Abstract
Secondary mutation of epidermal growth factor receptor (EGFR) resulting in drug resistance is one of the most critical issues in lung cancer therapy. Several drugs are being developed to overcome EGFR tyrosine kinase inhibitor (TKI) resistance. Here, we report that pyruvate kinase M2 (PKM2) stabilized mutant EGFR protein by direct interaction and sustained cell survival signaling in lung cancer cells. PKM2 silencing resulted in markedly reduced mutant EGFR expression in TKI-sensitive or -resistant human lung cancer cells, and in inhibition of tumor growth in their xenografts, concomitant with downregulation of EGFR-related signaling. Mechanistically, PKM2 directly interacted with mutant EGFR and heat-shock protein 90 (HSP90), and thus stabilized EGFR by maintaining its binding with HSP90 and co-chaperones. Stabilization of EGFR relied on dimeric PKM2, and the protein half-life of mutant EGFR decreased when PKM2 was forced into its tetramer form. Clinical levels of PKM2 positively correlated with mutant EGFR expression and with patient outcome. These results reveal a previously undescribed non-glycolysis function of PKM2 in the cytoplasm, which contribute to EGFR-dependent tumorigenesis and provide a novel strategy to overcome drug resistance to EGFR TKIs.
Similar content being viewed by others
Introduction
Epidermal growth factor receptor (EGFR) mutations have been shown to occur at high frequencies in patients with non-small cell lung cancer (NSCLC).1, 2, 3 Most of EGFR mutations are either exon 19 deletions (codons 746–750) or substitution of leucine with arginine on codon 858 (L858R). These activating mutations in EGFR drive lung carcinogenesis, and are believed to be responsible for the therapeutic success of the EGFR tyrosine kinase inhibitors (TKIs).4 Although administering EGFR TKIs was shown to lead to stabilization of lung cancer in a median of 10–14 months, acquired resistance eventually limits the effectiveness of the currently available TKIs.5, 6 Subsequent studies have found that a secondary mutation in the EGFR gene (T790M) and amplification of the MET proto-oncogene are the main resistance mechanisms involved. T790M, which is thought to cause steric hindrance and impair the binding of TKIs, accounts for about half of the cases of acquired resistance.7, 8, 9 Up to the present, many second- and third-generation inhibitors designed to improve effectiveness and interfere with TKI resistance continued to be developed and tested, but have yet to be proven clinically effective and need further evaluation.
Pyruvate kinase M2 (PKM2), which is usually alternatively spliced from a common mRNA precursor with M1 type pyruvate kinase (PKM1) during tumorigenesis, has been recognized as a key regulator of aerobic glycolysis.10, 11 PKM2 protein alternates between a high-activity tetramer form and a low-activity dimer form in normal tissue, while exhibiting a distinctive tendency to exist as a dimer in cancer cells.12 Dimeric PKM2 has low enzymatic activity, which results in an increase in anabolic glycolysis for marcromolecular biosynthesis, and thus promotes cancer cell proliferation. Highly enzymatic PKM2 can be formed by the binding of allosteric regulators like fructose-1,6-biphosphate and serine.12 This has led to the development of several enzyme activators that induce tetramer formation of PKM2 and suppress tumor formation in vivo and in vitro.13, 14
In addition to the metabolic advantages that cytosolic PKM2 provides for cancer cells, non-metabolic functions of PKM2 in the nucleus have also been identified. PKM2 acts as a protein kinase and phosphorylates STAT3, resulting in increased cell proliferation.15, 16 Recently, Yang et al.17, 18, 19 demonstrated that the activation of EGFR signaling leads to nuclear translocation of PKM2, resulting in transcriptional activation of cyclin D1 and Myc. Therefore, PKM2 is thought to be a downstream regulator of EGFR signaling and thus of the EGFR-mediated oncogenic pathway.
In light of these findings, we hypothesized that PKM2 might be functionally important in lung cancer cells with active EGFR. Herein, we identify a critical function of PKM2 in regulating the survivability of EGFR-mutated lung cancer cells, including lung cancer cells that carry L858R/T790M mutated EGFR. Mechanistic studies have shown that the protein level of PKM2 is directly associated with EGFR expression and that PKM2 prolongs the protein half-life of EGFR by stabilizing EGFR–HSP90 (heat-shock protein 90) protein complex. We thus identified PKM2-targeting as a novel therapeutic approach for combating EGFR-TKI resistance.
Results
PKM2 is essential for tumor growth in EGFR-mutant NSCLC cells
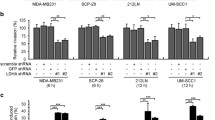
To determine the requirement of PKM2 for tumor growth of NSCLC cells, especially in those with EGFR mutations, we first silenced PKM2 in a panel of NSCLC cells with different EGFR genetic backgrounds and assayed for cell proliferation rates. Under a similar knockdown efficiency of PKM2 (Supplementary Figure S1a), loss of PKM2 in EGFR-mutant cells resulted in an almost 60% decrease of proliferation, while it only resulted in a 10% decrease of proliferation in wild-type cells (Figure 1a). This result was recapitulated in colony formation assays and soft agar assays (Figures 1b and c), suggesting that a critical role is played by PKM2 in regulating cell growth of EGFR-mutated NSCLC cells. Moreover, PKM2 depletion almost completely stopped the growth of PC9 xenografts that carried a TKI-sensitive EGFR mutation (exon 19 deletion), whereas H1355 xenografts still managed to grow to half the size of the control xenografts under PKM2 depletion (Figures 1d and f). PKM2 depletion also dramatically suppressed the growth of TKI-resistant xenografts (H1975) that carried L858R/T790M mutations (Figure 1e and Supplementary Figure S1b). Taken together, these results strongly suggest the essential role of PKM2 in EGFR-driven tumor growth of NSCLC cells.
