Abstract
Background
Maternal pre-pregnancy body mass index (BMI) has been linked to altered gut microbiota in women shortly after delivery and in their offspring in the first few years of life. But little is known about how long these differences persist.
Methods
We followed 180 mothers and children from pregnancy until 5-year postpartum in the Gen3G cohort (Canada, enrolled 2010–2013). At 5 years postpartum we collected stool samples from mothers and children and estimated the gut microbiota by 16 S rRNA sequencing (V4 region) using Illumina MiSeq, and assigning amplicon sequence variants (ASV). We examined whether overall microbiota composition (as measured by microbiota β diversity) was more similar between mother-child pairs compared to between mothers or between children. We also assessed whether mother-child pair sharing of overall microbiota composition differed by the weight status of mothers before pregnancy and of children at 5-year. Furthermore, in mothers, we examined whether pre-pregnancy BMI, BMI 5-year postpartum, and change in BMI between time points was associated with maternal gut microbiota 5-year postpartum. In children, we further examined associations of maternal pre-pregnancy BMI and child 5-year BMI z-score with child 5-year gut microbiota.
Results
Mother-child pairs had greater similarity in overall microbiome composition compared to between mothers and between children. In mothers, higher pre-pregnancy BMI and 5-year postpartum BMI were associated with lower microbiota observed ASV richness and Chao 1 index; in children’s gut microbiota, higher maternal pre-pregnancy BMI was weakly associated with lower microbiota Shannon index, whereas child’s 5-year BMI z-score was associated with higher observed ASV richness. Pre-pregnancy BMI was also linked to differential abundances of several microbial ASVs in the Ruminococcaceae and Lachnospiraceae families, but no specific ASV had overlapping associations with BMI measures in both mothers and children.
Conclusions
Pre-pregnancy BMI was associated with gut microbiota diversity and composition of mothers and children 5 years after birth, however, the nature and direction of most associations differed for mothers and children. Future studies are encouraged to confirm our findings and look into potential mechanisms or factors that may drive these associations.
Similar content being viewed by others
Introduction
Pre-pregnancy overweight or obesity (OW/OB) is associated with adverse health outcomes in both mothers and offspring. Women who are OW/OB before pregnancy have increased risks of pregnancy complications, including early pregnancy loss, hypertensive disorders, gestational diabetes, and post-partum hemorrhage [1, 2]. Offspring of mothers who are OW/OB are at elevated risks of adverse health consequences in metabolic and neurodevelopmental systems [1, 3, 4]. However, the etiologic underpinnings of these associations are still poorly understood. One hypothesis is that the effects may be mediated by an altered gut microbiome in both mothers and offspring [5,6,7]. However, although emerging evidence supports mother-to-infant vertical transmission of microbes [8,9,10,11,12,13,14,15,16], little is known about whether mother-offspring microbe sharing persists beyond infancy and if pre-pregnancy maternal weight status impacts mother-child microbial sharing.
Epidemiological studies have linked maternal pre-pregnancy body mass index (BMI) to altered gut microbiota structure and/or composition, separately for mothers during pregnancy and early postpartum [17,18,19], and for their offspring in early life [17,18,19,20,21]. However, it remains to be understood how long these differences persist beyond the first years postpartum because only a few studies followed mothers and offspring beyond the first years after birth. For example, Galley et al. [22] found that in 77 children aged 18 to 27 months in the U.S., those who were born to mothers of higher socioeconomic status with pre-pregnancy obesity versus those with normal pre-pregnancy weight had a gut microbiota profile with higher α diversity, altered beta diversity, higher relative abundances of genera Faecalibacterium and Oscillibacter, and lower relative abundances of Eubacterium and Blautia. In contrast, in another study of 169 children in Norway, maternal pre-pregnancy BMI was not associated with gut microbiota diversity or composition from birth to two years old [18]. To our knowledge, there have been no studies to simultaneously examine the microbiome mother-child dyads. As such, there is a need for studies on this topic, particularly studies that investigate the longer-term impacts of pre-pregnancy BMI on maternal and children’s gut microbiome.
Our study aimed to address this research gap in a longitudinal birth cohort of mothers and children in Canada that have been followed from early pregnancy until 5-year postpartum. Our specific research questions were (1) at 5-year postpartum, did related mother-child pairs have a similar gut microbiota and was the similarity impacted by their weight status? (2) in mothers, was BMI measured prior to pregnancy, at 5-year postpartum, as well as its change associated with maternal microbiota at 5-year postpartum? and (3) in children, was maternal pre-pregnancy BMI and child’s 5-year BMI associated with child’s microbiota at 5-year after birth?
