Abstract
Glucagon-like peptide 1 (GLP-1) regulates glucose homeostasis through the control of insulin release from the pancreas. GLP-1 peptide agonists are efficacious drugs for the treatment of diabetes. To gain insight into the molecular mechanism of action of GLP-1 peptides, here we report the crystal structure of the full-length GLP-1 receptor bound to a truncated peptide agonist. The peptide agonist retains an α-helical conformation as it sits deep within the receptor-binding pocket. The arrangement of the transmembrane helices reveals hallmarks of an active conformation similar to that observed in class A receptors. Guided by this structural information, we design peptide agonists with potent in vivo activity in a mouse model of diabetes.
Similar content being viewed by others
Main
Diabetes and obesity are serious public health concerns worldwide with increasing prevalence. GLP-1 is a hormone released primarily from the gastrointestinal tract in response to food intake and controls the secretion of insulin from pancreatic islets in a glucose-dependent manner1. Several GLP-1 agonists are now approved for the treatment of type 2 diabetes and, in addition to their insulinotropic action, also have beneficial effects on body weight and cardiovascular parameters2. Native GLP-1 is rapidly degraded by the circulating dipeptidyl peptidase 4 and therapeutic peptides have been modified to improve their stability and duration of action3,4. Current agents are given by injection and are costly to produce, and the search for small-molecule agonists has so far proved unsuccessful5. An alternative is to design smaller peptides and derivatives with oral bioavailability.
GLP-1 activates the GLP-1 receptor (GLP-1R), a class B G-protein-coupled receptor (GPCR) that is expressed on pancreatic islets as well as other organs including the heart, gastrointestinal tract and brain. GLP-1R exhibits a multi-domain architecture consisting of a large amino-terminal extra-cellular domain (ECD), which is required for binding the C-terminal portion of the GLP-1 peptide, and a transmembrane domain (TMD), which binds the N-terminal portion of the peptide6. Although X-ray structures of the ECD bound to peptides have been solved in isolation7,8, no structural data exist for the full receptor. TMD structures have been determined for the related class B glucagon receptor (GCGR)9,10; however, these did not include the ECD and were solved for receptors bound to antagonists. We designed potent truncated peptides with full agonist activity and determined the structure of one of these peptides (peptide 5) bound to the full-length GLP-1R.
Discovery of GLP-1 agonist nonapeptides
A prerequisite for obtaining an X-ray structure of GLP-1R was identification of a radiochemical suitable for generation of a GLP-1R thermostabilized receptor (StaR) in the agonist conformation. A series of truncated peptides has previously been reported11,12,13 and peptide 1 was selected as a closely related sequence (Extended Data Fig. 1 and Extended Data Table 1) for radiolabelling. Peptide 1 corresponds to residues 7–17 of the natural GLP-1 sequence: H7-A8-E9-G10-T11-F12-T13-S14-D15-V16-S17 (the first six residues are removed as part of post-translational processing of GLP-1). Peptide 1 contains the following substitutions: A8 with α-aminoisobutyric acid (Aib), F12 with α-methyl-Phe, V16 with 3-(4′-methoxy-2′-ethyl[1,1′-biphenyl]-4-yl)-L-alanine (referred to as X2) and S17 with 5-(3,5-dimethylphenyl)-L-norvaline, (referred to as X3). An olefin precursor was radiolabelled to produce the tritiated version of peptide 1. Focusing on optimization of the N terminus, the F12 position was further modified to α-methyl-o-fluoro-Phe, (referred to as X1) based on previous studies12. The H7–Aib8 N terminus was replaced with a ‘cap’ (Extended Data Table 1, referred to as Cap1), reversing the first peptide bond and removing the N-terminal amino group (see Supplementary Information). The resulting nonapeptide, peptide 2, has a significantly increased agonist potency compared to peptide 1 but no increased binding affinity (Extended Data Table 1). Next, E9 was substituted with Gln, N-linked tetrazolyl-Ala or C-linked tetrazolyl-Ala, generating peptides 3, 4 and 5, respectively (Extended Data Table 1). Of these, peptide 5 successfully recapitulated the affinity and potency of full-length peptides such as GLP-1 itself (Extended Data Fig. 1 and Extended Data Table 1) and was selected for structural studies.
Peptide 1 was used to generate a StaR that preferentially adopts the active conformation using an approach described previously14. The StaR construct contains 11 amino acid substitutions which substantially increase the thermal stability of the receptor while maintaining the same affinity towards peptide 1 as the wild-type receptor (Extended Data Fig. 2). In functional assays, the StaR shows reduced agonist potency compared with the wild-type receptor (Extended Data Fig. 2). This is consistent with other receptors stabilized in the agonist conformation15,16 and is indicative of the receptor being in a partially active conformation.
The crystallized construct includes both the ECD and TMD but lacks the last 31 residues (433–463). Crystals with peptide 5 were grown using the vapour diffusion method and the structure was solved by molecular replacement to 3.7 Å resolution, with one receptor molecule per asymmetric unit (Extended Data Table 2).
Overall architecture of GLP-1R
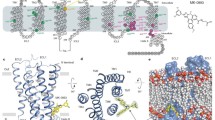
GLP-1R adopts an elongated conformation with the ECD extending away from the plane of the lipid bilayer (Fig. 1). The ECD adopts the same fold as previously reported for the isolated domain7 (PDB ID: 3IOL) with a root mean square deviation (r.m.s.d.) of 0.93 Å for main-chain atoms and is connected to the first helix of the TMD by a ten-residue linker sequence. In contrast to previously published data on the glucagon receptor, we do not observe a helical stalk linking ECD and TMD9,17. The helical stalk is thought to be stabilized by the peptide ligand traversing this region17 or by the N-terminal bRIL fusion and helix 8 of the adjacent unit cell9, both of which are absent in our structure. The TMD features the canonical seven transmembrane helices (TM1–TM7) arranged similarly to previously published class B GPCRs and displays the same V-shaped central cavity9,10,18. Continuous electron density was observed for all intra- and extracellular loops (ICLs and ECLs, respectively). All connecting loops lack regular secondary structure apart from ECL1, which forms a short helix–turn–helix. TM2 and ECL1 extend well outside the plane of the membrane and contact the ECD. ECL2 is anchored to TM3 by the conserved disulfide bond between Cys2263.29b and Cys296 (ECL2) (numbers in superscript refer to modified Ballesteros numbering system for class B GPCRs19). Residues 24 to 28 at the N terminus and residues 418 to 432 at the C terminus of the receptor are not resolved. The following regions of the structure show a high degree of flexibility as judged by the quality of the electron density map and the resultant refined B-factors: 127–136 (linker between ECD and the TMD), 206–224 (ECL1), 254–266 (ICL2), 291–307 (ECL2), 333–341 (ICL3), 362–380 (top of TM6 and ECL3) and 406–417 (helix 8) (Extended Data Fig. 3).
a, Ribbon representation of GLP-1R (TMD coloured cyan; ECD coloured brown), as viewed parallel to the membrane with approximate membrane boundaries indicated by grey boxes. The glycosylation modification resolved on the receptor ECD is shown in stick representation with carbon, nitrogen and oxygen atoms coloured grey, blue and red, respectively. b,The peptide 5 agonist is shown in stick representation with carbon, nitrogen and oxygen atoms coloured yellow, blue and red, respectively, Fo − Fc omit density map for peptide 5 is contoured at 2.5σ.