Depletion of PKM2 in EGFR-mutant cells diminished tumor growth. (a) EGFR wild-type and mutant cells were infected with control sh-RNA or sh-PKM2 lentiviruses. After 72 h, cell viability was measured by the Trypan blue exclusion method. *P<0.05, **P<0.01 versus control. (b) Cells were infected with control sh-RNA or sh-PKM2 and grown on the plates for long-term colony formation assays. Cells were fixed and stained after 10 days. Colony number was counted by ImageJ. (c) Cells infected with control sh-RNA or sh-PKM2 were grown on soft agar for 14 days. Scale bars: 100 μm. (d–f) NSG immunocompromised mice were subcutaneously injected with EGFR-mutant cells and wild-type cells. Before being implanted into mice, the cells were infected by control sh-RNA or sh-PKM2 for 48 h. When the tumor volume reached 50 mm3, gefitinib (10 mg/kg) or vehicle control was intraperitoneally injected three times per week into the mice. Tumor volume was measured every 2–3 days. Values are plotted as mean±s.e.m. A total of 8–10 mice were analyzed in each group. Data presented in (a–c) are representative of at least three independent experiments.
Cytosolic PKM2 modulates EGFR-driven cell proliferation
The EGF/EGFR signaling axis has been reported to induce nuclear translocation of PKM2 in glioma cells.17 We therefore sought to determine the functional importance of nuclear PKM2 in NSCLC cells carrying EGFR mutations. PKM2 nuclear translocation increased when EGFR activation was induced by EGF stimulation in EGFR wild-type NSCLC cells, but did not increase after EGF stimulation in EGFR-mutant cells (Figure 2a). This may be due to the highly active EGFR signaling already present in EGFR-mutant cells before EGF stimulation (Supplementary Figure S2a and b). Furthermore, PKM2 carrying N-terminal nuclear export signal peptide (NES-PKM2), which guarantees a cytosolic distribution of PKM2, restored cell proliferation and colony formation in PKM2 knockdown PC9 cells and in H1975 cells, as did the wild-type PKM2 (Figures 2b and c and Supplementary Figure S2c). In addition to these in vitro findings, our immunohistochemistry analysis revealed that only 17% of lung cancer patients possessed nuclear PKM2 (Supplementary Table S1), unlike the high distribution of nuclear PKM2 reported in glioma cell specimens.17 Even in the EGFR-mutant population, a small percentage of patients exhibited positive nuclear PKM2 staining (Supplementary Figure S2d). These results suggest that in NSCLC cells PKM2 does not regulate cell proliferation through other mechanism besides its known nuclear function.
Cytosolic PKM2 regulates EGFR-dependent cell proliferation, colony formation and signal transduction. (a) Nuclear fractions were prepared from EGFR wild-type (H1355 and H157) and mutant (PC9 and H1975) cells treated with EGF (100 ng/ml) for 6 h after 18 h of serum starvation. Glioma (U87) cells were used as a positive control. Nuclear lamin A/C was used as loading controls. (b) After seeding samples with the same number of cells for 72 h, the viability of indicated cells was measured by the Trypan blue exclusion method. (c) Colony formation assays for the cells expressing either 3’-UTR targeting sh-PKM2 or control shRNA alone or together with wild-type PKM2, NES-PKM2 or K367M-PKM2. Cells were fixed and stained after 10 days. Colony number was counted by ImageJ. (d) Lysates from the cells depleted of PKM2 by lentivirus infection within 48 h. Protein expression levels were analyzed by immunoblotting. (e) Restored expression of EGFR for 48 h in mutant cells after PKM2 depletion. Same cells were seeded on the first day and the viability of cells was measured by the Trypan blue exclusion method after 72 h. *P<0.05, **P<0.01 and ***P<0.001 versus sh-PKM2. Data presented in (a–e) are representative of at least three independent experiments; values in graphs represent means±s.d. Statistical analysis is performed by two-tailed paired Student’s t-test.
Depletion of PKM2 inhibits the expression of mutant EGFR protein and suppresses EGFR signaling
Since the EGF/EGFR signaling axis has been shown to drive NSCLC tumorigenesis,20 we next set out to determine the effect of PKM2 on EGFR signal transduction. In EGFR-mutant cells, activation of EGFR downstream signaling was inhibited by depletion of PKM2 (Figure 2d). Surprisingly, depletion of PKM2 substantially reduced EGFR expression and re-expression of PKM2 restored EGFR protein levels (Figure 2d and Supplementary Figure S2e). In EGFR wild-type cells, the decline was moderate and can only be observed 72 h after PKM2 depletion (Supplementary Figure S2f). Moreover, depletion of PKM1 did not result in reduction of EGFR protein (Supplementary Figure S2g), suggesting a highly specific role of PKM2 in EGFR regulation. To confirm the phenotype of PKM2 knockdown cells was due to reduction of EGFR expression, we carried out rescue experiments of mutant EGFR in the PKM2-depleted cells. Re-expressed EGFR protein abrogated the inhibition of cell proliferation (Figure 2e). These results indicate that cytosolic PKM2 sustains EGFR protein expression and thus promotes EGFR-dependent signaling and cell survival in NSCLC cells.