Materials and methods
Study population
We used data from the Genetics of Glucose regulation in Gestation and Growth (Gen3G) cohort. The Gen3G cohort is a prospective observational cohort that recruited pregnant women, representative of the general population of women of reproductive age, from the Estrie region in Québec, Canada [23]. Women were eligible to participate in the Gen3G cohort from 2010 to 2013 if they were between 5 and 16 weeks of gestation, older than 18 years of age, and did not have a history of diabetes. We prospectively followed women throughout pregnancy, at delivery, and postnatally until 5 years. We also followed their children from birth to 5 years of age. At the 5-year follow-up visit, Gen3G staff invited families to collect stool samples at home from mothers and children participants who did not use antibiotics within three months prior to the stool collection. For this study, we included 180 mothers and children that returned their stool samples and mothers reported their pre-pregnancy weight at enrollment in early pregnancy. All study protocols were approved by the ethics review board from the Centre Hospitalier Universitaire de Sherbrooke.
Measurements of maternal and child’s BMI
At enrollment, we collected women’s self-reported weight before pregnancy using standardized questionnaires. Research staff measured maternal height (to the near 0.5 cm) without shoes at enrollment using a wall stadiometer. During the follow-up visit at 5 years postpartum, trained research staff measured women’s weight (in kg) with a calibrated electronic scale with bare feet in light clothing. We calculated BMI as weight divided by squared height. We defined mothers with pre-pregnancy BMI ≥ 25 kg/m2 as having OW/OB. We further derived maternal BMI change as the difference between pre-pregnancy BMI and 5-year BMI. For all children presenting to the 5-year visit, we measured weight (in kg) using an electronic scale (Rice Lake Weighing systems), and measured height (in cm) using a calibrated stadiometer (Seca). We calculated child’s BMI z-score adjusted for sex and age using WHO AnthroPlus software based on WHO growth reference [24]. We defined children with BMI z-score ≥85th percentile as having OW/OB.
Microbial sampling and 16 s rRNA sequencing
We collected stool samples from both mothers and children at 5 years after delivery (henceforth referred to as the 5-year microbiome) [25]. We used the OMNIgene•Gut OM-200 collection tube from DNA Genotek (Canada), which stabilizes microbial DNA at the point of collection. We then sent the DNA Genotek tubes to the processing center within 2–3 days of collection, and the samples were frozen at −80 °C within 24 h of receipt. After thawing the stool samples, we extracted microbial DNA using the QIAamp Fast DNA stool mini-kit (QIAGEN) according to the manufacturer’s protocol.
We amplified the V4 hypervariable region of the 16 S rRNA gene using the 515 F (5’-GTGCCAGCMGCCGCGGTAA-3’) and 806 R (5’-GGACTACHVGGGTWTCTAAT-3’) primer pair initially designed by Caporaso et al. [26]. We performed multiplex sequencing using the Illumina MiSeq platform according to the protocol described by Kozich et al. [27] to obtain microbial information from the fecal samples. After demultiplexing using the sample-specific barcodes described by Kozich et al. [27], we performed quality read control and assigned amplicon sequence variants (ASV) using the R package DADA2 (v1.8.0) [28]. We trimmed forward reads at positions 10 (5’ end) and 240 (3’ end) and reverse reads at positions 10 (5’ end) and 210 (3’ end). We removed the reads predicted to have >2 expected errors based on quality scores, containing any ambiguous bases, or mapping to the PhiX genome using the function “filterAndTrim”, as recommended by DADA2 authors [28, 29].
We used a random subset of 103,129,700 bases from 448,390 reads in 17 samples to calculate error estimate for the forward reads, and 105,583,800 bases from 527,919 reads in 20 samples for the reverse reads using the function “learnErrors” in DADA2 [28]. We randomly included samples in models until 100,000,000 reads were reached. We dereplicated and pooled forward and reverse reads, denoised forward reads, and their reverse-complement, and merged the sequence pairs using default parameters in DADA2 [28]. With 86% of merged reads being non-chimeric, we removed chimeras from merged sequences using the pooled option in function “removeBimeraDenovo” in DADA2 [28]. This led to a final set of ASVs containing 78% of the raw reads (83% of filtered reads).
ASV taxonomic assignment and phylogenetic tree generation
We used the “assignTaxonomy” function in DADA2 with the HITdb v.1.00 16 S rRNA sequence database to classify gut microbial taxa [30]. Following recommendations by Murali et al. [31], we aligned ASVs using the R package DECIPHER to construct a preliminary neighbor-joining tree. We used the neighbor-joining tree to calculate a Generalized time-reversible with Gamma rate variation maximum likelihood phylogenetic tree rooted at the midpoint, using the R package phangorn [29, 32, 33]. We then combined the sample metadata, ASVs, taxonomy, and phylogenetic tree using the R package phyloseq [34].