Peptide agonist binding pocket
Strong additional electron density (Fig. 1) indicative of an α-helix was observed in the intra-helical cavity of GLP-1R. Peptide 5 was modelled in the density with the N terminus deepest in the binding pocket and the C terminus directed towards the ECD (Fig. 2a). The additional electron density branches near the C terminus of the peptide with X2 located between TM1 and TM2 and X3 located at the ECD–ECL1/ECL2 boundary. The contact surface area between peptide 5 and the receptor measures 1,584 Å2. The final refined model was subsequently used in ensemble refinement20, which indicated which parts of the structure are reliably interpretable (Extended Data Fig. 4).
a, Surface representation of GLP-1R as viewed parallel to the membrane plane, clipped to reveal peptide 5 (coloured as in Fig. 1). b–g, Interactions between the different groups of peptide 5 (moving from the N to C terminus) and GLP-1R (potential intra and intermolecular polar contacts depicted as dashed red lines); b–d, view from membrane, e–g, view from extracellular space. b, The amide of Cap1 and gem-dimethyl group of Cap1 which substitutes for A8 in peptide 5. c, The C-linked tetrazolyl-Ala (C-tet-Ala) in place of E9. d, G10, T11, S14 and D15. e, The α-methyl-o-fluoro-Phe substitution of F12 (X1). f, The 3-(4′-methoxy-2′-ethyl[1,1′-biphenyl]-4-yl)-l-alanine substitution of V16 (X2). g, The 5-(3,5-dimethylphenyl)-l-norvaline substitution of S17 (X3).
The amide of Cap1 is poised to interact with E3877.42b, which itself appears to interact stably with K3837.38b (Fig. 2b and Extended Data Fig. 4a). Charge reversal of E3877.42b results in significant loss of agonist peptide binding affinity21, and mutation of K3837.38b to alanine results in significant loss of peptide potency22. The gem-dimethyl group of Cap1 that substitutes for A8 is located in a hydrophobic pocket lined by L3847.39b and L3887.43b (Fig. 2b and Extended Data Fig. 4b). Consistently, mutating either of these residues significantly reduces agonist peptide binding affinity21,23. The C-linked tetrazolyl-Ala replacing E9 is within bonding distance of Y1521.47b and R1902.60b, both of which have been shown to be critical for GLP-1 binding affinity19,21,24,25. Moreover, the interaction of the C-linked tetrazolyl-Ala with R1902.60b is consistent with the predicted interaction of E9 from GLP-1 with R1902.60b (refs 22, 23). The side chain of R1902.60b also appears to interact with the side chains of both T3917.46b and N2403.43b (Fig. 2c and Extended Data Fig. 4c). G10 faces W3065.36b, which is poised to interact with N300 (ECL2; Extended Data Fig. 4d). Mutation of W3065.36b to alanine results in a significant affinity loss of GLP-1 to GLP-1R22. T11 is in close proximity to L3847.39b and D372 of ECL3, with the side chain of the latter also engaging with R3807.35b (Fig. 2d and Extended Data Fig. 4e). Mutating either D372 or R3807.35b to alanine has no effect on agonist affinity; instead, these mutations significantly reduce agonist potency22. The X1 substitution of F12 (Extended Data Table 1) is located in a hydrophobic pocket formed by L1411.36b, L1441.39b, Y1481.43b, L3847.39b and L3887.43b and is positioned favourably to π stack with the side chain of Y1481.43b (Fig. 2e and Extended Data Fig. 4f). The hydroxyl group of Y1481.43b points towards K1972.67b and D1982.68b on TM2 and mutation of these two residues leads to loss of agonist binding affinity21,24,25. This suggests that K1972.67b and D1982.68b hold the aromatic moiety of Y1481.43b in an optimal position for interaction with the peptide. T13 is located in a pocket formed by the side chains of K1972.67b, L2012.71b, F2303.33b, M2333.36b and the mutated T298 (ECL2; Extended Data Fig. 4g). Both L2012.71b and M2333.36b were shown to be critical for peptide binding21,23. In keeping with a previously reported homology model22, S14 is found within bonding distance of R299 and N300 (ECL2; Extended Data Fig. 4h) and D15 potentially interacts with R3807.35b (Fig. 2d and Extended Data Fig. 4i). R299, N300 and R3807.35b have previously been shown to contribute significantly to peptide binding and potency22,23,26,27,28. The X2 substitution of V16 exits the peptide binding pocket by inserting into a hydrophobic groove between TM1 and TM2 formed by the side chains of L1411.36b, L1421.37b, Y1451.40b, L2012.71b, the aliphatic portion of K2022.72b, Y205 (ECL1) and P1371.32b (Fig. 2f and Extended Data Fig. 4j). At a right angle from X2 lies the X3 substitution of S17 in a mainly hydrophobic groove created by L32 and W33 of the ECD, M204, Y205 and L217 of ECL1, the aliphatic portion of Q2213.24b and the mutated G295 and T298 of ECL2 (Fig. 2g and Extended Data Fig. 4k). Of these residues, M204 and Y205 have been demonstrated to contribute towards peptide binding21,23,29. Finally, the carbonyl group of the substituted S17 is poised to interact with S31 of the ECD and the terminal amine of peptide 5 with Y205 from ECL1 (Fig. 2f and Extended Data Fig. 4k).
In the absence of a crystal structure of the full-length GLP-1 peptide bound to the receptor, we have attempted to understand the binding mode of full-length GLP-1 peptide to the receptor by superposing the GLP-1 peptide from the ECD complex structure onto peptide 5. This shows that the position of the ECD in complex with the full-length peptide might be different from that observed in our structure (Extended Data Fig. 5). Assuming that the conformation of the GLP-1 peptide in complex with the full receptor remains unchanged, this difference could arise from the inherent flexibility of the ECD17,30. The position of the ECD in the peptide-5-bound structure is probably a consequence of the interaction of ECD residues L32 and W33 with X3, which is not present in the ECD–GLP-1 complex structure (Extended Data Fig. 5). It is possible that binding of different peptide agonists could alter the juxtaposition of the domains and this may contribute to the ability of different peptides to activate the receptor.
