Abstract
Breast cancer resistance to endocrine therapies such as tamoxifen and aromatase inhibitors is a significant clinical problem. Steroid receptor coactivator-1 (SRC-1), a coregulatory protein of the oestrogen receptor (ER), has previously been shown to have a significant role in the progression of breast cancer. The chromatin protein high mobility group box 2 (HMGB2) was identified as an SRC-1 interacting protein in the endocrine-resistant setting. We investigated the expression of HMGB2 in a cohort of 1068 breast cancer patients and found an association with increased disease-free survival time in patients treated with endocrine therapy. However, it was also verified that HMGB2 expression could be switched on in endocrine-resistant tumours from breast cancer patients. To explore the function of this poorly characterized protein, we performed HMGB2 ChIPseq and found distinct binding patterns between the two contexts. In the resistant setting, the HMGB2, SRC-1 and ER complex are enriched at promoter regions of target genes, with bioinformatic analysis indicating a switch in binding partners between the sensitive and resistant phenotypes. Integration of binding and gene expression data reveals a concise set of target genes of this complex including the RNA helicase DDX18. Modulation of DDX18 directly affects growth of tamoxifen-resistant cells, suggesting that it may be a critical downstream effector of the HMGB2:ER complex. This study defines HMGB2 interactions with the ER complex at specific target genes in the tamoxifen-resistant setting.
Similar content being viewed by others
Introduction
The majority of breast tumours that occur in women express the oestrogen receptor (ER), which is known to drive cell cycle and proliferation. While endocrine therapies such as tamoxifen and aromatase inhibitors are effective at inhibiting tumour progression in the majority of these patients, 30–40% of breast cancer patients will suffer from recurrence of disease.1 The p160 protein steroid receptor coactivator 1 (SRC-1) has classically been reported to act as a coactivator for ER in driving breast cancer.2 Recently, our group and others have demonstrated the importance of SRC-1 in disease progression and the development of metastasis. We reported that SRC-1 is an independent predictor of disease recurrence in a large cohort of breast cancer patients; while in women treated with tamoxifen, the interaction of SRC-1 with ER was enhanced upon the development of resistance to endocrine therapies.3 Qin et al.4 used a mouse model that develops spontaneous tumours to demonstrate that SRC-1 has a significant role in breast cancer invasiveness and metastasis. In aromatase inhibitor-resistant breast cancer, SRC-1 has been shown to mediate a more motile and aggressive phenotype.5
In this study we investigated high mobility group box 2 (HMGB2), which was identified as a novel SRC-1 interacting protein in the endocrine-resistant setting, using a liquid chromatography-mass spectrometry approach.6 HMGB2 is a chromatin protein containing 2 high-mobility boxes of 80 amino acids each that form three α-helices arranged in an L-shape, which binds the minor groove of the DNA. Although reported not to bind in a sequence-specific manner, this association alters the conformation of the DNA and enhances accessibility of other proteins.7 HMGB proteins have previously been reported to interact with nuclear receptors,8 Hox proteins9 and RNA polymerases.10 While considerable information is available about its closely related protein HMGB1, little is known about HMGB2 and only recent reports have focused on HMGB2 alone. Laurent et al.11 established that HMGB2 binds to the promoter of Gfi-1B and they propose that it regulates the GATA-1-dependent transcription involving Oct-1. The importance of HMGB proteins in cancer was recently highlighted in a report, which found a correlation between HMGB1 and HMGB2 overexpression with the progression and angiogenesis of human bladder cancer.12 In this study, we have investigated the HMGB2:SRC-1:ER complex in the endocrine-resistant setting using immunohistochemical, genomic and expression studies. Our results suggest a dynamic relationship between HMGB2 and the ER complex in the endocrine-resistant setting.
Results
HMGB2 associates with SRC-1 and ER in primary tissue from breast cancer patients and its expression can be upregulated in recurrent tumours
SRC-1 is involved in resistance to endocrine treatment and the development of metastasis in breast cancer3, 4, 5 and is overexpressed in a number of tamoxifen-resistant cell lines (Supplementary Figure 4C). We have previously investigated novel SRC-1 interacting proteins in the endocrine-resistant setting, using liquid chromatography-mass spectrometry analysis of immunoprecipitates from the endocrine-sensitive MCF-7 and endocrine-resistant LY2 cell lines after treatment with tamoxifen.6 One of the proteins identified as interacting with SRC-1 in the resistant, but not in the sensitive setting was the chromatin protein HMGB2. As HMGB2 has previously been reported to enhance binding of ER to DNA in in vitro systems, we decided to investigate this interaction further in a breast cancer setting.8,13 The interaction of HMGB2 with SRC-1 and ER in the LY2 cells under tamoxifen treatment was confirmed by immunoprecipitation and western blotting (Figure 1a and Supplementary Figure 1A). Proliferation of both cell lines was not perturbed by knockdown of HMGB2 alone (Supplementary Figures 1B and C).