Measurements of covariates
At enrollment in early pregnancy, we collected maternal age, race, education achievement, prior medical/obstetric history, and lifestyle habits (smoking status: never, past, current) using standardized questionnaires. At delivery, we collected offspring sex, birth weight, and any pregnancy/delivery adverse outcomes from electronic medical records. At the 5-year post-partum visit, mothers completed a brief diet survey (adapted from Canadian Community Health Surveys) from which we derived maternal and child intake of fruits and vegetables (servings per day). Mother reported child breastfeeding duration of any type (i.e., exclusive or mixed breastfeeding) at 5 years post-partum visits.
Statistical analysis
We estimated α and β diversity and ASV relative abundance of the 5-year gut microbiota of mothers and children. We used Shannon index, Chao 1 index, and observed unique ASVs to estimate α diversity (within-person diversity) via function “diversity” from vegan package [35] without rarefying the data. We used Bray-Curtis, weighted and unweighted UniFrac [36] distances as measures of β diversity (between-person diversity) after rarefying the data to the lowest sequencing depth. We calculated relative abundances at the ASV level.
We assessed the overall microbiota similarity by β diversity metrics. We examined the differences in microbiota similarity between pairs of mother-own child, mother-unrelated child, mother-mother, and child-child using Wilcoxon rank sum tests on the average β diversity distance metrics within each pair. Further, within mother-own child pairs, we created 4 groups according to the weight status of mothers prior to pregnancy and children at 5-year as follows: mothers and children both having normal weight, mothers and children both having OW/OB, mothers having normal weight but children having OW/OB, and mothers having OW/OB but children having normal weight. We combined OW and OB as a single weight status to ensure reasonable sample size for statistical inference within each grouping. We compared microbiota similarity across the four groups based on the diagonal values of each β diversity metric that represents the distance between mother and own child.
We conducted analyses to examine associations of BMI measures with 5-year microbiota separately for mothers and children. In mothers, we examined associations of (a) maternal pre-pregnancy BMI, (b) 5-year BMI, and (c) BMI change with maternal 5-year microbiota. In children, we examined associations of (d) maternal pre-pregnancy BMI and (e) child 5-year BMI z with child 5-year microbiota. We modeled all BMI measures by tertiles to facilitate comparisons between maternal and child specific associations and across the different BMI measures.
We used linear regression for the examination of BMI measures and α diversity and plotted the associations based on restricted cubic spline regressions. We used permutational multivariate analysis of variance (PERMANOVA) with 999 permutations for the examination of BMI measures and β diversity using function “adonis2” from vegan package [35]. We then used principal coordinates analysis (PCoA) to visualize the dissimilarity in β diversity metrics according to BMI tertiles. Finally, we applied analysis of composition of microbiomes with bias correction (ANCOM-BC) to identify individual microbial taxa that were differentially abundant (in log scale) according to BMI measures using package ANCOMBC [37]. In differential abundance analysis, we filtered out taxa with mean relative abundances of ≤0.5% in mothers and children, separately. We used a Benjamini-Hochberg false discovery rate (FDR) adjusted P < 0.05 to denote statistical significance for differential abundance analysis [38], and a two-sided P < 0.05 otherwise.
We ran regression models before and after inclusion of covariates considered to be potential confounders or prognostic factors. We considered covariates as confounders in our models if they are hypothesized to have an association with the exposure (pre-pregnancy BMI or 5-year BMI for mothers or children) and the 5-year microbiota for mothers or children. Model 1 included maternal age at 5 years postpartum and parity; Model 2 additionally included fruits and vegetables intake (in both mother and child models; servings per day) and breastfeeding duration (in child models only; months). When maternal BMI change was the exposure variable in mother models, we constructed a Model 3 that additionally included maternal pre-pregnancy BMI as a covariate. When pre-pregnancy BMI was the exposure variable in child models, Model 3 additionally included child sex and age.
Results
Participant characteristics
Of the 180 mothers included in our study, 97.2% (n = 175) were white and 55.0% (n = 99) had a college degree or higher. Mothers had a mean BMI of 24.5 kg/m2 (SD: 5.4) prior to pregnancy and 25.6 kg/m2 (SD: 5.9) at 5-year postpartum (Table 1). In total, 48.3% (n = 87) of the mothers were nulliparous, 33.9% (n = 61) ever smoked prior to pregnancy, and their mean age was 30.1 years (SD: 4.1) at delivery. Mothers reported that they consumed on average 2.4 servings of fruits and vegetables per day at 5-year postpartum. Of the children contributing data to this study, 57.2% (n = 103) were male and 81.7% (n = 147) were vaginally delivered. At birth, their mean gestational age was 39.2 weeks (SD: 1.3) and their mean weight was 3.4 kg (SD: 0.5). When the children were 5 years old, on average they had a BMI of 15.7 kg/m2 (SD: 1.5), a BMI z-score of 0.2 (SD: 0.9), a history of breastfeeding duration of 8.6 months (SD: 6.4), and consumed 2.5 servings of fruits and vegetables per day (SD: 0.7).