Depletion of PKM2 induces proteasome-dependent protein degradation of EGFR
To further confirm the involvment of PKM2 in modulating EGFR expression, we detected the mRNA level and protein half-life of mutant EGFR. The mRNA level of EGFR did not change in the absence of PKM2 (Figure 3a), whereas the protein half-life of EGFR was shortened in EGFR-mutant cells (Figure 3b), suggesting that PKM2 modulates EGFR by post-transcriptional regulation. In contrast to the EGFR-mutant cells, the protein half-life of wild-type EGFR was not altered by depletion of PKM2 (Supplementary Figure S3a). To verify whether PKM2 enhances EGFR protein stability and to assess whether PKM2 enzymatic activity is required for EGFR protein stability, we measured EGFR expression by co-transfecting with EGFR and wild-type PKM2 or kinase-dead mutant PKM2 (K367M).21, 22 Increasing either wild-type or kinase-dead PKM2 raised EGFR protein levels in a dose-dependent manner (Figure 3c). Even after glucose deprivation, the upregulation of EGFR protein by PKM2 was still observed (Supplementary Figures S3b and c), suggesting that PKM2-regulated EGFR expression may be a glycolysis-independent event. Moreover, the kinase-dead PKM2 mutant (K367M) also reversed the effect of PKM2 depletion, including EGFR expression levels (Figures 2b and c and Supplementary Figure S2e), suggesting that enzymatic activity of PKM2 may not be involved in EGFR regulation. Ectopically expressed PKM1 did not increase EGFR protein levels, confirming that the observed increase in EGFR stability was PKM2 specific (Supplementary Figure S3d). Furthermore, treatment of MG-132 rescued the reduction of EGFR after PKM2 depletion (Figure 3d and Supplementary Figure S3e), suggesting that PKM2 may prevent EGFR degradation resulting from proteasomal degradation. Also consistent with this interpretation, ubiquitination of EGFR increased after PKM2 depletion (Figure 3e). Collectively, these observations suggest that PKM2 contributes to EGFR protein stability by preventing proteasomal degradation.
Decrease of EGFR after PKM2 depletion is a post-transcriptional event. (a) Analysis of EGFR mRNA levels in PKM2-depleted H1355, PC9 and H1975 cells. The EGFR mRNA levels were quantified by RT-qPCR. (b) Cells were treated with cycloheximide to compare the degradation of mutant EGFR in the absence and presence of PKM2. 10 μM cycloheximide was added and degradation was measured at indicated intervals from hours 0 to 10. Protein expression levels were normalized with an internal control and quantified as a ratio compared with hour 0. (c) Co-expression of 2 μg EGFR from lysates of 293 T cells with wild-type or kinase-dead PKM2 at the indicated doses. (d) PC9 cells treated with 20μM MG-132 for indicated times after depletion of PKM2. Protein expression levels were normalized with an internal control and quantified as a ratio compared with control cells. *P<0.05 versus DMSO treated. (e) H1975 cells treated with or without MG-132 in the absence and presence of PKM2 in H1975 cells transiently transfected with c-Myc-ubiquitin-expressing plasmids. Cell lysates were immunoprecipitated with anti-Myc and immunoblotted with EGFR antibody. Internal controls were performed with c-Myc total protein. Data presented in (a–e) are representative of at least three independent experiments.
Cytosolic PKM2 stabilizes EGFR through direct physical interaction
To investigate whether PKM2-mediated stabilization of EGFR involves the physical interaction of PKM2 and EGFR, we first examined the endogenous interaction between them. We observed that both mutant and wild-type EGFR proteins were co-immunoprecipitated with PKM2 in NSCLC cells (Figure 4a and Supplementary Figure S4a). Colocalization of EGFR and PKM2 in cells was distributed at both the cell surface and in the cytoplasm (Supplementary Figure S4b). To evaluate whether there is a difference between the binding of PKM2 to wild-type or to mutant EGFR, we ectopically expressed HA-tagged PKM2 and wild-type or mutant EGFR in 293 T cell. Our results show that there was a stronger interaction between PKM2 and active EGFR mutants than between PKM2 and wild-type EGFR (Figure 4b). Furthermore, recombinant PKM2 was pulled down by GST-tagged intracellular domains of EGFR, suggesting a direct interaction between them. Also consistent with this interpretation was the finding that PKM2 interacted more strongly with the intracellular domain of EGFR-L858R than with wild-type EGFR (Figure 4c). We also observed similar bindings between kinase-dead (K367M-PKM2) and wild-type PKM2 to EGFR (Supplementary Figures S4c). Moreover, the similar pattern of association occurred between PKM1 and EGFR, although in a relatively weaker manner compared with PKM2 (Supplementary Figures S4d).
Dimeric PKM2 interacts with mutant EGFR. (a) Lysates of PC9 cells were prepared for immunoprecipitation with anti-PKM2 or anti-EGFR antibody followed by immunoblotting with anti-EGFR and anti-PKM2. (b) Immunoprecipitation of EGFR with anti-HA in 293 T cells co-transfected with HA-PKM2 and wild-type or mutant EGFR as indicated. (c) Purified recombinant intracellular domain of wild-type or L858R EGFR (GST-EGFR or GST-L858R) was mixed with purified His-PKM2. A GST pull-down assay was performed. Coomassie blue staining served as the loading control for GST-EGFR and GST-L858R. (d) Immunoprecipitation of EGFR with anti-HA in 293 T cells co-transfected with HA-PKM2 and wild-type EGFR. After 18 h of serum starvation, cells were treated with or without EGF (100 ng/ml, 15 min). (e) Purified recombinant L858R EGFR (GST-L858R) was incubated with phosphatase to de-phosphorylate the protein and mixed with purified His-PKM2. A GST pull-down assay was performed. (f) Immunoprecipitation of mutant ICM with anti-HA in 293 T cells co-transfected with HA-PKM2 and mutant EGFR-ICM. (g) Immunoprecipitation of exon 19 deletion EGFR with anti-HA in 293 T cells co-transfected with HA-PKM2 and mutant EGFR. Cells were incubated with F-1,6-BP (200 μM) for 2 h before being lysed. (h) H1975 cells were treated with cycloheximide to compare the degradation of mutant EGFR in the absence and presence of PKM2 activator, TEPP-46. 10μM cycloheximide was added and degradation was measured at indicated intervals from hours 0 to 12. Protein expression levels were normalized with internal control and quantified as a ratio compared with hour 0. Data presented in (a–h) are representative of at least three independent experiments.