HMGB2 expression in primary patient tissue predicts extended disease-free survival time; however, resistant tissue and tissue from LY2 xenografts also exhibit HMGB2 positivity. (a) Coimmunoprecipitation of SRC-1/ER and HMGB2 confirmed the interaction of these proteins in the LY2 tamoxifen-resistant cell line upon treatment with tamoxifen (left panel). Western blot illustrating basal protein levels of SRC-1, HMGB2 and ER in the MCF-7 and LY2 cell lines (right panel). (b) HMGB2 IHC in breast cancer patient TMA, illustrating positive staining (left, 10x, bottom right, 40x) and negative staining (top right). (c) Kaplan–Meier estimates of disease-free survival in tamoxifen-treated patients according to HMGB2 expression. HMGB2 positivity correlated with prolonged disease-free survival (Wilcoxon test, P=0.0069). (d) HMGB2 IHC in primary and matched resistant tumours. A number of patients with no staining in their primary tumour had expression in their resistant tumour biopsied after treatment. (e) Xenografts from LY2-luc cells were visualized using the IVIS system (left panel). HMGB2 IHC staining of xenograft tumours derived from the LY2 cell line (right panel). Animals treated with both oestrogen (n=5) and tamoxifen (n=5) expressed HMGB2.
The expression of HMGB2 protein in primary tumour specimens was assessed by immunohistochemistry (IHC) using a tissue microarray (TMA) with samples from 1068 patients. This revealed expression of HMGB2 in 36% of this breast cancer population (Figure 1b). HMGB2 positivity correlated with ER (P<0.001) and SRC-1 expression (P=0.017). With detailed follow-up for this patient population, using univariate analysis we determined that HMGB2 positivity predicted good outcome in tamoxifen-treated patients (P=0.0069, Figure 1c) but not in untreated patients (P=0.5922). Multivariate analysis was also performed taking into account tamoxifen treatment, size, node and grade; HMGB2 had a hazard ratio of 0.859 (P-value=0.317). Notably, in the ER-positive population, patients who went on to suffer a recurrence had a significant association between HMGB2 and SRC-1 expression (P=0.008, n=138, Fisher’s exact test) while in those patients who did not have disease recurrence, the association was lost (P=0.263, n=333, Fisher’s exact test), indicating a potential role for this chromatin protein in the more aggressive disease states (Supplementary Figure 1D).
To investigate this role in more advanced disease, HMGB2 expression was examined in matched primary and resistant tumours from six breast cancer patients. Two patients had HMGB2 expression in both the primary and resistant samples. Notably, HMGB2 expression was activated in the recurrent lesions of three patients whose primary tumours did not express the chromatin protein (Figure 1d). Xenograft tumours were established from the resistant LY2 cells and visualized using the IVIS system (Figure 1e, left). HMGB2 expression was examined by IHC in tumours of mice administered oestrogen alone (n=5) or a combination of oestrogen and tamoxifen (n=5) (Figure 1e). While there was increased expression of HMGB2 in the tumours from mice treated with tamoxifen, this result did not reach significance (Supplementary Figure 1E). These findings would indicate that despite the absence of HMGB2 in some primary tumours before endocrine treatment, there could be an induction of expression in the resistant disease. These data suggest a potential functional role for HMGB2 in the endocrine-resistant setting. To evaluate this, we investigated the functional properties of HMGB2 in both our resistant and sensitive cell line models by performing genome-wide binding assays.
HMGB2 ChIPseq analysis reveals enrichment at the promoter region of genes
HMGB2 ChIPseq was performed in the MCF-7 and LY2 cell lines upon treatment with tamoxifen. To our knowledge, this is the first time ChIPseq of an HMG protein has been completed and these proteins have been described as difficult candidates for transcription factor mapping approaches.14 Successful ChIPseq was achieved through modification of the standard ChIPseq protocol to include a longer cross-linking period at a lower temperature, to enhance cross-linking of the transient interactions of HMGB2 with the DNA. Two replicates were performed and a total of 647 high-confidence binding sites were identified in the MCF-7 cell line and 925 high-confidence binding sites in the LY2 cell line (Figure 2a). An example of a common peak in both the MCF-7 and LY2 cell line was found at the ZNF335 promoter region (Figure 2b). A direct comparison of HMGB2 binding sites in the two cell lines revealed notably different binding patterns. The majority of the HMGB2 peaks were unique to the individual cell lines; the shared peaks between the MCF-7 and LY2 cell lines accounted for 19 and 14% of the total peaks, respectively (Figures 2a and c). Interestingly, HMGB2 peaks in the resistant LY2 cells have a stronger enrichment at the promoter region of genes in comparison with the MCF-7 peaks (Figure 2d). The GREAT (Genomic Regions Enrichment of Annotations Tool) Transcription Factor Targets ontology tool allows comparison with ChIPseq data that links transcription factor regulators to downstream target genes. This tool revealed a significant overlap of HMGB2 binding events with ER regulated genes in the LY2 cell line, which was not significant in the MCF-7 cell line, supporting the theory of a switch in interacting partners from the sensitive to the resistant setting (Figure 2e).