5-year microbiota similarity between mothers and children
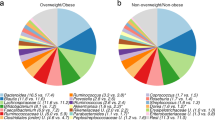
At 5-year postpartum, the overall microbiota composition between pairs of mothers and their children was more similar than the microbiota between pairs of mother and unrelated child, pairs of mothers, and pairs of children (Fig. 1 panel A). This observation was held regardless of the choice of β diversity metrics. Moreover, within pairs of mothers and their own children, 5-year microbiota β diversity did not differ according to the weight status of mothers prior to pregnancy and children at 5-year old (Fig. 1 panel B).
BMI measures and microbiota α diversity
For mothers, higher pre-pregnancy BMI, 5-year postpartum BMI, and greater between time point BMI increase tended to be associated with lower 5-year postpartum microbiota α diversity metrics (Supplement Fig. 1 panel A). Specifically, pre-pregnancy BMI and BMI change had non-linear associations with ASV richness, with the 2nd tertiles of each having a higher number of observed ASVs (i.e., richness) and the 3rd tertile had a lower number of observed ASVs compared to the 1st tertile (Table 2). Maternal 5-year BMI was associated with ASV richness in a relatively linear fashion, such that each 1-SD increment was associated with a −3.79 (95% CI: −5.61, −1.97) lower observed ASV richness after full adjustment. Although the associations of BMI measures with Chao 1 and Shannon indices were relatively linear, they were not statistically significant.
In the gut microbiota of children at 5 years, maternal pre-pregnancy BMI and child 5-year BMI z-score tended to be positively associated with microbiota observed ASV richness, but negatively associated with Chao 1 index and Shannon index in an overall non-linear fashion (Supplementary Fig. 1 panel B). We also detected statistically significant associations between pre-pregnancy BMI and Shannon index (Table 3). As compared with children born to mothers being in the 1st tertile of pre-pregnancy BMI, those born to mothers being in the 2nd and 3rd tertiles had a −0.31 (−0.51, −0.11) and −0.22 (−0.41, −0.02) lower Shannon index, respectively. In contrast, we detected a nominal association between higher child 5-year BMI z-score and higher observed ASVs: each 1-SD increment in child 5-year BMI z-score was associated with a 10.50 (1.92, 19.09) higher number of observed ASVs.
BMI measures and microbiota β diversity
None of the BMI measures in mothers or children were associated with β diversity metrics of 5-year gut microbiota in mothers (Supplementary Fig. 2) or children (Supplementary Fig. 3). Furthermore, maternal pre-pregnancy BMI, 5-year postpartum BMI, and BMI change explained a similar percentage of variance in maternal 5-year postpartum gut microbiota β diversity metrics (Fig. 2). In children microbiota analyses, maternal pre-pregnancy BMI explained a similar amount of variance in child gut microbiota β diversity metrics as child 5-year BMI z-score (Fig. 2).
BMI measures and microbiota composition
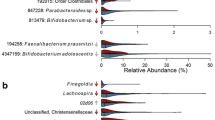
In the gut microbiota of mothers 5 years postpartum, 20 ASVs were differentially abundant according to pre-pregnancy BMI, 28 ASVs were differentially abundant according to 5-year postpartum BMI, and 23 ASVs were differentially abundant according to BMI change between those two time points (all FDR-adjusted P < 0.05; Fig. 3). Pre-pregnancy BMI and 5-year BMI were associated with 5 ASVs in the same direction (Fig. 3 highlighted in light grey): specifically, they were associated with lower relative abundances of Ruminococcaceae Ruminococcus (species unknown), Lachnospiraceae Coprococcus (sp. unknown), Lachnospiraceae Blautia (sp. unknown), and Peptostreptococcaceae (genus, sp. unknown), and higher relative abundance of Veillonellaceae Megasphaera (sp. unknown). Similarly, 5-year BMI and BMI change were associated with another 8 ASVs in the same direction (Fig. 3 highlighted in dark grey), including lower abundances of Ruminococcaceae Ruminococcus (sp. unknown), Ruminococcaceae (g., sp. unknown), Eubacteriaceae Eubacterium (sp. unknown), Pasteurellaceae Aggregatibacter (sp. unknown), and higher abundance of Lachnospiraceae Dorea (sp. unknown).
