Abstract
Glucagon is a 29-amino-acid peptide released from the α-cells of the islet of Langerhans, which has a key role in glucose homeostasis1. Glucagon action is transduced by the class B G-protein-coupled glucagon receptor (GCGR), which is located on liver, kidney, intestinal smooth muscle, brain, adipose tissue, heart and pancreas cells, and this receptor has been considered an important drug target in the treatment of diabetes. Administration of recently identified small-molecule GCGR antagonists in patients with type 2 diabetes results in a substantial reduction of fasting and postprandial glucose concentrations2. Although an X-ray structure of the transmembrane domain of the GCGR3 has previously been solved, the ligand (NNC0640) was not resolved. Here we report the 2.5 Å structure of human GCGR in complex with the antagonist MK-0893 (ref. 4), which is found to bind to an allosteric site outside the seven transmembrane (7TM) helical bundle in a position between TM6 and TM7 extending into the lipid bilayer. Mutagenesis of key residues identified in the X-ray structure confirms their role in the binding of MK-0893 to the receptor. The unexpected position of the binding site for MK-0893, which is structurally similar to other GCGR antagonists, suggests that glucagon activation of the receptor is prevented by restriction of the outward helical movement of TM6 required for G-protein coupling. Structural knowledge of class B receptors is limited, with only one other ligand-binding site defined—for the corticotropin-releasing hormone receptor 1 (CRF1R)—which was located deep within the 7TM bundle5. We describe a completely novel allosteric binding site for class B receptors, providing an opportunity for structure-based drug design for this receptor class and furthering our understanding of the mechanisms of activation of these receptors.
Similar content being viewed by others
Main
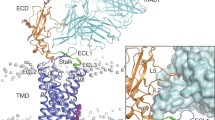
To obtain a high-resolution structure of the human GCGR transmembrane domain (TMD), a thermostabilized receptor (StaR) was generated6,7,8 containing 11 amino-acid substitutions. To facilitate crystallization further, the extracellular domain was removed from the N terminus (residues 2–135), and the C terminus was truncated by 60 residues (residues 418–477). Finally, T4-lysozyme (T4L) was inserted into intracellular loop (ICL)2 between Leu255 and Pro259 (Fig. 1a), resulting in the construct designated GCGR-StaR(136–417)–T4L. The structure was solved in the presence of the antagonist MK-0893 (Fig. 1b and Extended Data Table 1). The construct modifications did not alter the antagonist-binding properties of the receptor compared with wild type (Extended Data Table 2).
a, Crystallization construct showing stabilizing mutations (green), binding site residues (pink), disordered residues not located (grey), and the disulfide bond between Cys2243.29b and Cys294 (yellow line). b, Chemical structure of MK-0893. c, d, Ribbon representation of GCGR (blue), viewed parallel to the membrane (c) and from extracellular space (d). The position of the T4L insertion to ICL2 is indicated. MK-0893 in stick representation with carbon, nitrogen, oxygen and chlorine atoms coloured yellow, blue, red and green, respectively, Fo − Fc OMIT density contoured at 2.5σ. e, Surface representation of GCGR embedded within the membrane (20 ns molecular dynamics simulation), lipids in proximity of MK-0893 removed for clarity.
The core fold of the receptor features the canonical 7TM helices (TM1–TM7) (Fig. 1c) in a similar conformation to the previously published GCGR structure3. Continuous density is observed for intracellular loops ICL1 and ICL3, while extracellular loop (ECL)2 adopts a conformation capping the entrance to the orthosteric site. In contrast with the previously published GCGR structure3, the N terminus of TM5 unwinds by one helical turn, permitting ECL2 to stretch across to the central axis of the TM helical bundle, mediating interactions from TM3 across to TM6 and TM7 while maintaining the conserved disulfide bond between Cys2243.29b and Cys294 (numbers in superscript refer to the modified Ballesteros numbering system for class B GPCRs9,10,11). The highly conserved sequence motif GWGxP in TM4 of class B receptors has an important structural role supplying interactions stabilizing the configuration of TM2, TM3 and TM4 (ref. 5). In this GCGR structure, TM4 bulges at Gly2714.49b and, along with Pro2754.53b, they disrupt intra-helical hydrogen bonding and result in positioning of Trp2724.50b towards TM2 and TM3, which forms a hydrogen bond with Asn1792.52b on TM2 in an analogous fashion to CRF1R. In addition, a hydrogen bond from the side chain of Tyr2333.38b to the backbone carbonyl of Trp2724.50b further strengthens the TM4–TM3 interaction.