As PKM2 has been reported to directly bind to phosphotyrosine peptides,11 we next investigated whether the stronger PKM2 association of EGFR mutants resulted from their higher phosphorylation status. As anticipated, EGF treatment increased the level of wild-type EGFR in the PKM2 immunoprecipitate (Figure 4d). In reverse, the direct binding of phosphorylated EGFR protein to PKM2 was decreased after de-phosphorylation by phosphatase (Figure 4e). In accordance with this result, a phospho-tyrosine-binding PKM2 mutant,11 K433E-PKM2, exhibited diminished association with EGFR (Supplementary Figure S4e), suggesting that phosphorylation of EGFR increases PKM2 association.
Since there are several phosphorylation sites on EGFR protein, we have first confirmed that the PKM2-interaction site of EGFR is located in the intracellular domain (ICM). Furthermore, the interaction of EGFR with PKM2 is limited to the kinase activity (ICM-KA) region (Supplementary Figures S4f and g). We have next mutated the tyrosine sites of 703, 845, 891 and 920 in the ICM-KA region into phenylalanine, which is a non-phosphorylatable mutation, and examined whether the interaction of PKM2 and EGFR-ICM would be modulated. As shown in Figure 4f, while PKM2 association seems to decline as tyrosine phosphorylation level of EGFR-ICM decreased, mutants containing Y845F lost majority of their PKM2-binding capacity. These data suggested that the Y845 phosphorylation, which is important for constitutive activation of mutant EGFR,23 is the most critical phosphorylation site that mediates PKM2 and EGFR interaction. However, since the Y845F mutant did not completely abolish the interaction of PKM2 and EGFR, we hypothesized that other tyrosine sites may also help to further increase or stabilize PKM2 association. Collectively, these results were also consistent with our observation that PKM2 could preferably bind to phosphorylated EGFR.
Previous studies have shown that tetrameric PKM2 participates in glycolysis in the cytosol, while dimeric PKM2 acts as a protein kinase in the nucleus.15, 16 We, therefore, set out to examine which form of PKM2 interacts with EGFR proteins. After treating 293 T cells with the PKM2 tetrameric inducer, F-1,6-BP, the interaction of PKM2 and EGFR obviously decreased (Figure 4g and Supplementary Figures S4h and i). In contrast, the dimeric mutation of PKM2, PKM2-R399E,15 co-immunoprecipitated more EGFR protein than wild-type PKM2. On the other hand, the association of EGFR to K305Q mutant of PKM2, which tended to form a monomer but to form dimer in the presence of F-1,6-BP,24 was increased when PKM2-K305Q was induced to form dimer (Supplementary Figures S4j–l). We further examined whether the protein half-life of EGFR would decrease in response to tetramer inducer treatment. As expected, EGFR protein half-life was decreased after treatment with TEPP-4614 and F-1,6-BP in EGFR-mutant cells (Figure 4h and Supplementary Figure S4l). These findings suggest that PKM2 associates with EGFR and stabilizes EGFR protein expression mainly in its dimeric form.
PKM2 associates with HSP90 and co-chaperones to modulate EGFR stability
Cells harboring mutated EGFR have been reported to be more sensitive than wild-type cells to HSP90 inhibitors.25, 26 To determine whether PKM2 is involved in HSP90-mediated EGFR protein stabilization, we first tested for the presence of HSP90 and co-chaperones in PKM2 immunoprecipitates.27 As shown in Figure 5a, HSP90, HSP90-organizing protein and p23 were all associated with endogenous PKM2, suggesting that PKM2 may exist in the EGFR–chaperone protein complexes found in NSCLC cells. When we abrogated PKM2-induced EGFR degradation with MG-132 and examined the association of EGFR and chaperones, we found that PKM2 depletion decreased levels of EGFR-associated HSP90 and co-chaperones (Figure 5b). However, PKM2 depletion did not abrogate the formation of HSP90/co-chaperone complex (Figure 5c). Furthermore, the HSP90 inhibitor, 17-AAG, did not change the binding of PKM2 to EGFR (Figure 5e). These observations suggest that PKM2 may be required for EGFR–HSP90 interaction. Our observations that recombinant PKM2 also directly interacted with His-tagged HSP90 also support this hypothesis (Figure 5d and Supplementary Figure S5a). Of note, PKM2-induced EGFR protein stability was reduced after silencing of HSP90α or treating with HSP90 inhibitor (Figure 5e, Supplementary Figures S5b and c). Moreover, the reduction of EGFR protein half-life was similar in HSP90α-knockdown cells with or without PKM2 silencing, which further confirms that PKM2-mediated EGFR stabilization was HSP90 dependent (Figure 5f).
PKM2 stabilized EGFR through HSP90/co-chaperone. (a) Immunoprecipitation of EGFR and HSP90/co-chaperone proteins by anti-PKM2 antibody in H1975 cells. (b, c) Immunoprecipitation of HSP90 and co-chaperone proteins by anti-EGFR (b) or anti-HSP90 antibody (c) in the H1975 cells with or without PKM2 depletion. Before collection of cell lysates, cells were treated with 20 μM MG-132 for 2 h to inhibit proteasomal degradation. (d) Purified His-HSP90 and His-PKM2 were mixed overnight at 4 °C. PKM2 was pulled down by anti-PKM2 antibody, which was conjugated with agarose A beads. Rabbit IgG was used as a negative control. (e) Transient ectopic expression of mutant EGFR in lysates from 293 T cells with or without wild-type PKM2. Immunoprecipitation of EGFR and HSP90 by anti-Flag beads in cell expression of mutant EGFR and wild-type PKM2. Cells were treated with 100 nM 17-AAG for 16 h to inhibit HSP90 activity. (f) H1975 cells were treated with cycloheximide to compare the degradation of mutant EGFR in the absence and presence of HSP90 and PKM2. 10 μM cycloheximide was added and degradation was measured at indicated intervals from hours 0 to 12. Protein expression levels were normalized with internal control and quantified as a ratio compared with hour 0. Data presented in (a–f) are representative of at least three independent experiments.