ChIPseq of HMGB2 in tamoxifen conditions in the MCF-7 and LY2 cell lines reveals distinct binding patterns with enrichment at promoter regions and distinct gene targets in each setting. (a) Venn diagram indicating overlap between HMGB2 binding sites in MCF-7 and LY2 cell lines. (b) A common peak for HMGB2 in both cell lines was found at the promoter region of the ZNF335 gene. (c) Heatmap of HMGB2 ChIPseq peaks categorized by cell line. The majority of the peaks found were unique to each cell line. (d) Cis-regulatory element annotation system (CEAS) analysis of ChIPseq binding sites. In the LY2 cells, HMGB2 was enriched at the promoter region of genes. (e) Genomic regions enrichment of annotations tool (GREAT) transcription factor targets ontology analysis of ChIPseq data. Analysis of known transcription factor targets revealed binding of HMGB2 near ER targets in the LY2-resistant cell line but not in the MCF-7 sensitive cell line.
The HMGB2/ER/SRC-1 complex acts at promoter regions of target genes in the LY2 cell line
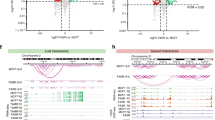
ER binding was mapped for the first time in the LY2 endocrine-resistant cell line and the data combined with that of HMGB2 and with previously published SRC-1 ChIPseq results from our group.15 The majority of the HMGB2 binding sites (52%) in the LY2 cell line overlapped with SRC-1 binding regions and 29% were common with ER peaks (Figure 3a). Moreover, 179 (19%) of the HMGB2 binding sites were common binding sites for both ER and SRC-1, an example of which is shown in Figure 3b, in the region of two genes, IPO4 and TM9SF1. Despite the fact that silencing HMGB2 did not effect ER binding (Supplementary Figure 2A), heatmaps and peak intensity plots from a random sample of peaks revealed that, on average, the ER binding sites that are cobound by HMGB2 and SRC-1 are considerably stronger binding sites than the ER only peaks (Figures 3c and d). Analysis of the genomic locations where the peaks were found revealed that the common HMGB2/SRC-1/ER binding sites were considerably enriched at promoters (29.2%), in comparison with those peaks where ER alone was bound (5% at promoters) (Figure 3e). Motif analysis of the ER only sites predictably revealed the ESR1 (ER) and Forkhead (FoxA1) motifs (Figure 3f). In contrast, the regions which had all three proteins bound did not have a strong presence of these nuclear receptor related motifs but instead had a significant presence of a number of Ets motifs. This would suggest that HMGB2 interacts with SRC-1 and ER on the promoters of genes, along with other transcription factors such as Ets proteins, in comparison with distal enhancer regions where ER appears to associate with a different set of binding partners (Figure 3g).
HMGB2 binding in the LY2 endocrine-resistant cell line, after tamoxifen treatment, overlaps with SRC-1 and ER at non-classic ER binding sites, and this complex is enriched at promoter regions. (a) Venn diagram of HMGB2, SRC-1 and ER binding sites. The majority of HMGB2 binding sites overlap with SRC-1 and/or ER. (b) An example of binding events for HMGB2, SRC-1 and ER. (c) Heatmap illustrating a selection of common binding sites for HMGB2, SRC-1 and ER and sites with ER only. (d) Intensity plots of binding events for each protein. (e) Cis-regulatory element annotation system (CEAS) analysis revealed the HMGB2:ER:SRC-1 common sites were enriched at promoter regions. (f) Motif analysis using the CentDist website indicated that the common sites had an enrichment for Ets motifs in comparison with ESR1 and Forkhead motifs at the ER-only sites. (g) Model depicting the interaction of HMGB2, SRC-1 and ER at the promoter regions, involving other transcription factors such as Ets proteins. At distal enhancer regions, ER generally binds directly to the DNA with FOXA1.