In gut microbiota of the children at 5 years, pre-pregnancy BMI was associated with 10 ASVs and child’s 5-year BMI z-score was associated with 8 ASVs (all FDR-adjusted P < 0.05; Fig. 3). Most of these associations were with ASVs in the families of Ruminococcaceae and Lachnospiraceae. There were 2 ASVs in the Ruminococcaceae family that were positively associated with both pre-pregnancy BMI and child 5-year BMI, but the genus and species were unclassified (Fig. 3 highlighted in light blue). In addition, though belonging to different species, three additional ASVs in the Faecalibacterium genus were also positively associated with pre-pregnancy BMI and child 5-year BMI.
Discussion
In the Gen3G pre-birth cohort, mothers and their children shared a more similar gut microbiota profile than non-related participants (mothers or children) at 5-year postpartum, and the mother-child pair microbiota similarity was not modified by mother or child weight status. In mothers, higher pre-pregnancy BMI, higher 5-year BMI, and increase in BMI were each associated with lower richness as well as differential abundances of many taxa in the families of Ruminococcaceae and Lachnospiraceae, measured at 5-year postpartum. In children at 5-year of age, maternal pre-pregnancy BMI was associated with lower Shannon index, whereas child’s 5-year BMI z-score was associated with higher ASV richness. Several taxa in the Ruminococcaceae family of child microbiota were associated with both maternal pre-pregnancy BMI and child 5-year BMI z-score consistently in the positive direction, suggesting these microbial taxa could play a role in the intergenerational association of obesity.
Our study uniquely complements prior studies [8,9,10,11,12,13,14,15,16] of mother-infant microbe sharing by providing evidence that at 5-year postpartum, mothers and their children still had a more similar gut microbiota profile than unrelated individuals. Our findings align with the hypothesis that gut microbes can be transmitted or co-acquired within mother-child pairs due to shared environmental and/or genetic factors. However, our study does not provide direct evidence that the microbes shared between mother and child are indeed the same because we used 16 S rRNA amplicon sequencing. Furthermore, in our study, the 5-year microbiota similarity of mother-own child pairs did not differ according to their obese phenotypes. This merits further investigation, ideally one that assesses the impacts of pre-pregnancy BMI on mother-child microbe sharing longitudinally using repeated measurements of both maternal and child gut microbiota from birth onward.
Our study adds to a limited number of studies that have examined the association of pre-pregnancy BMI with maternal gut microbiota years after pregnancy. Most of the previous studies that have examined maternal BMI with gut microbiota were conducted cross-sectionally without establishing temporality [39], or focused on microbiota measured during pregnancy, as summarized by three review articles published in recent years [17, 19, 21]. In the study that measured gut microbiota in 169 Norwegian women at 4 days postpartum, higher pre-pregnancy BMI was associated with a lower number of observed species, which was consistent with our study [18]. But the association became non-significant after adjusting for maternal age, education, Norwegian ethnicity, parity, twins, and smoking during pregnancy. The authors also detected a significant association with Shannon index that remained after adjustment [18]. In contrast, we did not find that Shannon index was associated with pre-pregnancy BMI. Regarding differentially abundant taxa, both our study and the Norwegian study found that higher pre-pregnancy BMI was associated with lower abundances of taxa in the family Ruminococcaceae (especially in the genus Ruminococcus), but our study and their study findings did not agree on the directions of associations (negative in our study whereas positive in the Norwegian study) with taxa in the family Lachnospiraceae (especially in the genus Blautia). More studies are needed to investigate the long-term impacts of pre-pregnancy BMI on the postpartum gut microbiota of mothers.
Our findings that pre-pregnancy BMI, 5-year BMI, as well as BMI change between the two timepoints, were associated with maternal 5-year microbiota composition in similar patterns suggest that certain microbial taxa could be obesity-related markers. Lachnospiraceae Dorea (sp. unknown), Lachnospiraceae Blautia (sp. unknown), and some taxa in the family Ruminococcaceae that were consistently associated with BMI measures in our study have also been reported by studies of non-pregnanct populations [40, 41]. The observation in our study that change in weight may itself lead to altered microbiome profiles has also been found in several weight-loss intervention studies [42,43,44,45]. Conversely, fecal microbial transfer experiments in germ-free murine models support the hypothesis that the direction of association goes from alterations in gut microbiome composition to changes in weight [46,47,48,49]. Taken together, our findings support the arguments that the interplay between weight and microbiota is complex and could be bidirectional [50].