The relationship between ligand binding affinity and complementary interactions between peptide 5 and lipophilic hotspots on GLP-1R was analysed as for other GPCRs31. The X2 and X3 groups form energetically favourable interactions with lipophilic hotspots on GLP-1R32,33 (Extended Data Fig. 6). The dimethyl phenyl group of X3 overlaps with a substantial region of lipophilicity while the X2 side chain places the pendant ethyl group of the biaryl group into another lipophilic patch. These interactions provide a rationale for the high affinity of the truncated peptides in the absence of an interaction with the ECD.
Comparison to other class B GPCRs
Superposition of the transmembrane helices (TM1–TM7) of GLP-1R with those of GCGR (PDB ID: 5EE7) and CRF1R (PDB ID: 4K5Y) results in r.m.s.d. values for main-chain atoms of 2.7 Å and 3.4 Å, respectively. Considerable differences are observed between the extracellular surfaces of these three receptors, where the extracellular ends of TM1, TM6 and TM7 of GLP-1R are rotated clockwise (as viewed from extracellular space) around the central TMD axis by 4.7 Å, 5.1 Å and 3.4 Å, respectively, compared to GCGR (Fig. 3a), and 7.4 Å, 10.6 Å and 10.9 Å, respectively, compared to CRF1R (Fig. 3c), where distances are measured between the Cα atoms of residues at positions 1.37b, 6.58b and 7.35b.
a, Structural superposition of GLP-1R TMD with GCGR TMD in ribbon representation, as viewed from extracellular space. b, Representation as in a rotated to view from the cytoplasm. c, Structural superposition of GLP-1R TMD with CRF1R TMD as viewed from extracellular space. d, Representation as in c rotated to view from the cytoplasm. Differences in transmembrane helix positions are denoted by red arrows. e, Superposition of eight class A and B GPCR structures focusing only on TM6 and covering the known range of activation states. PDB accession codes for the structures are as follows: A2A (red): 3PWH; GCGR: 5EE7; CRF1R: 4K5Y; Rho: 4ZWJ; GLP-1R: 5NX2; β2AR–Nb (nanobody): 4LDE; A2A (green): 5G53; β2AR–Gs: 3SN6.
The conformations of ECL1 differ between the three receptors. ECL1 in GCGR, though not resolved completely, lacks secondary structure and ECL1 in CRF1R is short and adopts an α-helical structure. However, in GLP-1R, TM2 exhibits an extended α-helical structure ending at residue S206, where the helix unwinds and then forms a short α-helix between residues W214 and L217. The extended conformation of TM2 positions ECL1 upwards, and further out into the extracellular space where it interacts with the ECD (residues E207–W214 of ECL1, and residues E127–R131 of the ECD) (Fig. 1). In GLP-1R, ECL2 is retracted and adopts a conformation similar to that of CRF1R (Fig. 3c). However, ECL2 in GCGR10 stretches across the central axis of the transmembrane helical bundle mediating interactions from TM3 across to TM6 and TM7 (Fig. 3a).
The antagonist-bound structures of GCGR and CRF1R revealed interactions between the intracellular portions of TM6 and those of TM3 and TM7 (Fig. 3b, d). In the agonist-bound GLP-1R structure, the intracellular portion of TM6 swings away from the central axis of the TMD by over 10 Å (distances measured between the Cα atoms of residues at position 6.35b in each receptor; GLP-1R–GCGR, 11.7 Å; GLP-1R–CRF1R, 10.8 Å) (Fig. 3b, d). When TM6 and TM7 are excluded from the superposition of GLP-1R with GCGR and CRF1R, the structures are in much closer agreement (r.m.s.d. values of 1.5 Å and 1.4 Å for main-chain atoms, respectively), demonstrating the substantial rearrangements of TM6 and TM7 relative to TM1–TM5 between agonist-bound GLP-1R and antagonist-bound GCGR and CRF1R.
Peptide 5 intersects the central axis of the TMD at an angle of approximately 25°, affecting different regions of GLP-1R (Figs 1 and 2). In broad mechanistic terms, the buried N terminus of peptide 5 holds the middle of TM1, TM2, TM3 and the extracellular half of TM7 together to permit the observed movement of TM6, while the C terminus holds in the extracellular halves of TM1, TM2, TM3 and the ECLs. In comparison to the fully activated β2AR–Gs complex34, TM6 in GLP-1R is not as far from the central TMD axis (Fig. 3e), indicating that the agonist-peptide-bound GLP-1R structure is only partially activated. Yet this ‘opening’ of the intracellular GLP-1R surface probably facilitates G-protein coupling and β-arrestin signalling, in a manner similar to that observed for the inactive to active receptor state transitions in class A GPCR structures, indicating a common activation mechanism across class A and B receptors35,36.
Design of potent GLP-1 agonist peptides
As part of the ongoing peptide program, peptides 6, 7 and 8 (Extended Data Table 1) were synthesized, further modifying the N-terminal peptide residues and addressing metabolic stability and pharmacokinetic properties. In peptide 6, Cap1 is replaced by Cap2 where the gem-dimethyl group is modified to a cyclobutyl ring, introducing an additional constraint into this part of the peptide. This modification had no negative impact on binding or efficacy (Extended Data Table 1) and can be rationalized in light of the GLP-1R structure by an increase in hydrophobic contacts involving L388 and L384 (Fig. 2b and Extended Data Fig. 4b). In peptide 7, Cap1 is replaced by Cap3 where the imidazole is replaced by a bulkier phenol, proposed to retain the ability to hydrogen bond to residues in this part of the binding pocket. In accordance with the structure (Fig. 2b and Extended Data Fig. 4a), this peptide showed somewhat reduced affinity, emphasizing the steric constraints existing in the deepest part of the binding pocket, but notably retained similar functional potency and efficacy (Extended Data Table 1). Finally, in an effort to increase in vivo half-life, a PEG (polyethylene glycol) group was incorporated at X2 (peptide 8). PEG–X2 is predicted to exit the binding site through the hydrophobic groove formed between TM1 and TM2 (Fig. 2a, f and Extended Data Fig. 4j) without making any specific or further interactions with the receptor, thereby explaining the comparable functional potency of peptide 8 to peptide 5.
A major target for GLP-1 is the pancreatic β cell where, in the presence of increased glucose levels, it activates GLP-1R, resulting in the initiation of signalling cascades which ultimately lead to insulin secretion37. Consistently, the insulinotropic activities of our peptide analogues demonstrated dose-dependent responses in isolated rat pancreatic islets (Extended Data Fig. 7a–d). In vivo, GLP-1 is rapidly inactivated and cleared from the plasma with a half-life of around two minutes38. Optimization to peptide 5 removes the dipeptidyl peptidase 4 proteolytic cleavage site following A8, resulting in a significant increase in plasma exposure after intravenous administration (area under the curve, 3,176 ± 206 versus 1,464 ± 232 for peptide 2; Extended Data Fig. 7e and Extended Data Table 3). In addition, the PEGylated linker at position X2 in peptide 8 was predicted to further reduce proteolysis and increase plasma stability. This hypothesis was confirmed following intravenous administration to male Sprague Dawley rats (Extended Data Fig. 7e and Extended Data Table 3) and subcutaneous administration of peptide 8 demonstrated dose-proportional increases in plasma exposure (Extended Data Fig. 7f).