The ER/HMGB2/SRC-1 complex regulates a concise set of target genes in the endocrine-resistant setting
To identify gene targets of this ER complex, we performed gene expression microarray analysis in the LY2 cell line following treatment with Fulvestrant/ICI 182780 (ICI), which degrades ER. This perturbation of ER levels by ICI significantly reduces growth of the LY2 cell line (Figure 4a), suggesting that a functional ER complex is still required for growth of the endocrine-resistant LY2 cells. This drug exposure resulted in differential expression of 632 genes, with 441 downregulated and 191 upregulated (Figure 4b, Supplementary Table 2). A number of classic ER target genes were affected, including ErbB2, XBP1 and Myc. GeneGO pathway analysis confirmed significant perturbation of the ER regulation of G1/S transition, along with changes to the PI3K and ErbB signalling networks (Figure 4c). To identify genes that were regulated by the promoter binding of HMGB2:SRC-1:ER complex, the gene expression data were merged with the LY2 ChIPseq data. Genes that had binding of HMGB2, SRC-1 and ER within a window of 3 kb either side of the transcription start site (TSS) were compared with the gene expression data set. This resulted in a concise list of 12 genes which are regulated by binding within a very short distance form the TSS, including DDX18, Myc and TFAP4 (Figure 4d, Supplementary Table 3). Regulation of a number of these genes by ER was confirmed by RT-qPCR (Figure 4e), although HMGB2 alone did not regulate their expression (Supplementary Figure 2B).
In the endocrine-resistant setting, the ER/SRC-1/HMGB2 complex is involved in transcriptional regulation of a number of genes in the LY2 cell line. (a) Proliferation assay in the LY2 cell line with ICI treatment. (b) Heatmap of differentially expressed genes was determined by gene expression microarray analysis in the LY2 cell line after treatment with vehicle (Veh) or 100 nM ICI. Degradation of ER by ICI resulted in the downregulation of 441 genes and the upregulation of 191 genes. (c) GeneGO Pathway analysis of the differentially regulated genes revealed significant perturbations of a number of pathways involved in breast cancer cell growth and endocrine resistance. (d) Overlapping the gene expression microarray data with the common HMGB2:SRC-1:ER ChIPseq binding sites gave a shortlist of 12 genes, 8 of which were positively regulated by ER. (e) Regulation of these genes by ER was confirmed by knockdown and qPCR.
DDX18 is required for cell growth in endocrine resistance
DDX18 is a DEAD-box protein with putative RNA helicase activity, which is highly conserved across species.16 It has previously been reported to be essential for hematopoietic cell-cycle progression in zebrafish,17 although little is known about its function in humans. Binding of HMGB2, ER and SRC-1 at the TSS of DDX18 was evident from the ChIPseq data (Figure 5a). DDX18 was required for growth of the tamoxifen-resistant cell lines LY2 and MMU2R (Figure 5b, Supplementary Figure 2C) as well as the HER2-positive BT474 and the endocrine-sensitive cell line MCF-7 (Supplementary Figure 3). Analysis of the expression of DDX18 across this panel of cell lines revealed little difference in levels (Supplementary Figure 4B). Furthermore, overexpression experiments confirmed that an increase in the levels of DDX18 caused a significant increase in cell proliferation in the LY2 cell line (Supplementary Figure 4A). We also explored the possibility that DDX18 was involved in cell-cycle regulation in the breast cancer-resistant phenotype. Silencing of DDX18 in the LY2 cell line resulted in a significant increase in the percentage of cells in the G1 phase of the cell cycle, suggesting a role for DDX18 in the G1/S transition (Figure 5c). Using a publically available data set,18 survival analysis in a population of 2239 patients revealed DDX18 mRNA expression to be a significant predictor of poor outcome in ER-positive patients (P=2.3E−11, Figure 5d), with no significance in the ER-negative population.
DDX18 is required for endocrine-resistant cell growth. (a) ChIPseq peaks for HMGB2, SRC-1 and ER at the promoter of the DDX18 gene. (b) Knockdown of DDX18 in the LY2 and MMU2R endocrine-resistant cell lines significantly perturbed proliferation in the presence of 10 nM tamoxifen. Student’s t-test, *P<0.019, **P=0.037. Error bars represent±s.d. from three replicate experiments. (c) Cell-cycle analysis by propidium iodide staining after knockdown of DDX18 resulted in a significant increase in the cell population in the G1 phase of cell cycle in the presence of 10 nM tamoxifen. Student’s t-test, ***P=0.017. Error bars represent±s.d. from three replicate experiments. (d) Kaplan–Meier estimates of disease-free survival in tamoxifen-treated patients according to DDX18 expression. The upper and lower quartile of expression was used to group these patients. High levels of DDX18 mRNA correlated with reduced disease-free survival (Wilcoxon test, P=2.3E−11).