Our study on the associations between maternal pre-pregnancy BMI and child 5-year gut microbiota extends previous investigations that usually did not measure offspring gut microbiota beyond infancy. According to a recent meta-analysis, maternal pre-pregnancy overweight or obesity was barely associated with Shannon index of infant gut microbiota (−0.01; 95% CI: −0.19, 0.17) with considerable heterogeneity across studies [21]. We also detected a weak association in a small magnitude between maternal pre-pregnancy BMI and 5 year child gut microbiota Shannon index. Also consistent with a prior study that measured gut microbiota in toddlers (aged 18 to 27 months) [22], we found that higher pre-pregnancy BMI was associated with a lower abundance of genus Blautia (sp. unknown) and a higher abundance of genus Faecalibacterium (sp. unknown) in the 5-year child gut microbiota. Taken collectively, findings in our study join findings from prior infant studies [19, 21] to suggest that maternal pre-pregnancy BMI could have not only transient but also long-lasting impacts on child gut microbiota from birth to early childhood. Given that we did not detect similar associations of pre-pregnancy BMI with genera Blautia (sp. unknown) and Faecalibacterium (sp. unknown) in maternal microbiota, the observed long-lasting associations of pre-pregnancy BMI with these two taxa in child microbiota are unlikely to be explained by shared environmental or genetic factors between mothers and their offspring.
Similarly, although both maternal pre-pregnancy BMI and child 5-year BMI z-score had positive associations with several taxa in the 5-year child gut microbiota within the family Ruminococcaceae, including three species of genus Faecalibacterium, these taxa in the maternal gut microbiota were not associated with any of maternal BMI measures. It is possible that these taxa are specifically linked to BMI in children (but not in mothers) and that it is due to a correlation between child and maternal BMI that we concurrently found these taxa in children (but not in mothers) also linked to maternal pre-pregnancy BMI. Another plausible explanation is that higher pre-pregnancy BMI impacts in-utero fetal programming [51, 52] and breast milk components such as microbes and fatty acids [53, 54], which predisposes offspring to higher BMI and possibly more Faecalibacterium. Future studies with both maternal and children’s microbiome measures are encouraged to confirm the child-specific associations of Ruminococcaceae Faecalibacterium with maternal and child BMI and look into possible physiological explanations.
The complex associations between weight and gut microbiota, as well as the contribution of specific microbial taxa to other health outcomes are yet to be fully understood. It has been speculated that several taxa of families Ruminococcaceae and Lachnospiraceae link to BMI through physiological mechanisms including the production of short-chain fatty acids and inflammation. For example, Dorea and Blautia are considered pathological bacteria, under some conditions, with pro-inflammatory effects and thus could be contribute to inflammation-driven obesity. On the other hand, Ruminococcus and Faecalibacterium can produce butyrate (a short-chain fatty acid), which is involved in intestinal homeostasis and energy metabolism, with controversial effects on obesity development [55]. It should be noted that the health effects of microbes may vary across species of the same genus and can be influenced by the coexistence of other microbes and host factors such as general health and diet [56, 57]. Much needs to be done to elucidate causality, direction, and mechanisms of the association between BMI and gut microbiota to facilitate the development of microbiota-focused interventions in combating the obesity pandemic [58].
Strengths of our study include the long follow-up that enabled the investigation of potential long-lasting links between pre-pregnancy BMI on both maternal and children’s gut microbiota. We also adjusted for a variety of confounding variables, including breastfeeding and fruit and vegetable intake, to enhance the robustness of our findings. Furthermore, the relatively large sample size afforded us reasonable statistical power when controlling for confounding. Our study has several limitations. First, we are unable to perform a microbial strain tracing analysis to determine the potential impacts of pre-pregnancy BMI on microbial transmission from mothers to children because the maternal microbiome during pregnancy was not available in Gen3G, and because we did not have whole genome shotgun metagenomic sequencing data. Second, we only measured gut microbiota at one time point; having multiple measures including during pregnancy and within a few years after delivery could have afforded more insights into the dynamic changes in gut microbiota. Third, we sequenced microbiota via 16S rRNA sequencing, which brings the possibility of misclassification of the taxonomic assignment and excludes the examination of microorganisms other than bacteria and archaea. Fourth, we cannot rule out the possibility of residual or unmeasured confounding that may have biased the observed associations. Fifth, we were not able to perform the primary analysis based on the weight classification (normal weight, OW, OB) because of the relatively small proportions of mothers and children being in the OW or OB categories in our study.