The in vivo efficacy of peptide 8 was confirmed using an oral glucose tolerance test (OGTT) in overnight fasted mice, where it generated comparable effects to those of exendin-4 when administered subcutaneously 30 min before the glucose challenge (Fig. 4a, b). To further probe the benefits of PEGylation in peptide 8 the OGTT was repeated 5 h after peptide 8 administration. Consistent with the improved pharmacokinetic profile, peptide 8 demonstrated significant glucose-lowering effects at all three doses tested (Fig. 4c, d). By contrast, exendin-4 had no effect when dosed at this earlier time point, in keeping with its relatively short duration of action.
a, Dose-dependent effects of peptide 8 versus exendin-4 administered subcutaneously 30 min before oral glucose administration. b, Glucose AUC0–120. c, Dose-dependent effects of peptide 8 versus exendin-4 administered subcutaneously 5 h before oral glucose administration. d, Glucose AUC0–120. Results are presented as mean ± s.e.m. (n = 6 each group) and analysed using a one-way analysis of variance followed by Dunnett’s multiple comparison test. Significant differences from vehicle responses are indicated with an asterisk (**P < 0.01, ***P < 0.001).
Conclusions
The X-ray structure of an agonist-bound full-length GLP-1R unveils the remarkably complex network of interactions between peptide ligand and receptor protein and explains why it has been so difficult to mimic this effect with a small molecule. This research demonstrates that structure-based methods can be applied to the design of optimized peptide therapeutics as an alternative approach. Finally, the structure of GLP-1R increases our understanding of the mechanism of activation of the class B secretin group of GPCRs which includes many other important drug targets.
Methods
Synthesis of peptides 1–9
The peptides were synthesized using solid-phase synthesis in an analogous manner to similar peptides reported previously11,12,13. The full experimental conditions and synthesis of caps and unnatural amino acids (if not commercially available) are reported in UK Patent Application No. 1522431.4 (publication date June 2017) and the key details are described in the Supplementary Information. The peptides described herein are as follows; peptides 2–8 are patent examples 1, 2, 6, 3, 18, 20 and 15, respectively. Peptides 1 and 9 were synthesized by analogous methods. Peptide 9 is the radiochemical precursor described in the Supplementary Information, which was converted to the radiolabelled version of peptide 1.
The synthesis of peptides was carried out by Cambridge Research Biochemicals (http://www.crbdiscovery.com/home/) and can be briefly described as follows. Peptides were assembled manually on a roller-mixer by Fmoc SPPS (solid-phase peptide synthesis) using polypropylene columns equipped with a filter disc. Rink Amide AM resin (0.4 mmol, Novabiochem, loading 0.6 mmol g−1) was swollen in CH2Cl2 for 15 min. Removal of the resin-bound Fmoc protecting group was achieved using 20% piperidine in DMF for 5 min and then 10 min. After six washes of the resin with DMF (30 s), HATU, Fmoc-protected amino acid solution and DIEA were added to the resin as a solution in DMF. Natural amino acids were coupled using five molar equivalents (eq) of the Fmoc amino acid with 4.5 eq HATU and 10 eq DIEA for 1 h (typical concentration 1 M in DMF). Unnatural amino acids were coupled using 1.5–2 eq of the Fmoc-amino acid with 1.4–1.8 eq HATU and 5 eq of DIEA (typical concentration 0.4 M in DMF). Completion of coupling reactions was confirmed by TNBS resin test (2,4,6-trinitrobenzene sulfonic acid solution). Threonine was triple-coupled (that is, the resin was subjected to the reaction conditions for 1 h each time and the solution was drained and the resin was re-subjected to fresh reagents each time). After completion of the synthesis, the resin was washed three times with DMF, three times with CH2Cl2 and three times with diethylether. Cleavage from the resin was performed using 95% TFA with 2.5% water and 2.5% triisopropylsilane. After 3 h, the resin was filtered and discarded; the filtrate was evaporated to dryness. Cold diethylether was added to precipitate the peptide and the solid was triturated. The supernatant was discarded and the process was repeated. Residual diethylether was evaporated and the peptide was obtained as an off-white powder. Final purification was performed by preparative HPLC on a Gilson system at 230 nm using a 250 × 21.2 mm ACE 10 μm C18 300 Å column at a flow rate of 15 ml min−1. Peptide crudes were dissolved in 2–4 ml of 25–30% eluent B and loaded onto a 5 ml loop. Eluents were 0.1% TFA in milli-Q water (A) and 0.1% TFA in acetonitrile (B). Purities are reported in the Supplementary Table 1.1.
Cell culture
HEK293T cells (ATCC) were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS). Cells were transfected using GeneJuice (Merck Millipore) according to manufacturer’s instructions and harvested after 48 h.
Thermostability measurement and StaR generation
Full-length human GLP-1R was used as background for the generation of the conformationally thermostabilized receptor using a scanning mutagenesis approach described previously14. Following transient transfection 2 × 106 HEK293T cells expressing wild-type GLP-1R or mutant variants were resuspended in 500 μl of assay buffer (50 mM HEPES pH 7.5, 150 mM NaCl and EDTA-free cOmplete Protease Inhibitor (Roche) tablets) containing 200 nM 3H-peptide 1. Ligand binding was allowed to reach equilibrium for 2 h at room temperature followed by incubation at 4 °C for 5 min. Next, the cells were solubilized with 1% (w/v) n-dodecyl-β-d-maltopyranoside (DDM) supplemented with 0.01% (w/v) cholesteryl hemisuccinate for 1 h rotating at 4 °C and crude lysates were cleared by centrifugation at 16,000g for 15 min. Thermostability was measured by incubation of lysate samples at different temperatures for 30 min, followed by separation of unbound radioligand using IMAC purification. Levels of ligand-bound receptor were determined using a liquid scintillation counter. Thermal stability (Tm) is defined as the temperature at which 50% ligand binding is retained following plotting the radioligand binding data against temperature using the sigmoidal dose–response (variable slope) equation in GraphPad Prism.