Discussion
Tamoxifen resistance remains a clinical problem for a considerable number of breast cancer patients today. Progression of disease while on this treatment has been reported to be due to a number of different mechanisms of action, including upregulation of growth factor pathways and deregulation of coregulatory proteins.19 We have previously demonstrated that the coactivator protein SRC-1 has an important role in breast cancer progression.3,5 Using mass spectrometry, HMGB2 was identified as an interactor of SRC-1 in the endocrine-resistant, but not in the endocrine-sensitive, setting.6 In the study presented here, analysis of HMGB2 expression in our patient population revealed this protein to be a significant predictor of prolonged disease-free survival in tamoxifen-treated patients. Further analysis of tissue samples from resistant tumours revealed that HMGB2 expression could in fact be switched on in the resistant setting. In a xenograft model using LY2 cells, HMGB2 was strongly expressed upon treatment with tamoxifen. As a result we decided to investigate the role for HMGB2 in the endocrine-resistant phenotype, considering the presence of HMGB2 protein in the primary tumour being a marker for good prognosis, but with a shift in some resistant tumours to also express HMGB2. This conflicting expression is similarly reported for the pioneer factor FOXA1; expression of this protein correlates with good outcome in breast cancer patients20,21 but expression is also present in the majority of metastatic samples.22
HMGB proteins are known to interact with DNA in a transient manner8 and so obtaining ChIPseq data for this protein required optimization and modifications to capture these interactions between HMGB2 and the DNA. ChIPseq of HMGB2 revealed considerably different binding patterns in the endocrine-resistant and -sensitive cell lines. In the resistant LY2 cells, there was strong enrichment at the promoter region of genes and at previously reported targets of ER. HMGB proteins have previously been reported to facilitate binding of nuclear receptors to the DNA in an in vitro system8 and to act as coactivator proteins for Ets1 and 2 in prostate cells.23 They have also been shown to stimulate transcription in vitro by RNA polymerase II and III.10 This study is the first to elucidate the genomic positioning for HMGB2 in a cellular context.
Comparison of the binding of HMGB2 with the nuclear receptor ER and the coactivator protein SRC-1 in the resistant setting revealed that the majority of the HMGB2 binding sites overlapped with one or both of these factors. At the common binding regions, ER had stronger binding intensity than at those with ER alone, while analysis of these genomic sites revealed a propensity for this complex to bind at the promoter region and in the vicinity of Ets motifs. This would suggest that ER might bind to these promoter regions via other transcription factors. Classic ER binding sites are found at enhancers, which are generally at distal regions far from TSSs.24 However, a recent report suggests that activation of ER by protein kinase A redirects the receptor to the promoter region.25 The results presented here could suggest a switch from more classic ER biology in the endocrine-sensitive setting to a resistant system more reliant on other transcription factors including those of the MAPK pathway. This is consistent with previous reports highlighting a role for growth factor pathways in endocrine resistance.26,27
To determine the influence of the promoter-bound ER in the resistant phenotype, we determined whether these genes were influenced by the loss of ER after Fulvestrant treatment of the LY2 cell line. This drug degrades the ER and has a significant effect on growth of this cell line, with a concomitant modulation of a considerable number of genes, demonstrating the dependence of this cell line on the receptor. To define the genes regulated by the ER:HMGB2:SRC-1 complex at the promoter region, we overlapped the microarray data with the list of genes that had all three proteins at a single binding site within a 3-kb window either side of the TSS. This produced a concise list of 12 genes including DDX18, the oncogene Myc and the transcription factor TFAP4. Regulation of DDX18 by ER has not been previously reported.
In zebrafish, DDX18 has previously been shown to have a role in the regulation of cell-cycle progression.17 There is minimal information about its function in humans but it has been reported to be regulated by Myc in lymphocytes.16 Other members of the DDX family of proteins, DDX5 and DDX17, have previously been reported to act as ER coactivators.28 The results here could indicate a positive feedback loop of the ER complex upregulating DDX18 to then utilize this protein as a coactivator. Notably, silencing of DDX18 in three tamoxifen-resistant cell lines and one tamoxifen-sensitive cell line resulted in significant inhibition of growth. Given the report of cell-cycle regulation by DDX18 in zebrafish, the role in endocrine-resistant cell growth was analysed, with a significant increase in the cell population in the G1 phase upon silencing of this protein. The expression of DDX18 in breast cancer patient tumours was found to be significantly associated with outcome, with high DDX18 mRNA levels correlating with poor outcome.
This study illustrates the dynamic factors involved in breast cancer and the development of drug resistance. HMGB2 protein levels in primary tumours, before any hormonal treatment, predict prolonged disease-free survival times in a large cohort of breast cancer patients who went on to be treated with tamoxifen. However, expression of this protein may increase in resistant tumours after treatment with this drug and expression was evident in the presence of tamoxifen in xenograft tumours. Our data indicate that in the resistant setting, a complex including HMGB2, the coactivator SRC-1 and the nuclear receptor ER bind at the promoter of DDX18. These findings reveal a complex system of gene expression changes in the development of resistance to therapies; more detailed investigation is needed to understand the intricacies of this process and to elucidate further potential targets for drug treatments.