In conclusion, our study addressed a knowledge gap by showing that maternal pre-pregnancy BMI was associated with the gut microbiota of mothers and their children until at least 5-year postpartum. Given that most of the differentially abundant ASVs in relation to BMI were not overlapping between mothers and children, it is unlikely that shared genetic or environmental factors explain our findings. Future studies are warranted to meticulously measure maternal and children’s gut microbiota over multiple time points, from pregnancy onward, to better unravel both the short- and long-term impacts of pre-pregnancy BMI.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
Vats H, Saxena R, Sachdeva MP, Walia GK, Gupta V. Impact of maternal pre-pregnancy body mass index on maternal, fetal and neonatal adverse outcomes in the worldwide populations: a systematic review and meta-analysis. Obes Res Clin Pract. 2021;15:536–45.
Poston L, Caleyachetty R, Cnattingius S, Corvalan C, Uauy R, Herring S, et al. Preconceptional and maternal obesity: epidemiology and health consequences. Lancet Diabetes Endocrinol. 2016;4:1025–36.
Sanchez CE, Barry C, Sabhlok A, Russell K, Majors A, Kollins SH, et al. Maternal pre-pregnancy obesity and child neurodevelopmental outcomes: a meta-analysis. Obes Rev. 2018;19:464–84.
Wang H, Zhang Z, Liu Y, Yang J, Zhang J, Clark C, et al. Pre-pregnancy body mass index in mothers, birth weight and the risk of type I diabetes in their offspring: a dose-response meta-analysis of cohort studies. J Gynecol Obstet Hum Reprod. 2021;50:101921.
Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–5.
Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42.
Calatayud M, Koren O, Collado MC. Maternal microbiome and metabolic health program microbiome development and health of the offspring. Trends Endocrinol Metab. 2019;30:735–44.
Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–45.e5.
Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, et al. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018;28:561–8.
Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018;24:146–54.e4.
Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–21.
Korpela K, Helve O, Kolho KL, Saisto T, Skogberg K, Dikareva E, et al. Maternal fecal microbiota transplantation in cesarean-born infants rapidly restores normal gut microbial development: a proof-of-concept study. Cell. 2020;183:324–34.e5.
Li W, Tapiainen T, Brinkac L, Lorenzi HA, Moncera K, Tejesvi MV, et al. Vertical transmission of gut microbiome and antimicrobial resistance genes in infants exposed to antibiotics at birth. J Infect Dis. 2021;224:1236–46.
Song SJ, Wang J, Martino C, Jiang L, Thompson WK, Shenhav L, et al. Naturalization of the microbiota developmental trajectory of Cesarean-born neonates after vaginal seeding. Med (N Y). 2021;2:951–64.e5.
Wilson BC, Butler EM, Grigg CP, Derraik JGB, Chiavaroli V, Walker N, et al. Oral administration of maternal vaginal microbes at birth to restore gut microbiome development in infants born by caesarean section: a pilot randomised placebo-controlled trial. EBioMedicine. 2021;69:103443.
Valles-Colomer M, Bacigalupe R, Vieira-Silva S, Suzuki S, Darzi Y, Tito RY, et al. Variation and transmission of the human gut microbiota across multiple familial generations. Nat Microbiol. 2022;7:87–96.
Singh S, Karagas MR, Mueller NT. Charting the maternal and infant microbiome: what is the role of diabetes and obesity in pregnancy? Curr Diab Rep. 2017;17:11.
Stanislawski MA, Dabelea D, Wagner BD, Sontag MK, Lozupone CA, Eggesbo M. Pre-pregnancy weight, gestational weight gain, and the gut microbiota of mothers and their infants. Microbiome. 2017;5:113.
Dreisbach C, Prescott S, Alhusen J. Influence of maternal prepregnancy obesity and excessive gestational weight gain on maternal and child gastrointestinal microbiome composition: a systematic review. Biol Res Nurs. 2020;22:114–25.
Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, Becker AB, et al. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. Jama Pediatr. 2018;172:368–77.
Grech A, Collins CE, Holmes A, Lal R, Duncanson K, Taylor R, et al. Maternal exposures and the infant gut microbiome: a systematic review with meta-analysis. Gut Microbes. 2021;13:1–30.
Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One. 2014;9:e113026.
Guillemette L, Allard C, Lacroix M, Patenaude J, Battista MC, Doyon M, et al. Genetics of glucose regulation in gestation and growth (Gen3G): a prospective prebirth cohort of mother-child pairs in Sherbrooke, Canada. BMJ Open. 2016;6:e010031.
de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85:660–7.
Differding MK, Doyon M, Bouchard L, Perron P, Guerin R, Asselin C, et al. Potential interaction between timing of infant complementary feeding and breastfeeding duration in determination of early childhood gut microbiota composition and BMI. Pediatr Obes. 2020;15:e12642.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 2012;6:1621–4.
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res. 2016;5:1492.
Ritari J, Salojarvi J, Lahti L, de Vos WM. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genomics. 2015;16:1056.