Unexpectedly, strong and unambiguous density is observed for the MK-0893 antagonist outside the 7TM helical bundle (Fig. 1c–e), straddling TM6 from within the lipid bilayer. TM6 then acts to divide the binding site into two distinct regions: a hydrophobic interface with TM5, and a polar cleft towards TM7. The different physicochemical properties of this bipartite antagonist pocket correspond to the dual hydrophilic/hydrophobic nature of the ligand. The apolar methoxynaphthalene moiety of the small molecule makes hydrophobic contacts in the TM5–TM6 interface with Leu3295.61b, Phe3456.36b, Leu3526.43b, Thr3536.44b and the alkyl chain of Lys3496.40b (Fig. 2a). On the opposite site, within the TM6–TM7 cleft, MK-0893 participates in a network of polar contacts: the ligand amide group hydrogen bonds with Lys3496.40b and Ser3506.41b, while the carboxyl group forms a salt bridge with Arg3466.37b. This moiety also makes additional polar interactions with Asn4047.61b, the backbone of Lys405 (located between TM7 and H8) and a water-mediated hydrogen bond with Ser3506.41b and Leu3997.56b (Fig. 2a). The phenylethylpyrazole core of the molecule makes further interactions with TM6, in particular with Thr3536.44b and Lys3496.40b. The position of the pyrazole moiety parallel to the membrane provides the two ligand vectors towards the bipartite antagonist sub-pockets. A third vector starting from the ligand pyrazole ring leads to the dichlorophenyl group, which makes a crystal contact to TM4 of a symmetry mate (see Extended Data Fig. 1). Molecular dynamics simulations of MK-0893 binding to the wild-type receptor (outside the constraints of the crystal system) demonstrate that the receptor–ligand interaction is stable, and MK-0893 remains at the cytoplasmic membrane boundary with the carboxyl moiety able to interact with intracellular solvent and with interactions between the ligand and receptor, involving Arg3466.37b, Lys3496.40b, Asn4047.61b and Lys405, maintained (Extended Data Fig. 2). Sequence conservation analysis demonstrates that, with the exception of Thr3536.44b and Phe3456.36b, the other key binding-site residues show good conservation across other members of human class B receptors (Extended Data Fig. 5).
a, Diagram of ligand interactions in the MK-0893-binding site. Hydrogen bonds are depicted as dashed red lines with distances between heavy atoms in Å. GCGR in ribbon representation is coloured blue, MK-0893 in stick representation is coloured as per Fig. 1. b, Comparison of pKd of wild type (WT) with the mutants. Data are average of three independent experiments and error bars represent standard error of the mean (s.e.m.). P values are calculated from a two-tailed t-test. NS, not significant. The data set for R346E and N404A did not fit the one-site binding unambiguously due to near complete loss of specific binding. ND, not determined.
Tritium-labelled MK-0893 was prepared and used to characterize the ligand-binding site. Membrane fractions prepared from HEK293T cells expressing the wild-type GCGR were used in saturation binding analysis. Saturation binding was monophasic and best fitted to a one-site model with a linear Scatchard plot, consistent with the presence of a single high-affinity binding site (Fig. 3a). Competition with [3H]MK-0893 was used to characterize the binding of a number of reported GCGR antagonists (see Extended Data Fig. 3 for details). These compounds were selected to represent molecules that exhibit chemical similarity to MK-0893 (for example, NNC0640, Cpd-01, 02, 03 and 04), as well as those that are chemically distinct (for example, Cpd-05 and 06). Consistently, NNC0640, Cpd-01, 02, 03 and 04 were able to fully compete with [3H]MK-0893 binding, indicating that these compounds share the same binding site (Fig. 3b–f). By contrast, Cpd-05 and Cpd-06 were not competitive with [3H]MK-0893, indicating that these compounds bind to a different site (Fig. 3g, h). Furthermore, [3H]-MK-0893 was not displaced by glucagon or the related peptide antagonist des-His1-[Glu9]-glucagon (Fig. 3i, j), which bind at the orthosteric site. Interestingly, NNC0640 was the ligand used in the crystallization of the first reported GCGR structure3; however, the position of the ligand was not resolved. A clear peak is observed in the electron density map of GCGR–NNC0640 between TM6 and TM7 towards the intracellular side of the receptor. Although this was modelled as a polyethylene glycol (PEG) molecule in the reported structure, superposition with the GCGR structure reported here demonstrates that the amide moiety and carboxyl function of MK-0893 directly overlays with this peak (Extended Data Fig. 4). Given the chemical similarity of these ligands, coupled with our competition data, it is likely that this constitutes residual signal from NNC0640 binding in an analogous position to MK-0893 on GCGR rather than being a PEG molecule.
a, Saturation binding of [3H]MK-0893 to membranes containing wild-type GCGR. Inset shows the Scatchard plot. b–j, Representative competitive binding data for inhibition of [3H]MK-0893 binding to membranes containing wild-type GCGR in the presence of indicated compounds (for chemical structures, see Extended Data Fig. 3). Values represent an average of at least three independent experiments ± s.e.m. Kd, dissociation constant. pKi, negative logarithm of the inhibition constant. Bmax, total concentration of receptors. B and F in the Scatchard plot denote bound and free ligand concentrations, respectively.