Materials and methods
Cell lines, treatments and proliferation assays
Endocrine-sensitive MCF-7 (American Type Culture Collection (ATCC)), endocrine-resistant BT474 (American Type Culture Collection (ATCC)), LY2 cells (kind gift from R Clarke, Georgetown, Washington DC) and MMU2R cells (kind gift from V Speirs, University of Leeds) were grown as previously described.26,29,30 Cells were maintained in steroid-depleted medium for 72 h before treatment with hormones (estradiol (Est) 10−8 mol/l, 4-hydroxytamoxifen (4-OHT) 10−8 mol/l; Sigma-Aldrich, St Louis, MO, USA) over varying time periods. All cells were incubated at 37 °C in 5% CO2 in a humidified incubator. For proliferation assays, cells were seeded into 24-well dishes at a cell density of 2 × 104 per well and growth was monitored using the Essen Bioscience INCUCYTE machine (Ann Arbor, MI, USA).
Coimmunoprecipitation and western blotting
Protein was immunoprecipitated with anti-SRC-1 and anti-ER and blotted for HMGB2. Western blotting was carried out using rabbit anti-SRC-1 (sc-8995, Santa Cruz, Santa Cruz, CA, USA), rabbit anti-ER (sc-543, Santa Cruz), rabbit anti-HMGB2 (ab67282, Abcam) or mouse anti-β-actin (ab6276, Abcam, Cambridge, UK) antibodies.
Patient information and construction of TMA
Following ethical approval, two TMAs were constructed using breast tumour samples from two separate clinical institutions. The TMA construction was conducted as previously described.31 The first TMA contained samples from archival cases at St. Vincent’s University Hospital, Dublin Ireland from 1987 to 1999 (n=560). The second TMA was constructed using samples from archival cases at Beaumont Hospital, Dublin, Ireland from 2000 to 2008 (n=508). Data on the patients for both TMAs included tumour size, grade, lymph-node status, HER2 status and ER status (Supplementary Table 4).
Mouse xenograft model
All mouse experiments were performed in accordance with the European Communities Council Directive (86/609/EEC) and were reviewed and approved by the Research Ethics Committee of the Royal College of Surgeons in Ireland (REC #406) under license from the Department of Health, Dublin, Ireland. Six-week-old female BALB/c SCID mice (Harlan, Indianapolis, IN, USA) were implanted with either 17-B-estradiol (0.36 mg/pellet, 60-day release) pellets alone or with both 17-B-estradiol (n=5 animals, 0.36 mg/pellet, 60-day release) and tamoxifen (n=5 animals, 5 mg/pellet, 60-day release) pellets from Innovative Research of America. One week later, 1 × 106 LY2-luc cells mixed with 50% Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) were implanted into the fourth inguinal mammary gland by injection. Five weeks after implantation, tumours were imaged using an IVIS whole body imaging system (Caliper Life Sciences, Hopkinton, MA, USA) to detect luciferase activity of the LY2-luc cells and primary tumours were surgically removed, formalin fixed and paraffin embedded.
Immunohistochemistry
Staining of tissue samples and scoring was performed as described previously.3 Xenograft images were analysed using the Aperio software (Leica, Milton Keynes, UK) and a positivity score was determined for each sample. The primary antibody used was mouse anti-HMGB2 (H00003148-M05, Novus, Littleton, CO, USA) at a concentration of 5 μg/ml incubated overnight at 4 °C.
ChIP-sequencing experiments
ChIP experiments were performed as described previously.32 Antibodies used were anti-HMGB2 (ab67282, Abcam) and anti-ER (sc-543, Santa Cruz). Cells were treated for 45 min and harvested for ChIPseq. This entailed crosslinking proteins to the chromatin by incubation with 1% formaldehyde for 60 min at 4 °C (HMGB2) or 10 min at room temperature (ER), followed by quenching with glycine (0.1 M). Cells were scraped into PBS with protease inhibitors. Chromatin was extracted from the cells and the protein of interest, along with the DNA attached, was immunoprecipitated using an antibody attached to Dynal beads. The proteins were then removed from the DNA by reverse crosslinking overnight and the DNA purified and amplified before being sequenced using the Illumina Genome Analyzer-II system (Illumina, San Diego, CA, USA). Single end 36-bp ChIP-seq data were generated by the Illumina analysis pipeline version 1.6.1 (Illumina), and reads were aligned to the Human Reference Genome (assembly hg18, NCBI Build 36.6, March 2008) using BWA version 0.5.5.33 Reads were filtered by alignment quality, with all reads with a score of <15 removed. ER and SRC-1 peaks were called using MACS, version 1.3.7.1,34 HMGB2 peaks were called using SPP,35 then consensus peaks were chosen according to the IDR statistic.36 Peaks were visualized using the University of California, Santa Cruz genome browser (http://genome.ucsc.edu). Two biological replicates were performed. The Cistrome platform,37 Genomic Regions Enrichment of Annotations Tool (GREAT) website38 and CentDist website 39 were used to analyse ChIPseq peaks data sets.