We also observed the endogenous interaction of PKM2, HSP90 and EGFR in EGFR wild-type cells. However, the downregulation of wild-type EGFR upon silencing of PKM2 was much slower than mutant EGFR in NSCLC cells (Supplementary Figures S2f and d), which was consistent with previous reports that protein stability of mutant EGFRs is more dependent on HSP90 than wild-type EGFR.25, 26 Although the protein expression levels of other HSP90 clients, MET and Her2, were not altered after PKM2 silencing (Supplementary Figure S5e), it is worth to investigate whether there are other proteins require PKM2 for maintaining their protein stability.
Depletion of PKM2 induces apoptosis in EGFR-mutant lung cancer cells
The above results demonstrate that EGFR protein stability and cell viability were reduced due to depletion of PKM2 in EGFR-mutant cancer cells. Given that active mutations of EGFR have been shown to lead to aberrant EGFR-related oncogenic cassettes, which contribute to cell proliferation and survival,28 we next investigated whether PKM2 contributes to the survival benefit of EGFR signaling. After PKM2 depletion was induced, apoptotic cells were significantly increased in PC9 and H1975 cells, but that no conspicuous apoptotic cell death in EGFR wild-type cells occurred (Figure 6a and Supplementary Figure S6a). In addition, cleaved caspase 3 also obviously increased after PKM2 depletion in EGFR-mutant cells but not in wild-type cells (Figure 6b). Similar phenomena were also observed in EGFR knockdown cells, implying that PKM2 might mediate the EGF/EGFR regulation of cell survival. Moreover, the pro-apoptotic gene, Bim, and the anti-apoptotic gene, Bcl-xl, were upregulated and downregulated, respectively, after PKM2 inhibition (Figures 6c and d). Both Bim and Bcl-xl were reported to be key regulators of cell survival after EGFR depletion,29 as we also confirmed in EGFR knockdown cells (Figures 6c and d). In addition, since the EGFR signaling axis is also thought to propagate cell-cycle progression, we also tested for and found a reduction in cyclin D1 and Myc expression in both PC9 and H1975 cells (Figures 6c and d), which are critical downstream regulators of mutant EGFR signaling.30, 31 These results show that EGFR-related signaling pathways were downregulated, resulting in reduction of cell viability, after PKM2 silencing.
Depletion of PKM2 caused apoptotic cell death in EGFR-mutant cells. (a) After PKM2 or EGFR depletion for indicated times, cells were stained with annexin V and apoptosis status was measured by flow-cytometry analysis. (b) After PKM2 or EGFR was depleted for indicated times, protein expression was analyzed by immunoblotting. (c, d) Proliferation and apoptosis of EGFR-related downstream genes were downregulated both in PKM2-depleted cells and in EGFR-depleted cells. Cells were collected after knockdown of PKM2 and EGFR by lentivirus infection within 72 h. The mRNA expression levels (c) were quantified by real-time RT–PCR. Protein expression (d) was analyzed by immunoblotting. *P<0.05, versus sh-control. Data presented in (a–d) are representative of at least three independent experiments.
PKM2 levels highly correlate with the expression of mutant EGFR and patient survival in NSCLC patients
Having established in our in vitro studies that PKM2 expression highly correlated with the expression and function of mutated EGFR protein in NSCLC cells, we sought to assess whether such a correlation exists among PKM2 and mutated EGFR protein expression levels in lung cancer specimens in vivo. We collected 67 NSCLC specimens with EGFR mutations and 74 NSCLC specimens with wild-type EGFR and carried out an immunohistochemistry analysis of EGFR and PKM2 expression. Our result shows that PKM2 expression was significantly correlated with EGFR levels in the EGFR-mutant cohort, but not in the EGFR wild-type cohort (Figure 7a, Supplementary Tables S2 and S3). Since PKM2 expression has been found to be a prognostic marker for different kinds of cancer,32, 33 we then examined the correlation between PKM2 protein levels and disease-free survival within this 141 patient cohort. Of note, protein expression levels of PKM2 significantly correlated with shorter survival in the EGFR-mutant cohort but not in the EGFR wild-type cohort (Figures 7b and c). Similar observations were also obtained from analysis of a publicly available microarray data set, which included 226 patients with primary lung adenocarcinoma (GSE31210). The correlation of PKM2 mRNA expression levels and patient disease-free survival was more significant in the EGFR-mutant cohort (P=0.0005) than in the EGFR wild-type cohort (P=0.0084) (Supplementary Figures S7a and b). Collectively, these findings strengthen the linkage between PKM2 and EGFR protein expression and also suggest that PKM2 has prognostic value in treating patients with EGFR-mutated NSCLC.