Construct design, expression and purification
A modified construct of GLP-1R was produced comprising residues 24–432, containing 11 thermostabilizing mutations with GP67 signal sequence and a double Strep-tag II on the N terminus and apocytochrome b562 (bRIL) followed by a deca-histidine tag on the C terminus. The receptor and bRIL were separated by a PreScission protease cleavage site. The construct was expressed in Sf9 cells in ESF921 medium (Expression systems) supplemented with 10% heat-inactivated FBS and 1% (v/v) penicillin/streptomycin using the Bac to Bac Expression System (Invitrogen). Cells were infected at a density of 4.0 × 106 cells ml−1 and an approximate multiplicity of infection of 0.5. Expression was carried out over 72 h at 27 °C with constant shaking. Cells were pelleted at 2,500 relative centrifugal force (r.c.f.) for 10 min at 4 °C. Cell pellets were resuspended in PBS supplemented with protease inhibitors tablets and 0.1 μM of peptide 5. The resuspended cell paste was then incubated at room temperature for 2–3 h and 10% (v/v) glycerol added just before freezing the pellet at −80 °C.
All subsequent steps were carried out at 4 °C unless otherwise stated. Cell pellets were thawed and then resuspended in a buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, protease inhibitors tablets and 0.1 μM of peptide 5. The cell suspension was left to stir at room temperature for one hour. Next, cells were homogenized for 90 s at 9,500 r.p.m. using a VDI 25 (VWR) and then lysed using a microfluidizer at 60 p.s.i. (M-110L Pneumatic, Microfluidics). After lysis, the mixture was centrifuged at 335 r.c.f. for 10 min and the supernatant was ultra-centrifuged at 158,420 r.c.f. for 90 min at 4 °C. The pellets were resuspended in a buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol (v/v) supplemented with protease inhibitors tablets and 0.1 μM peptide 5. The suspension was then homogenized for 90 s at 9,500 r.p.m. and the resulting membranes stored at −80 °C.
To purify the receptor, membranes were thawed and incubated with 0.2 μM peptide 5 for 60 min at room temperature. The membranes were then treated with 100 μM TCEP and 2 mg ml−1 iodoacetamide before solubilization with 2% (w/v) DM (n-decyl-β-d-maltopyranoside; Anatrace) for 1.5 h with constant stirring. The solubilized material was then clarified by ultra-centrifugation for 40 min at 98,834 r.c.f. The clarified solubilized material was loaded onto a 5 ml Strep-Tactin Superflow Plus Cartridge (Qiagen) pre-equilibrated in buffer A: 50 mM HEPES pH 7.5, 150 mM NaCl, 100 μM TCEP, 0.15% (w/v) DM, and 0.1 μM peptide 5 at a flow rate of 1 ml min−1 overnight. The column was washed with 125 ml of buffer A before elution in buffer A supplemented with 5 mM d-desthiobiotin. Elution fractions were pooled and incubated for 2 h at 4 °C with 10 μl PNGase F (NEB) and 10 μl PreScission protease (GE Healthcare Life Sciences). Next, the protein was concentrated using a 50 kDa cut-off PES concentrator (Sartorius), ultra-centrifuged at 355,040 r.c.f. for 10 min and injected onto a Superdex 200 Increase 10/300 GL size-exclusion chromatography column (GE Healthcare Life Sciences) pre-equilibrated at 12 °C in a buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 100 μM TCEP, 0.3% (w/v) OTG (n-octyl-β-d-thioglucoside; Anatrace) and 0.1 μM peptide 5. Receptor purity was analysed by SDS–PAGE and LC-MS and receptor monodispersity was assayed by analytical size-exclusion chromatography. Fractions containing the pure, monomeric receptor were pooled together and then POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol); Avanti Polar Lipids) was added at an 8:1 molar ratio (POPG:GLP-1R) from a 2% (w/v) stock solution prepared in DMSO and incubated for 30 min at room temperature. The mixture was then concentrated using a 50 kDa cut-off PES concentrator (Sartorius) at 12 °C to no higher than 3.5 mg ml−1.
Crystallization
Purified GLP-1R StaR was crystallized using the vapour diffusion method at 10 °C. A 0.1 μl sample of the concentrated protein (approximately 3.0 mg ml−1) was mixed with 0.1 μl of mother liquor in MRC 2-drop 96-well plates (Molecular Dimensions) using a Mosquito from TTP Labtech. 70–150 μm rod-like crystals of GLP-1R StaR were grown in 100 mM Tris-HCl pH 8.0–9.0, 32–44% (v/v) polyethylene glycol 200. Typically, crystals grew within 4 h but took 3–4 days to reach their maximum size. Single crystals were cryo-protected in 0.1 M Tris-HCl pH 8.4, 40% PEG200, 12.5 mM HEPES pH 7.5, 75 mM NaCl, 0.005% POPG, 0.3% OTG and 0.1 μM peptide 5, mounted using nylon cryo loops (Hampton Research), flash-frozen and then stored in liquid nitrogen until data collection.
Diffraction data collection and processing
X-ray diffraction data were measured on a Pilatus 6M detector at Diamond Light Source beamline I24 using a beam size of 10 μm × 10 μm. Crystals displayed diffraction initially out to approximately 3.6 Å following exposure to a non-attenuated beam for 0.2 s per 0.2° of oscillation. It was possible to collect about 12° of useful data from each crystal before radiation damage became too severe. Data from individual crystals were integrated using XDS39. A complete dataset to 3.7 Å was obtained by merging diffraction data from eight crystals belonging to the trigonal space group P3121. Data merging and scaling were carried out using the program AIMLESS from the CCP4 suite of programs40. Data collection statistics are reported in Extended Data Table 2.
Structure solution and refinement
The structure was solved by molecular replacement using the program Phaser41 and the GLP-1R ECD structure (PDB ID: 3IOL) and glucagon receptor TMD structure (PDB ID: 5EE7) as the input models searching for one copy of each in the asymmetric unit. Initial rigid body refinement was carried out with Refmac542 where each transmembrane helix was set as an independent rigid body. This led to the repositioning of TM6. Then 50 cycles of jelly body refinement were run using Refmac5 with external constraints from the GLP-1R ECD structure (PDB ID: 3IOL) applied during refinement using ProSMART43. Manual model building was performed in COOT44 using σA-weighted 2Fo – Fc and Fo – Fc maps and using maps from Prime and Switch generated using PHENIX45. Map sharpening was performed in COOT. Refinement was performed using PHENIX with external constraints from the GLP-1R ECD structure, secondary structure and Ramachandran restraints applied throughout. Ensemble refinement was performed in PHENIX with the following optimized parameters: pTLS = 0.8, TLS groups: residues 29–138, 139–336 and 337–417, Tx automatically selected based on resolution (in this case Tx = 0.1) and wxray_coupled_tbath_offset = 2.5. Refinement statistics are presented in Extended Data Table 2. Model quality was assessed using MolProbity46.