Microarray analysis
Cells were maintained in steroid-depleted medium for 72 h before treatment with vehicle or 100 nM ICI 182780 for 6 h. RNA was collected from four biological replicates. The Illumina BeadChIP (HumanWG-12 version 4) bead-level data were preprocessed, log2-transformed and quantile normalized using the beadarray package40,41 in Bioconductor.42 Differential expression analysis was performed using limma eBayes43 with a Benjamini and Hochberg multiple test correction procedure44 to identify statistically significant differentially expressed genes (false discovery rate 0.01). Gene Ontology pathway enrichment was determined using GeneGO Metacore.
siRNA and overexpression assays
Cells were transfected with ER or DDX18 siRNA (ON-TARGETplus individual siRNA J-013451, Dharmacon, Lafayette, CO, USA) using Lipofectamine 2000 (Invitrogen, Paisley, UK). AllStars Negative Control siRNA (Qiagen, Manchester, UK) was used as a negative control. Dual transfections of DDX18 siRNA and DDX18 overexpression vector (SC115885 Origene) were performed using Lipofectamine 2000 (Invitrogen).
Gene expression by RT-qPCR
Cells were transfected with siRNA. Total RNA was collected 48 h later and RT-qPCR performed using primers listed in Supplementary Table 1.
Flow cytometry
Cells were transfected with siRNA and harvested 72 h later, fixed with ethanol, treated with RNAse A and stained with propidium iodide. Samples were run on the BD Biosciences FACSCalibur flow cytometer and cell-cycle populations analysed using the FlowJo software (FlowJo, Ashland, OR, USA).
Statistical analysis
Statistical analyses for proliferation and cell-cycle experiments were performed using the two-tailed Student’s t-tests. Only tests with a P-value of <0.05 were considered as statistically significant. Error bars represent standard deviations. Univariate statistical analysis was carried out using Fisher's Exact test for categorical variable and Wilcoxon test for continuous variables. Multivariate analysis was conducted using the STATA 10 data analysis software (Stata Corp. LP, Stata Corp, College Station, TX, USA) and Cox’s proportional hazard model, using the Breslow method for ties.
Data access
The microarray data and ChIPseq data from this study have been deposited in ArrayExpress with accession numbers E-MTAB-1870 and E-MTAB-1865, respectively.
References
Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 2011; 378: 771–784.
Onate SA, Tsai SY, Tsai MJ, O'Malley BW . Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 1995; 270: 1354–1357.
Redmond AM, Bane FT, Stafford AT, McIlroy M, Dillon MF, Crotty TB et al. Coassociation of estrogen receptor and p160 proteins predicts resistance to endocrine treatment; SRC-1 is an independent predictor of breast cancer recurrence. Clin Cancer Res 2009; 15: 2098–2106.
Qin L, Liu Z, Chen H, Xu J . The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res 2009; 69: 3819–3827.
McBryan J, Theissen SM, Byrne C, Hughes E, Cocchiglia S, Sande S et al. Metastatic progression with resistance to aromatase inhibitors is driven by the steroid receptor coactivator SRC-1. Cancer Res 2012; 72: 548–559.
Walsh CA, Bolger JC, Byrne C, Cocchiglia S, Hao Y, Fagan A et al. Global gene repression by the steroid receptor coactivator SRC-1 promotes oncogenesis. Cancer Res 2014; 74: 2533–2544.
Stros M . HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 2010; 1799: 101–113.
Melvin VS, Harrell C, Adelman JS, Kraus WL, Churchill M, Edwards DP . The role of the C-terminal extension (CTE) of the estrogen receptor alpha and beta DNA binding domain in DNA binding and interaction with HMGB. J Biol Chem 2004; 279: 14763–14771.
Zappavigna V, Falciola L, Helmer-Citterich M, Mavilio F, Bianchi ME . HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J 1996; 15: 4981–4991.
Tremethick DJ, Molloy PL . High mobility group proteins 1 and 2 stimulate transcription in vitro by RNA polymerases II and III. J Biol Chem 1986; 261: 6986–6992.
Laurent B, Randrianarison-Huetz V, Marechal V, Mayeux P, Dusanter-Fourt I, Dumenil D . High-mobility group protein HMGB2 regulates human erythroid differentiation through trans-activation of GFI1B transcription. Blood 2010; 115: 687–695.
Wang W, Jiang H, Zhu H, Zhang H, Gong J, Zhang L et al. Overexpression of high mobility group box 1 and 2 is associated with the progression and angiogenesis of human bladder carcinoma. Oncol Lett 2013; 5: 884–888.
Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol 1998; 18: 4471–4487.
Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P et al. DNA-binding specificities of human transcription factors. Cell 2013; 152: 327–339.