In NSCLC tumors, expression of PKM2 correlated with EGFR levels and with poor prognosis in EGFR-mutant patients. (a) Representative pictures of IHC staining with anti-PKM2 or anti-EGFR antibody on EGFR-mutant tumor tissues. Cases 1–3 are the patients with both high PKM2 and EGFR. Cases 4–6 are the patients with both low PKM2 and EGFR. Scale bars, 50 μm. (b, c) Kaplan–Meier disease-free survival rate analysis of (b) 67 patients with mutated EGFR and (c) 74 patients with wild-type EGFR, stratified by IHC PKM2 level. Each patient was classified as high or low PKM2 by a cutoff value of 160, measured by adding each staining intensity (0–3) multiplied by its percentage of coverage (0–100). The log-rank P-value is indicated. (d) Dimeric PKM2 accompanies HSP90 and co-chaperones to stabilize mutant EGFR, which harbors higher phosphorylation status. Disassociation of dimeric PKM2, followed by release of HSP90/co-chaperones, caused accumulation of ubiquitination on EGFR and accelerated its degradation. Degradation of EGFR resulted in apoptosis and reduction of tumor growth in vivo and in vitro.
Discussion
PKM2 is a key regulator of aerobic glycolysis, also known as the Warburg effect, in tumor cells.34 Overexpression of PKM2 has been found in different cancers and has been associated with aggressive tumor progression and poor prognosis.32, 33 There is accumulating evidence suggesting that diverse PKM2-related mechanisms exist in cancer cells, all of which contribute to cancer initiation and progression.10, 35 In addition to its well-known metabolic activity, PKM2 has also been found to act as a protein kinase in the nucleus upon EGFR activation, hypoxia or glucose depletion and to be required for the proliferation and survival of some cancer cells.10, 35, 36 However, although PKM2 is generally considered to be oncogenic, a contradictory role of PKM2 in tumorigenesis has also been observed.37, 38 In the absence of functional PKM2, the tumorigenesis process of a Brca1-loss-driven breast cancer model has even been accelerated.37 Furthermore, since the structure and activity of PKM2 is regulated by a variety of signals,12, 16, 39 the oncogenic function of PKM2 may be different among cancer types and dependent on the genetic background of cancer cells. In this study, we identified a specific role of PKM2 in promoting cell proliferation of EGFR-mutant NSCLC cells. PKM2 stabilized the association of protein chaperones with EGFR expression by direct interaction with both EGFR and HSP90 proteins, resulting in the maintenance of EGFR expression. Reduction of the association of dimeric PKM2 and EGFR will resulted in the degradation of EGFR, which was followed by apoptosis and reduction of tumor growth in vivo and in vitro (Figure 7d). Importantly, EGFR protein stability and cell viability of gefitinib-resistant EGFR-T790M mutant cells were also highly dependent on PKM2 expression. Our data reveal a unique role of PKM2 in the regulation of EGFR protein stability and highlight the potential therapeutic benefit of targeting PKM2 in NSCLC, especially in those carrying EGFR mutations.
In addition to the metabolic function of PKM2 in the cytoplasm, recent studies have reported the non-metabolic function of PKM2 in the nucleus, where it works as a protein kinase and transcriptionally regulates gene expression to promote cell-cycle progression and metabolic reprogramming in response to mitogen activation or hypoxia stress.18, 35, 40 For instance, under hypoxia PKM2 interacts with p300 and HIF-1α and stimulates the expression of metabolic genes that contain hypoxia response element.40 The nuclear translocation of PKM2 is also believed to rely on its conversion between tetramer and dimer forms by different post-translational modifications of PKM2.15, 16 Although the importance of PKM2 to metabolic function in the cytosol and non-metabolic function in the nucleus have been well studied in the past few years, whether or not PKM2 has an important non-metabolic function in the cytosol remains unclear. Here, we identify a novel function of PKM2 in the cytosol, which is not related to its known metabolic activity. Dimerized PKM2 acts like a co-chaperone to stabilize HSP90–EGFR interaction in the cytosol, and thus maintains EGFR expression in EGFR-mutant NSCLC cells. Recent studies have also discovered in the cytosol several putative phosphorylated protein substrates acted on by PKM2,36 strengthening evidence of the importance of PKM2 in non-metabolic functions in the cytosol. Furthermore, our data demonstrated that EGFR regulation of PKM2 did not require its enzymatic activity, suggesting that the PKM2 protein may regulate cellular functions through more diverse ways.
Previous studies indicated that PKM2 is one of the downstream transcriptional coactivators of the EGF/EGFR signaling pathway that promotes tumor progression in glioma cells17, 18, 19, 41 and that PKM2 might even be transcriptionally upregulated through the EGFR-dependent NF-κB activating pathway.41 Here, we point out a critical and exclusive role of PKM2 in maintaining protein stability of mutant EGFR in the cytosol. This challenges the view that PKM2 is only a downstream regulator of EGFR signaling. In fact, recent evidence has shown that PKM2 directly phosphorylates Erk1/2 and forms a positive feedback loop to promote EGF/MAPK signaling.36 Our finding further suggests that a positive feedback loop between EGFR and PKM2 may exist in EGFR-mutant NSCLC cells. The role of PKM2 in the EGFR-dependent signaling pathway may be more active than previously thought. Although we have demonstrated the critical role of cytosolic PKM2 in maintaining the growth of EGFR-mutant cells, the possible importance of nuclear PKM2 in these cells was not eliminated. Whether the accumulation of nuclear PKM2 stimulates the genes involved in lung tumor formation of EGFR-mutant NSCLC needs further investigation.
One novel finding of our study is to identify PKM2 in the HSP90 chaperone complex. In our opinions, PKM2 may persistently associate with HSP90, active or inactive state. The binding of PKM2 to active HSP90 may be stronger than inactive HSP90, since our results suggest that the interaction between PKM2 and HSP90 was slightly decreased in the presence of HSP90 inhibitor (Figure 5e). On the other hand, PKM2 may still associate with inactive HSP90 because in our directly binding experiment with bacterial purified HSP90, which mostly inactive due to the lack of post-translational modifications, PKM2–HSP90 interaction was also observed (Figure 5d). However, the detail mechanism by which PKM2 act in the HSP90 complex remains unclear and needs further investigation.