3H-peptide 1 radioligand binding assay
Membranes fractions of cells expressing GLP-1R were prepared as previously described10 and stored in small aliquots at −80 °C. On the day of the experiment frozen aliquots were thawed slowly and then resuspended in the binding buffer (50 mM HEPES, 150 mM NaCl, 0.05 mg ml−1 bacitracin, protease inhibitor tablets (Roche), pH 7.4; 0.15% CHAPS, 0.1% BSA) to a final assay concentration of 40 μg protein per well. The affinity (Kd) and receptor number (Bmax) of the radioligand binding to membranes were determined by homologous competitive binding, using the cold version of the radioligand. Three concentration response curves of cold ligand (1 μM to 0.1 pM) were generated and a different single concentration of radioligand was added to each. The mixture was allowed to reach equilibrium in a total reaction volume of 500 μl for 2 h at room temperature. Non-specific binding (NSB) was defined by 1 μM exendin-4 (Tocris bioscience, catalogue number 1933). Incubation was terminated by rapid filtration using a Tomtec harvester, onto a 96-well GF/C filter plates (Perkin Elmer) pre-soaked with 0.3% polyethylenimine. Filters were washed eight times with 1 ml cold wash buffer (PBS + 0.15% CHAPS) and allowed to dry before addition of 50 μl scintillation fluid (Ultima Gold F, PE) to each well; radioactivity on the filters was counted on a Trilux Microbeta counter. Data were analysed in GraphPad PRISM 6 by global-fitting to a homologous competitive binding equation. For competition binding experiments, membranes were incubated with 3H-peptide 1 at a concentration equal to the Kd value of the radioligand and eight concentrations of the inhibitory compound. IC50 values were derived from the inhibition curves and corrected using the Cheng–Prusoff equation to obtain the equilibrium dissociation constants (Ki).
Functional GLP-1R cAMP agonist activity
HEK-293 cells transiently transfected with the human wild-type GLP-1R were harvested using cell dissociation solution (Gibco), centrifuged and resuspended in assay buffer (HBSS (Lonza) supplemented with 0.5 mM IBMX (Tocris)) to deliver 20,000 cells per well in 20 μl. Following addition of test peptide dilutions (0.1 μM to 0.01 pM final assay concentrations) prepared in assay buffer, plates were incubated for 30 min at room temperature. The levels of cAMP generated were measured using Cisbio cAMP HTRF kit according to the manufacturer’s recommendations. Data were analysed in GraphPad PRISM 6 and the in vitro potency of agonists was quantified by determining the concentrations that caused 50% activation of maximal response (EC50). The intrinsic efficacy of compounds was expressed as a percentage effect of the maximal GLP-1(7–36) peptide (Tocris) response.
Rat pancreatic islet insulin secretion (performed at Jubilant Biosys, India)
The effects of peptides on glucose stimulated insulin secretion were performed on rat islets of Langerhans. The pancreas was isolated from Sprague Dawley rats killed under anaesthesia and digested with type V collagenase (Sigma C9263) followed by purification of islets with Histopaque (Sigma 10771/11191) layering. Islets were cultured overnight in RPMI 1640 complete media supplemented with GutaMax (Gibco 61870-010) and the next day starved in low glucose (2.8 mM) Earle’s Balanced salt solution (EBSS: Gibco 08-0045DJ) buffer for 30 min at 37 °C with 5% CO2. After starvation approximately four equal-sized islets were picked and transferred into low glucose EBSS buffer (2.8 mM glucose) and incubated at 37 °C for 1 h. Islets were then treated with peptide agonists as described in the presence of alternative glucose concentrations (up to 15.7 mM) for 90 min. The culture supernatant was collected and stored at −80 °C and the insulin content is measured using the Mercodia rat ELISA kit (cat. no: 10-1250-01) according to manufacturer’s protocol. Low (11.2 mM) and high (15.7 mM) glucose incubations in addition to 100 nM GLP-1 were included as controls on each plate and the amount of insulin secreted was normalized to the basal secretion in the presence of 11.2 mM glucose. Statistical differences between the groups were calculated for each sample using the fold difference between the basal glucose stimulated insulin secretion at 11.2 mmol l−1 glucose and stimulated insulin secretion at 15.7 mmol l−1 glucose in the different study groups. Data were analysed using a one-way analysis of variance (ANOVA) with Bonferroni’s post hoc test for comparison. Data are represented as means ± s.e.m.
In vivo mouse oral glucose tolerance test (performed at Fidelta, Croatia)
All animal related research was conducted in accordance with 2010/63/EU and National legislation regulating the use of laboratory animals in scientific research. Male C57BL/6 mice were purchased from Charles River Laboratories (Italy) at 12 weeks of age and acclimated for a minimum of 1 week before experiments commenced. Animals were housed under controlled ambient conditions at 22 ± 2 °C following a 12:12 h light/dark cycle, with the lights on at 6:00, and received standard chow (VRF1 (P), Special Diets Services (SDS)) ad libitum. At day −1, before the start of the experiment, animals were randomly allocated into groups (n = 6) based on body weight. The group size was determined based on previous experience of running this model. A Grubbs’ test for outlier detection was used with α = 0.05 and no data was excluded from any part of this study. The investigators were not blinded to allocation during experiments and outcome assessment.
In part 1 of the study animals were fasted for 16 h before glucose tolerance tests with free access to drinking water. At −30 min, mice were dosed via subcutaneous injection (5 ml kg−1 body weight) with vehicle, exendin-4 or peptide 8. At time = 0 mice received an oral gavage of glucose containing 2 g kg−1 body weight. Glucose was measured in blood collected from the tip of the tail vein using AccuChek glucose meters and strips (Roche) at baseline and at 0, 15, 30, 60 and 120 min.
In part 2 of the study animals were fasted for 12 h before glucose tolerance tests with free access to drinking water. At −300 min (that is, 5 hours before administration of glucose), mice were dosed via subcutaneous injection with vehicle, exendin-4 or peptide 8 in a volume of 5 ml kg−1 body weight. At time = 0 mice received an oral gavage of glucose containing 2 g kg−1 body weight. Glucose was measured in blood collected from the tip of the tail vein using AccuChek glucose meters and strips (Roche) at baseline and at 0, 15, 30, 60 and 120 min.
The peptides for part 1 and part 2 were administered subcutaneously at 30–3,000 μg kg−1 and were formulated in phosphate buffered saline (pH 7.4). All peptides were blinded to the experimenter using unique identifier codes. The mice were dosed at a fixed volume of 5 ml kg−1. Blood was collected into Li-heparin Microtainer tubes (BD, ref. no. 365966) and centrifuged at 3,500 r.p.m. for 15 min and plasma samples stored at −80 °C until analyses. To determine the integrated glucose response to the oral glucose challenge the blood glucose excursion profile from t = 0 to t = 120 min was used to integrate an area under the curve (AUC) for each treatment group. Per cent lowering in glucose is calculated from the AUC data of the compounds with respect to the AUCs of the vehicle group. To examine main effects statistical analyses were made using two-way analysis of variance (ANOVA) and a Dunnett’s multiple comparison post hoc analysis to determine differences from the control group. P < 0.05 was defined as statistically significant. Data are expressed as mean ± s.e.m.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Co-ordinates and structure factors have been deposited in the Protein Data Bank under accession number 5NX2.