McCartan D, Bolger JC, Fagan A, Byrne C, Hao Y, Qin L et al. Global characterization of the SRC-1 transcriptome identifies ADAM22 as an ER-independent mediator of endocrine-resistant breast cancer. Cancer Res 2012; 72: 220–229.
Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN . Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J 1996; 15: 4344–4357.
Payne EM, Bolli N, Rhodes J, Abdel-Wahab OI, Levine R, Hedvat CV et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood 2011; 118: 903–915.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731.
Musgrove EA, Sutherland RL . Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 2009; 9: 631–643.
Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM et al. FOXA1 expression in breast cancer–correlation with luminal subtype A and survival. Clin Cancer Res 2007; 13: 4415–4421.
Hisamatsu Y, Tokunaga E, Yamashita N, Akiyoshi S, Okada S, Nakashima Y et al. Impact of FOXA1 expression on the prognosis of patients with hormone receptor-positive breast cancer. Ann Surg Oncol 2012; 19: 1145–1152.
Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012; 481: 389–393.
Shiota M, Izumi H, Miyamoto N, Onitsuka T, Kashiwagi E, Kidani A et al. Ets regulates peroxiredoxin1 and 5 expressions through their interaction with the high-mobility group protein B1. Cancer Sci 2008; 99: 1950–1959.
Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 2006; 38: 1289–1297.
de Leeuw R, Flach K, Bentin Toaldo C, Alexi X, Canisius S, Neefjes J et al. PKA phosphorylation redirects ERalpha to promoters of a unique gene set to induce tamoxifen resistance. Oncogene 2013; 32: 3543–3551.
Al-azawi D, Ilroy MM, Kelly G, Redmond AM, Bane FT, Cocchiglia S et al. Ets-2 and p160 proteins collaborate to regulate c-Myc in endocrine resistant breast cancer. Oncogene 2008; 27: 3021–3031.
Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res 2008; 68: 826–833.
Wortham NC, Ahamed E, Nicol SM, Thomas RS, Periyasamy M, Jiang J et al. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene 2009; 28: 4053–4064.
Limer JL, Parkes AT, Speirs V . Differential response to phytoestrogens in endocrine sensitive and resistant breast cancer cells in vitro. Int J Cancer 2006; 119: 515–521.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–527.
Dillon MF, Stafford AT, Kelly G, Redmond AM, McIlroy M, Crotty TB et al. Cyclooxygenase-2 predicts adverse effects of tamoxifen: a possible mechanism of role for nuclear HER2 in breast cancer patients. Endocr Relat Cancer 2008; 15: 745–753.
Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT . ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 2009; 48: 240–248.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137.
Kharchenko PV, Tolstorukov MY, Park PJ . Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 2008; 26: 1351–1359.
Li Q, Brown JB, Huang H, Bickel PJ . Measuring reproducibility of high-throughput experiments. Ann Appl Stat 5: 1752–1779.
Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 2011; 12: R83.
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28: 495–501.
Zhang Z, Chang CW, Goh WL, Sung WK, Cheung E . CENTDIST: discovery of co-associated factors by motif distribution. Nucleic Acids Res 2011; 39: W391–W399.
Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG . BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics 2008; 24: 2921–2922.
Dunning MJ, Smith ML, Ritchie ME, Tavare S . beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 2007; 23: 2183–2184.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol: 2004; 3; Article 3.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995; 57: 289–300.
Acknowledgements
We would like to acknowledge the support of Irish Research Council for Science Engineering and Technology co-funded by Marie Curie Actions under FP7, Breast Cancer Ireland, Health Research Board, The University of Cambridge, Cancer Research UK, Hutchison Whampoa Limited and European Molecular Biology Organisation. We thank the members of the genomics core facility and Stuart MacArthur, former member of the bioinformatics core facility at Cancer Research UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Redmond, A., Byrne, C., Bane, F. et al. Genomic interaction between ER and HMGB2 identifies DDX18 as a novel driver of endocrine resistance in breast cancer cells. Oncogene 34, 3871–3880 (2015). https://doi.org/10.1038/onc.2014.323
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.323
- Springer Nature Limited
This article is cited by
-
Identification of HMGB2 associated with proliferation, invasion and prognosis in lung adenocarcinoma via weighted gene co-expression network analysis
BMC Pulmonary Medicine (2022)
-
Crucial role of high-mobility group box 2 in mouse ovarian follicular development through estrogen receptor beta
Histochemistry and Cell Biology (2022)
-
Identification of differentially expressed proteins and clinicopathological significance of HMGB2 in cervical cancer
Clinical Proteomics (2021)
-
Molecular markers associated with the outcome of tamoxifen treatment in estrogen receptor-positive breast cancer patients: scoping review and in silico analysis
Discover Oncology (2021)
-
HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer
Cell Communication and Signaling (2018)