Murali A, Bhargava A, Wright ES. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome. 2018;6:140.
Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3.
Wright ES, Yilmaz LS, Noguera DR. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microb. 2012;78:717–25.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217.
Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30.
Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35.
Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat Commun. 2020;11:3514.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Series B (Methodological). 1995;57:289–300.
Haddad EN, Ferro LE, Russell KE, Sugino KY, Kerver JM, Comstock SS. Fecal bacterial communities differ by lactation status in postpartum women and their infants. J Hum Lactation. 2022;38:270–80.
Zeng Q, Li D, He Y, Li Y, Yang Z, Zhao X, et al. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci Rep. 2019;9:13424.
Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow STE, et al. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol. 2018;2018:4095789.
Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23:859–68.
Lee CJ, Florea L, Sears CL, Maruthur N, Potter JJ, Schweitzer M, et al. Changes in gut microbiome after bariatric surgery versus medical weight loss in a pilot randomized trial. Obes Surg. 2019;29:3239–45.
Shen N, Caixas A, Ahlers M, Patel K, Gao Z, Dutia R, et al. Longitudinal changes of microbiome composition and microbial metabolomics after surgical weight loss in individuals with obesity. Surg Obes Relat Dis. 2019;15:1367–73.
Behari J, Graham L, Wang R, Schirda C, Borhani AA, Methe BA, et al. Dynamics of hepatic steatosis resolution and changes in gut microbiome with weight loss in nonalcoholic fatty liver disease. Obes Sci Pract. 2021;7:217–25.
Kulecka M, Paziewska A, Zeber-Lubecka N, Ambrozkiewicz F, Kopczynski M, Kuklinska U, et al. Prolonged transfer of feces from the lean mice modulates gut microbiota in obese mice. Nutr Metab (Lond). 2016;13:57.
Kang Y, Cai Y. Gut microbiota and obesity: implications for fecal microbiota transplantation therapy. Hormones (Athens). 2017;16:223–34.
Lee P, Yacyshyn BR, Yacyshyn MB. Gut microbiota and obesity: an opportunity to alter obesity through faecal microbiota transplant (FMT). Diabetes Obes Metab. 2019;21:479–90.
Perez-Matute P, Iniguez M, de Toro M, Recio-Fernandez E, Oteo JA. Autologous fecal transplantation from a lean state potentiates caloric restriction effects on body weight and adiposity in obese mice. Sci Rep. 2020;10:9388.
Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell Host Microbe. 2017;22:589–99.
Neri C, Edlow AG. Effects of maternal obesity on fetal programming: molecular approaches. Cold Spring Harbor Perspect Med. 2016;6:a026591.
Chang E, Hafner H, Varghese M, Griffin C, Clemente J, Islam M, et al. Programming effects of maternal and gestational obesity on offspring metabolism and metabolic inflammation. Sci Rep. 2019;9:16027.
Andreas NJ, Hyde MJ, Gale C, Parkinson JR, Jeffries S, Holmes E, et al. Effect of maternal body mass index on hormones in breast milk: a systematic review. PLoS One. 2014;9:e115043.
Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–35.e4.
Liu H, Wang J, He T, Becker S, Zhang G, Li D, et al. Butyrate: a double-edged sword for health? Adv Nutr. 2018;9:21–9.
Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–25.
Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362:776–80.
Sorbara MT, Pamer EG. Microbiome-based therapeutics. Nat Rev Microbiol. 2022;20:365–80.
Acknowledgements
NTM was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K01HL141589. Gen3G was supported by a Fonds de recherche du Québec – Santé (FRQS) operating grant (to MFH, grant #20697); a Canadian Institute of Health Research (CIHR) operating grant (to MFH grant #MOP 115071 and to LB #PJT-152989); and a Diabète Québec grant (to PP). LB is a senior research scholar from the FRQS. MFH was a recipient of an American Diabetes Association (ADA) Pathways To Stop Diabetes Accelerator Award (#1-15-ACE-26). EM was funded by an operating grant from the Canadian Institutes of Health Research (CIHR) #BMB 389354.
Author information
Authors and Affiliations
Contributions
TL and FJ analyzed and interpreted the data, and also drafted the manuscript. NTM and MFH developed the research question, designed the study, helped interpret the data, and contributed to writing the manuscript. MD, LB, PP, and MFH were involved with the conception and creation of the Gen3G cohort study. All authors edited, read, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, T., Jia, F., Differding, M.K. et al. Pre-pregnancy body mass index and gut microbiota of mothers and children 5 years postpartum. Int J Obes 47, 807–816 (2023). https://doi.org/10.1038/s41366-023-01322-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-023-01322-4
- Springer Nature Limited