Accession codes
References
Todd, J. F. & Bloom, S. R. Incretins and other peptides in the treatment of diabetes. Diabet. Med. 24, 223–232 (2007)
Chatterjee, S., Ghosal, S. & Chatterjee, S. Glucagon-like peptide-1 receptor agonists favorably address all components of metabolic syndrome. World J. Diabetes 7, 441–448 (2016)
Mentlein, R., Gallwitz, B. & Schmidt, W. E. Dipeptidyl-peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon-like peptide-1(7-36)amide, peptide histidine methionine and is responsible for their degradation in human serum. Eur. J. Biochem. 214, 829–835 (1993)
Deacon, C. F., Johnsen, A. H. & Holst, J. J. Degradation of glucagon-like peptide-1 by human plasma in vitro yields an N-terminally truncated peptide that is a major endogenous metabolite in vivo. J. Clin. Endocrinol. Metab. 80, 952–957 (1995)
Willard, F. S., Bueno, A. B. & Sloop, K. W. Small molecule drug discovery at the glucagon-like peptide-1 receptor. Exp. Diabetes Res. 2012, 709893 (2012)
Mann, R. et al. Peptide binding at the GLP-1 receptor. Biochem. Soc. Trans. 35, 713–716 (2007)
Underwood, C. R. et al. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 285, 723–730 (2010)
Runge, S., Thøgersen, H., Madsen, K., Lau, J. & Rudolph, R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 283, 11340–11347 (2008)
Siu, F. Y. et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature 499, 444–449 (2013)
Jazayeri, A. et al. Extra-helical binding site of a glucagon receptor antagonist. Nature 533, 274–277 (2016)
Haque, T. S. et al. Identification of potent 11mer glucagon-like peptide-1 receptor agonist peptides with novel C-terminal amino acids: homohomophenylalanine analogs. Peptides 31, 950–955 (2010)
Haque, T. S. et al. Exploration of structure-activity relationships at the two C-terminal residues of potent 11mer glucagon-like peptide-1 receptor agonist peptides via parallel synthesis. Peptides 31, 1353–1360 (2010)
Mapelli, C. et al. Eleven amino acid glucagon-like peptide-1 receptor agonists with antidiabetic activity. J. Med. Chem. 52, 7788–7799 (2009)
Serrano-Vega, M. J., Magnani, F., Shibata, Y. & Tate, C. G. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc. Natl Acad. Sci. USA 105, 877–882 (2008)
Lebon, G. et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474, 521–525 (2011)
White, J. F. et al. Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012)
Yang, L. et al. Conformational states of the full-length glucagon receptor. Nat. Commun. 6, 7859 (2015)
Hollenstein, K. et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499, 438–443 (2013)
Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl Acad. Sci. USA 110, 5211–5216 (2013)
Tom Burnley, B ., Afonine, P. V ., Adams, P. D. & Gros, P. Modelling dynamics in protein crystal structures by ensemble refinement. eLife 2012, 1–29 (2012)
Yang, D. et al. Structural determinants of binding the seven-transmembrane domain of the glucagon-like peptide-1 receptor (GLP-1R). J. Biol. Chem. 291, 12991–13004 (2016)
Dods, R. L. & Donnelly, D. The peptide agonist-binding site of the glucagon-like peptide-1 (GLP-1) receptor based on site-directed mutagenesis and knowledge-based modelling. Biosci. Rep. 36, e00285 (2015)
Wootten, D. et al. The extracellular surface of the GLP-1 receptor is a molecular trigger for biased agonism. Cell 165, 1632–1643 (2016)
Coopman, K. et al. Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor. Mol. Endocrinol. 25, 1804–1818 (2011)
Xiao, Q., Jeng, W. & Wheeler, M. B. Characterization of glucagon-like peptide-1 receptor-binding determinants. J. Mol. Endocrinol. 25, 321–335 (2000)
Koole, C. et al. Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation. J. Biol. Chem. 287, 3642–3658 (2012)
Wood, V. et al. The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880 (2002)
Moon, M. J. et al. Ligand binding pocket formed by evolutionarily conserved residues in the glucagon-like peptide-1 (GLP-1) receptor core domain. J. Biol. Chem. 290, 5696–5706 (2015)
López de Maturana, R., Treece-Birch, J., Abidi, F., Findlay, J. B. C. & Donnelly, D. Met-204 and Tyr-205 are together important for binding GLP-1 receptor agonists but not their N-terminally truncated analogues. Protein Pept. Lett. 11, 15–22 (2004)
Koth, C. M. et al. Molecular basis for negative regulation of the glucagon receptor. Proc. Natl Acad. Sci. USA 109, 14393–14398 (2012)
Mason, J. S. et al. High end GPCR design: crafted ligand design and druggability analysis using protein structure, lipophilic hotspots and explicit water networks. In Silico Pharmacol. 1, 23 (2013)
Goodford, P. J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 28, 849–857 (1985)
Sciabola, S. et al. High-throughput virtual screening of proteins using GRID molecular interaction fields. J. Chem. Inf. Model. 50, 155–169 (2010)
Rasmussen, S. G. F. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011)
Deupi, X. & Standfuss, J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 21, 541–551 (2011)
Tehan, B. G., Bortolato, A., Blaney, F. E., Weir, M. P. & Mason, J. S. Unifying family A GPCR theories of activation. Pharmacol. Ther. 143, 51–60 (2014)
Wheeler, M. B. et al. Functional expression of the rat glucagon-like peptide-I receptor, evidence for coupling to both adenylyl cyclase and phospholipase-C. Endocrinology 133, 57–62 (1993)
Vilsbøll, T., Agersø, H., Krarup, T. & Holst, J. J. Similar elimination rates of glucagon-like peptide-1 in obese type 2 diabetic patients and healthy subjects. J. Clin. Endocrinol. Metab. 88, 220–224 (2003)
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011)
Nicholls, R. A., Long, F. & Murshudov, G. N. Low-resolution refinement tools in REFMAC5. Acta Crystallogr. D Biol. Crystallogr. 68, 404–417 (2012)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010)
Diederichs. K. & Karplus, P. A. Better models by discarding data? Acta Crystallogr D Biol Crystallogr. 69, 1215–1222 (2013)
Acknowledgements
We thank various colleagues past and present who have helped with the project. In particular we would like to acknowledge the contribution of K. Hollenstein, M. Koglin and C. Larner. We thank G. Brown for his help with coordinating peptide synthesis and radio-labelling and C. Scully for his assistance with the GRID analysis. We are grateful to R. Owen, J. Waterman and D. Axford at I24, Diamond Light Source, Oxford, UK for technical support.