Disease stabilization of patients with EGFR-mutated lung cancer is typically achieved when they are initially given EGFR TKIs, but acquired resistance eventually results in disease recurrence. Currently, many drugs are being developed to target the altered structure of EGFR with T790M mutation,42 and some are already under clinical trials to compare their effects to standard first-line chemotherapy in metastatic lung adenocarcinoma patients with EGFR mutations.43, 44 Nevertheless, increased side effects, such as diarrhea, rash and nail disorders, are still of great concern and may dampen clinical demand.45 In addition, even though most tumors are responsive to a number of different TKIs, when newly administered, they almost inevitably become increasingly resistant to TKIs in general, even newly administered ones, as TKI treatment continues.46 Therefore, evaluation of the therapeutic strategy to combat tumor growth with second-generation TKIs is worth further investigation. Since mutated EGFR is highly dependent on HSP90-mediated protein stabilization,25 HSP90 inhibitors are being tested for their therapeutic effect on EGFR-TKI-resistant cells.47 Unfortunately, since HSP90 is an abundant protein in both tumor and normal cells and regulates stabilization and maturation of many proteins,27 the comprehensive side effects of HSP90 inhibitor therapy may limit further clinical applications.48, 49 Our results have shown that tumor-specific PKM2 stabilizes EGFR through coordination with HSP90 without affecting other HSP90 clients. Theoretically, this implies that fewer side effects should be expected from targeting the interaction of PKM2/EGFR or PKM2/HSP90 in tumor cells. Developing strategies to disrupt these direct interactions might provide therapeutic benefits to patients exhibiting EGFR-T790M mutation.
Here, we have identified an HSP90-dependent non-metabolic function of PKM2 that stabilizes mutant EGFR and facilitates lung cancer proliferation. Depletion of PKM2 reduced EGFR stability and diminished tumor formation in vivo and in vitro. Once PKM2 was stimulated to tetrameric form, binding of EGFR and PKM2 decreased, leading to a shortening of EGFR protein half-life. We have thus provided a novel application for targeting PKM2 to overcome resistance to EGFR TKIs in NSCLC cells.
Materials and methods
Cell culture
Lung cancer cell lines A549, H157, H1355, H1975 and HCC827 were purchased from American Type Culture Collection, and PC9 NSCLC cell line was kindly provided by Dr James Chih-Hsin Yang. All cells were routinely authenticated on the basis of morphologic and growth characteristics as well as by STR analysis and confirmed to be free of mycoplasma.
Xenograft studies
Cancer cells with/without PKM2 knockdown were injected subcutaneously into the right flank of the NSG (NOD scid gamma; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice at the age of 5–6 weeks. After transplantation, the tumor volume was calculated as 1/2 × length × width2 in mm3. In the gefitinib treatment experiments, mice were injected intraperitoneally with gefitinib (10 mg/kg) three times per week. In all, 30–70 days after implantation, animals were killed by cervical dislocation and dissected to obtain tumor samples.
Immunoprecipitation
Cells were lysed in NETN buffer (20 mM Tris, at pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40). In all, 1500 μg lysates were incubated with indicated antibody (5–10 μg) for 16 h at 4 °C followed by 1 h incubation with Protein A Sepharose beads. After washing the beads five times with NETN buffer, Immunoprecipitates were resolved by immunoblotting with indicated antibodies.
EGFR mutation status identification
Nucleotide mass spectrometry was used to determine EGFR mutation status.50 Matrix-assisted laser desorption ionization time-of-flight mass spectrometry was performed using the MassARRAY system (SEQUENOM). Analyses of the clinical material have been approved by the NTUH ethics committee (201304088RINB).
Statistical analysis
Comparison of the treatment group(s) versus the control group was performed using a Student's t test for studies with only two groups. Data were obtained from n>3 experiments for every figure. Data are presented as mean±s.e.m. or s.d. *P<0.05 and **P<0.01 were considered as statistically significant. Kaplan–Meier analysis was used for survival curves.
References
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–2139.
Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–1500.
Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 2004; 101: 13306–13311.
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–354.
Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947–957.
Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361: 958–967.
Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, Zakowski MF et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011; 17: 1169–1180.
Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005; 2: e73.
Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 352: 786–792.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008; 452: 230–233.
Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC . Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008; 452: 181–186.
Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 2012; 491: 458–462.
Parnell KM, Foulks JM, Nix RN, Clifford A, Bullough J, Luo B et al. Pharmacologic activation of PKM2 slows lung tumor xenograft growth. Mol Cancer Ther 2013; 12: 1453–1460.
Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol 2012; 8: 839–847.
Gao X, Wang H, Yang JJ, Liu X, Liu ZR . Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 2012; 45: 598–609.
Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell 2013; 52: 1–13.
Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 2011; 480: 118–122.
Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 2012; 150: 685–696.
Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol 2012; 14: 1295–1304.
Pao W, Chmielecki J . Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 2010; 10: 760–774.
Vollmer SH, Walner MB, Tarbell KV, Colman RF . Guanosine 5'-O-[S-(4-bromo-2,3-dioxobutyl)]thiophosphate and adenosine 5'-O-[S-(4-bromo-2,3-dioxobutyl)]thiophosphate. New nucleotide affinity labels which react with rabbit muscle pyruvate kinase. J Biol Chem 1994; 269: 8082–8090.
Le Mellay V, Houben R, Troppmair J, Hagemann C, Mazurek S, Frey U et al. Regulation of glycolysis by Raf protein serine/threonine kinases. Adv Enzyme Regul 2002; 42: 317–332.
Chung BM, Dimri M, George M, Reddi AL, Chen G, Band V et al. The role of cooperativity with Src in oncogenic transformation mediated by non-small cell lung cancer-associated EGF receptor mutants. Oncogene 2009; 28: 1821–1832.
