S, Thomas X, Delaunay J, Cahn JY, et al. Acute myeloid leukemia with translocation (8;21) or inversion (16) in elderly patients treated with conventional chemotherapy: a collaborative study of the French CBF-AML intergroup. J Clin Oncol. 2009;27:4747–53.
Kuchenbauer F, Schnittger S, Look T, Gilliland G, Tenen D, Haferlach T, et al. Identification of additional cytogenetic and molecular genetic abnormalities in acute myeloid leukaemia with t(8;21)/AML1-ETO. Br J Haematol. 2006;134:616–9.
Cui W, Bueso-Ramos CE, Yin CC, Sun J, Chen S, Muddasani R, et al. Trisomy 14 as a sole chromosome abnormality is associated with older age, a heterogenous group of myeloid neoplasms with dysplasia, and a wide spectrum of disease progression. J Biomed Biotechnol. 2010;2010:365318.
Kuwatsuka Y, Miyamura K, Suzuki R, Kasai M, Maruta A, Ogawa H, et al. Hematopoietic stem cell transplantation for core binding factor acute myeloid leukemia: t(8;21) and inv(16) represent different clinical outcomes. Blood. 2009;113:2096–103.
The 44th Annual Meeting of the European Society for Blood and Marrow Transplantation: Physicians Oral Session. Bone Marrow Transplantation. 2018; https://doi.org/10.1038/s41409-018-0318-y.
Ishiyama K, Takami A, Kanda Y, Nakao S, Hidaka M, Maeda T, et al. Allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia with t(6;9)(p23; q34) dramatically improves the patient prognosis: a matched-pair analysis. Leukemia. 2012;26:461–4.
Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–18.
Brunet S, Labopin M, Esteve J, Cornelissen J, Socie G, Iori AP, et al. Impact of FLT3 internal tandem duplication on the outcome of related and unrelated hematopoietic transplantation for adult acute myeloid leukemia in first remission: a retrospective analysis. J Clin Oncol. 2012;30:735–41.
Schmid C, Labopin M, Socie G, Daguindau E, Volin L, Huynh A, et al. Outcome of patients with distinct molecular genotypes and cytogenetically normal AML after allogeneic transplantation. Blood. 2015;126:2062–9.
Haferlach C, Alpermann T, Schnittger S, Kern W, Chromik J, Schmid C, et al. Prognostic value of monosomal karyotype in comparison to complex aberrant karyotype in acute myeloid leukemia: a study on 824 cases with aberrant karyotype. Blood. 2012;119:2122–5.
Pasquini MC, Zhang MJ, Medeiros BC, Armand P, Hu ZH, Nishihori T, et al. Hematopoietic cell transplantation outcomes in monosomal karyotype myeloid malignancies. Biol Blood Marrow Transplant. 2016;22:248–57.
Yanada M, Kurosawa S, Yamaguchi T, Yamashita T, Moriuchi Y, Ago H, et al. Prognosis of acute myeloid leukemia harboring monosomal karyotype in patients treated with or without allogeneic hematopoietic cell transplantation after achieving complete remission. Haematologica. 2012;97:915–8.
Poire X, Labopin M, Cornelissen JJ, Volin L, Richard Espiga C, Veelken JH, et al. Outcome of conditioning intensity in acute myeloid leukemia with monosomal karyotype in patients over 45 year-old: A study from the acute leukemia working party (ALWP) of the European group of blood and marrow transplantation (EBMT). Am J Hematol. 2015;90:719–24.
Kayser S, Zucknick M, Dohner K, Krauter J, Kohne CH, Horst HA, et al. Monosomal karyotype in adult acute myeloid leukemia: prognostic impact and outcome after different treatment strategies. Blood. 2012;119:551–8.
Fang M, Storer B, Estey E, Othus M, Zhang L, Sandmaier BM, et al. Outcome of patients with acute myeloid leukemia with monosomal karyotype who undergo hematopoietic cell transplantation. Blood. 2011;118):1490–4.
Strickland SA, Sun Z, Ketterling RP, Cherry AM, Cripe LD, Dewald G, et al. Independent Prognostic Significance Of Monosomy 17 and Impact of Karyotype Complexity in Monosomal Karyotype/Complex Karyotype Acute Myeloid Leukemia: Results from Four ECOG-ACRIN Prospective Therapeutic Trials. Leuk Res. 2017;59:55–64.
Armand P, Kim HT, Zhang MJ, Perez WS, Dal Cin PS, Klumpp TR, et al. Classifying cytogenetics in patients with acute myelogenous leukemia in complete remission undergoing allogeneic transplantation: a Center for International Blood and Marrow Transplant Research study. Biol Blood Marrow Transplant. 2012;18:280–8.
Medeiros BC, Othus M, Fang M, Appelbaum FR, Erba HP. Cytogenetic heterogeneity negatively impacts outcomes in patients with acute myeloid leukemia. Haematologica. 2015;100:331–5.
Acknowledgements
We thank all the European Group for Blood and Marrow Transplantation (EBMT) centers and national registries for contributing patients to the study and data managers for their excellent work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Canaani, J., Labopin, M., Itälä-Remes, M. et al. Prognostic significance of recurring chromosomal abnormalities in transplanted patients with acute myeloid leukemia. Leukemia 33, 1944–1952 (2019). https://doi.org/10.1038/s41375-019-0439-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0439-3
- Springer Nature Limited
This article is cited by
-
Matched related versus unrelated versus haploidentical donors for allogeneic transplantation in AML patients achieving first complete remission after two induction courses: a study from the ALWP/EBMT
Bone Marrow Transplantation (2023)
-
Non-T depleted haploidentical stem cell transplantation in AML patients achieving first complete remission after one versus two induction courses: a study from the ALWP/EBMT
Bone Marrow Transplantation (2022)
-
Cytogenetics analysis as the central point of genetic testing in acute myeloid leukemia (AML): a laboratory perspective for clinical applications
Clinical and Experimental Medicine (2022)
-
Indications for haematopoietic cell transplantation for haematological diseases, solid tumours and immune disorders: current practice in Europe, 2022
Bone Marrow Transplantation (2022)
-
Outcome of human umbilical cord blood stem cell transplantation (CBT) for acute myeloid leukemia in patients achieving first complete remission after one versus two induction courses: a study from the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation (EBMT)
Bone Marrow Transplantation (2022)