Author information
Authors and Affiliations
Contributions
J.K., N.J.R. and A.J. carried out the conformational thermostabilization of constructs and determined the stability of the StaR in a panel of reagents/additives. A.H.B. and I.T. carried out the in vitro pharmacology. A.J.H.B. managed the in vivo studies. M.C. and S.P.A. designed the novel peptides, aided by A.B. and J.S.M. who designed the homology models and carried out in silico analyses of peptide binding. M.R. and J.C.E. designed the crystallization construct, and with C.F.V. performed and optimized protein expression and purification. M.R. and C.F.V. performed and optimized protein crystallization. M.R. and A.S.D. harvested crystals, collected and processed X-ray diffraction data, and solved and refined the structure. Project management was carried out by A.J., R.M.C., F.H.M. and M.W. The manuscript was prepared by M.R., A.J., A.S.D., A.J.H.B., M.C., R.M.C. and F.H.M. All authors contributed to the final editing and approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors are employees of Heptares Therapeutics Ltd and are shareholders in Sosei Group Corporation.
Additional information
Reviewer Information Nature thanks T. Schwartz, C. Siebold and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Peptide 5 and in vitro pharmacology of wild-type GLP-1R.
a, Two-dimensional chemical plot of peptide 5 used in this study. b, Pharmacological characterization of 3H-peptide 1 at the wild-type GLP-1R. Affinity of peptide 1 for wild-type GLP-1R construct was measured using homologous competition experiments against three different concentration of 3H-peptide 1. cpm, counts per minute. Data are representative of four independent experiments and the Kd values are calculated as the arithmetic mean and s.e.m. c, Heterologous competition binding of exendin-4 and exendin-3 at the wild-type GLP-1R determined using 3H-peptide 1. Affinity constants (Ki) were calculated from IC50 values, using the Cheng–Prusoff equation and results are given as the arithmetic mean ± s.e.m.
Extended Data Figure 2 Stability, pharmacological characterization and functional activity of GLP-1R StaR.
a, Thermal stability (Tm) comparison of GLP-1R wild-type and StaR (containing the following point mutations: T207E, Q211A, D215R, L232F, G295A, T298A, C329A, P358A, G361A, H363V and V405A). Thermal stability was measured following solubilization in n-dodecyl-β-d-maltopyranoside supplemented with cholesteryl hemisuccinate (see Methods). Data representative of two independent experiments with Tm values calculated as the arithmetic mean and the standard deviation of the mean. b, Pharmacological characterization of GLP-1R wild-type and StaR. Affinity of peptide 1 for wild-type and StaR constructs was measured using homologous competition experiments against three different concentrations of 3H-peptide 1. Data representative of three independent experiments and the Kd values calculated as the arithmetic mean and the standard deviation of the mean. The difference in the means is not statistically significant as analysed by two-tailed t-test. c, d, cAMP response of GLP-1R wild-type and StaR in the presence of the indicated peptide agonists. e, Reported pEC50 values for each peptide agonist. Data presented is the arithmetic mean of three independent experiments. Error bars represent s.e.m. P values are calculated by multi-parametric two-way ANOVA.
Extended Data Figure 3 The GLP-1R StaR B-factors and electron density.
a, b, B-factor putty representation of the GLP-1R–peptide-5 structure (rainbow spectrum, blue to red = lowest to highest B-factors). c, d, representative 2Fo − Fc electron density contoured at 1.0σ across the orthosteric peptide-binding pocket of GLP-1R.
Extended Data Figure 4 Walkthrough of peptide 5 interactions with GLP-1R StaR.
a–k, Views moving from the N to C terminus of peptide 5. Four overlayed models generated by PHENIX ensemble refinement are shown in each panel to demonstrate the confidence that can be assigned to interactions with the receptor described in this study.
Extended Data Figure 5 Structural superposition of the GLP-1R peptide 5 crystal structure with the crystal structure of the isolated GLP-1R extracellular domain in complex with the GLP-1 peptide.
a, Tilted view from membrane of the GLP-1R represented as cartoon (cyan) with the peptide 5 agonist in stick representation and carbon, nitrogen and oxygen atoms coloured yellow, blue and red respectively; the ECD solved in isolation from the TMD of GLP-1R in cartoon representation is coloured orange, with the GLP-1 peptide coloured magenta. The superposition was achieved using equivalent residues from peptide 5 and the GLP-1 peptide. b, Rotation of the superposed structures in a to view from extracellular space, the relative difference in orientation of the ECD is denoted.
Extended Data Figure 6 Evaluation of the lipophilic hotspots on GLP-1R and interactions with peptide 5.
a, GRID hotspot analysis of the binding mode of peptide 5 showing the overlap of the Cap1, X2 and X3 groups of peptide 5 with lipophilic regions of the GLP-1R. GLP-1R is represented as cartoon (cyan) with the ECD coloured brown. The peptide 5 agonist is shown in stick representation with carbon, nitrogen and oxygen atoms coloured yellow, blue and red respectively. The c1 = GRID map is represented as mesh (orange) and contoured at −2.5 kcal mol−1. b, View as in a rotated by 180°.
Extended Data Figure 7 In vitro pharmacological and pharmacokinetic profiles of selected peptides.
a–d, Insulinotropic activities of GLP-1 and selected peptides on isolated rat pancreatic islets. Results are presented as mean ± s.e.m. (n = 6 each group) and analysed using a one-way analysis of variance and Dunnett’s post hoc test. Significant differences from basal responses are indicated with an asterisk (*P < 0.05). e, Pharmacokinetics of peptide 2 (1 mg kg−1), peptide 5 (1 mg kg−1) and peptide 8 (0.5 mg kg−1) following intravenous administration in male Sprague Dawley rats. Results presented as mean ± s.e.m. (n = 3). f, Pharmacokinetics of peptide 8 following subcutaneous administration in male CD1 mice. Results presented as mean ± s.e.m. (n = 3).
Supplementary information
Supplementary information
This file contains Supplementary Text and Data, 2 Supplementary Tables and Supplementary references. (PDF 413 kb)
Rights and permissions
About this article
Cite this article
Jazayeri, A., Rappas, M., Brown, A. et al. Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546, 254–258 (2017). https://doi.org/10.1038/nature22800
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22800
- Springer Nature Limited
This article is cited by
-
Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with Gs proteins
Cell Discovery (2024)
-
Insights into the structure and activation mechanism of some class B1 GPCR family members
Molecular Biology Reports (2024)
-
Recent advances in chemical protein synthesis: method developments and biological applications
Science China Chemistry (2024)
-
Applications and prospects of cryo-EM in drug discovery
Military Medical Research (2023)
-
GLP-1R signaling neighborhoods associate with the susceptibility to adverse drug reactions of incretin mimetics
Nature Communications (2023)