Wang P, Sun C, Zhu T, Xu Y . Structural insight into mechanisms for dynamic regulation of PKM2. Protein Cell 2015; 6: 275–287.
Shimamura T, Lowell AM, Engelman JA, Shapiro GI . Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res 2005; 65: 6401–6408.
Yang S, Qu S, Perez-Tores M, Sawai A, Rosen N, Solit DB et al. Association with HSP90 inhibits Cbl-mediated down-regulation of mutant epidermal growth factor receptors. Cancer Res 2006; 66: 6990–6997.
Taipale M, Jarosz DF, Lindquist S . HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 2010; 11: 515–528.
Ciardiello F, Tortora G . EGFR antagonists in cancer treatment. N Engl J Med 2008; 358: 1160–1174.
Nyati MK, Morgan MA, Feng FY, Lawrence TS . Integration of EGFR inhibitors with radiochemotherapy. Nat Rev Cancer 2006; 6: 876–885.
Kobayashi S, Shimamura T, Monti S, Steidl U, Hetherington CJ, Lowell AM et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer Res 2006; 66: 11389–11398.
Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res 2010; 70: 8822–8831.
Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH et al. Overexpression of the M2 isoform of pyruvate kinase is an adverse prognostic factor for signet ring cell gastric cancer. World J Gastroenterol 2012; 18: 4037–4043.
Li J, Yang Z, Zou Q, Yuan Y, Li J, Liang L et al. PKM2 and ACVR 1C are prognostic markers for poor prognosis of gallbladder cancer. Clin Transl Oncol 2014; 16: 200–207.
Warburg O . On the origin of cancer cells. Science 1956; 123: 309–314.
Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y et al. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell 2014; 53: 75–87.
Keller KE, Doctor ZM, Dwyer ZW, Lee YS . SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol Cell 2014; 53: 700–709.
Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 2013; 155: 397–409.
Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc Natl Acad Sci USA 2013; 110: 489–494.
Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2009; 2: ra73.
Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011; 145: 732–744.
Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L et al. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell 2012; 48: 771–784.
Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov 2013; 3: 1404–1415.
Yang JC, Hirsh V, Schuler M, Yamamoto N, O'Byrne KJ, Mok TS et al. Symptom control and quality of life in LUX-Lung 3: a phase III study of afatinib or cisplatin/pemetrexed in patients with advanced lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013; 31: 3342–3350.
Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014; 4: 1046–1061.
Haspinger ER, Garassino MC, Torri V, Cinquini M, De Braud F, Gelsomino F . Do we really need another epidermal growth factor receptor tyrosine kinase inhibitor in first-line treatment for patients with non-small-cell lung cancer and EGFR mutations? J Clin Oncol 2014; 32: 859–863.
Pirazzoli V, Nebhan C, Song X, Wurtz A, Walther Z, Cai G et al. Acquired resistance of EGFR-mutant lung adenocarcinomas to afatinib plus cetuximab is associated with activation of mTORC1. Cell Rep 2014; 7: 999–1008.
Kobayashi N, Toyooka S, Soh J, Yamamoto H, Dote H, Kawasaki K et al. The anti-proliferative effect of heat shock protein 90 inhibitor, 17-DMAG, on non-small-cell lung cancers being resistant to EGFR tyrosine kinase inhibitor. Lung Cancer 2012; 75: 161–166.
Chiosis G, Dickey CA, Johnson JL . A global view of Hsp90 functions. Nat Struct Mol Biol 2013; 20: 1–4.
Solit DB, Ivy SP, Kopil C, Sikorski R, Morris MJ, Slovin SF et al. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. Clin Cancer Res 2007; 13: 1775–1782.
Yuan S, Yu SL, Chen HY, Hsu YC, Su KY, Chen HW et al. Clustered genomic alterations in chromosome 7p dictate outcomes and targeted treatment responses of lung adenocarcinoma with EGFR-activating mutations. J Clin Oncol 2011; 29: 3435–3442.
Acknowledgements
We thank Dr Zhimin Lu for the Flag-tagged K367M, K433E and wild-type PKM2, Dr Zhi-Ren Liu for HA-tagged R399E and wild-type PKM2, Dr Mien-Chie Hung for myc-tagged EGFR-ECM and -ICM, Dr Ann-Lii Cheng and Dr James Chih-Hsin Yang for helpful discussion, Integrated Core Facility for Functional Genomics of the National Core Facility Program for Biotechnology, the Microarray Core Facility of the National Taiwan University Center of Genomic Medicine and Dr Sung-Liang Yu for technical support. This study was supported by grants from National Science Council, Taiwan (NSC 104-2321-B-002-006, NSC-104-2911-I-002-302 and NSC 104-2923-B-002-003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, YC., Cheng, TY., Huang, SM. et al. Cytosolic PKM2 stabilizes mutant EGFR protein expression through regulating HSP90–EGFR association. Oncogene 35, 3387–3398 (2016). https://doi.org/10.1038/onc.2015.397
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.397
- Springer Nature Limited
This article is cited by
-
Sex differences in colonic gene expression and fecal microbiota composition in a mouse model of obesity-associated colorectal cancer
Scientific Reports (2024)
-
AKIP1 accelerates glioblastoma progression through stabilizing EGFR expression
Oncogene (2023)
-
An intrinsic purine metabolite AICAR blocks lung tumour growth by targeting oncoprotein mucin 1
British Journal of Cancer (2023)
-
HELZ2 promotes K63-linked polyubiquitination of c-Myc to induce retinoblastoma tumorigenesis
Medical Oncology (2022)
-
A small-molecule compound D6 overcomes EGFR-T790M-mediated resistance in non-small cell lung cancer
Communications Biology (2021)