To confirm the allosteric pocket identified in the structure, single point mutations were made to residues in the binding site and the binding of [3H]MK-0893 was subsequently assessed. The mutations were introduced in the full-length wild-type receptor with a C-terminal enhanced green fluorescent protein (eGFP) tag. The presence of the eGFP tag has no impact on MK-0893 binding or glucagon activation of the receptor (data not shown). For this analysis, Arg3466.37b was mutated to alanine or glutamic acid. Lys3496.40b was mutated to either alanine or methionine. Ser3506.41b and Asn4047.61b were changed to alanine. These residues were selected as they participate in polar interactions with the ligand and thus were considered more likely to have a measurable effect on ligand binding (Fig. 2a). In addition, these residues interact with the amide and carboxyl functions of MK-0893, which have been demonstrated to be critical for the antagonist activity of the ligand4. Fluorescence-activated cell sorting (FACS) analysis using an antibody to the extracellular surface of receptor was performed to compare cell surface expression of mutants with the wild-type receptor. None of the mutations caused any reduction in the cell surface expression levels of the receptor nor had any effect on the binding affinity of glucagon peptide for the orthosteric site (Extended Data Fig. 6 and Extended Data Table 2). Mutation of Arg3466.37b to glutamic acid or Asn4047.61b to alanine reduced binding levels to close to undetectable above non-specific binding, while mutation of Lys3496.40b to alanine or methionine, and mutation of Arg3466.37b to alanine, significantly reduced the binding affinity of [3H]MK-0893 (Fig. 2b and Extended Data Fig. 7). Consistently, these mutations also reduced the ability of MK-0893 to antagonize the glucagon-mediated increase in cAMP (Extended Data Fig. 8).
The MK-0893 binding mode suggests that the ligand acts as a clamp holding TM6 in the inactive state and hampering receptor conformational changes required for G-protein coupling (Fig. 4a, c). In the case of class A receptors, activation results in a rotation of TM6 in response to agonist binding, which is transmitted through a rigid-body movement amplified along TM6, thereby altering the interface between TM5 and TM6 and leading to an outward movement of the cytoplasmic end of TM6 to enable G-protein binding12. So far, no active structures of class B receptors have been reported; however, the ability of MK-0893 to block signalling of GCGR points to a similar critical role of TM5 and TM6 in class B receptor activation. This is consistent with the observation that, despite divergence in the extracellular arrangement of TM helices between class A and B receptors, very good structural conservation exists on the intracellular sides in the inactive conformation5,11. An extra-helical binding site was recently described for 1-(2-(2-(tert-butyl)phenoxy)pyridin-3-yl)-3-(4-(trifluoromethoxy)phenyl)urea (BPTU), an allosteric antagonist of the P2Y1 receptor13, although in this case the binding site was located between TM1, 2 and 3 (Fig. 4b). It is likely that BPTU inhibits the movement of TM3 that is also critical in receptor activation14. Together, these structures demonstrate that modulation of helical movements from membrane-proximal surfaces represents an alternative way of modifying the activity of GPCRs.
a, b, Ribbon representation of GCGR (blue) and P2Y1 receptor (P2Y1R; yellow) respectively, viewed parallel to the membrane. MK-0893 in stick representation is coloured as per Fig. 1. BPTU is shown in stick representation with carbon, nitrogen, oxygen and fluorine atoms coloured yellow, blue, red and grey respectively. Potential restrictions on TM movements are indicated. c, View of cytoplasmic side of GCGR (rotated 90° from a) superposed with the β2-adrenoceptor (β2-AR) in complex with carazolol (grey) (Protein Data Bank (PDB) accession 3NY9) and the β2-AR–Gs complex (orange) (PDB accession 3SN6). d, Schematic overview of known binding positions of class A, B and C GPCR ligands.
The identification of an allosteric binding site in the class B GCGR located outside the canonical helical bundle further adds to the diversity of interactions now known to occur between GPCRs and their ligands in modifying receptor activation states (Fig. 4d). The GCGR–MK-0893 structure provides insight into the activation mechanism of class B receptors, as well as facilitating the application of structure-based drug design strategies to discover compounds with improved qualities. Strong conservation of this binding site across human class B receptors indicates that the structural information provided here can be applied to other members of this medically relevant family of GPCRs. In future, drug design paradigms must consider that, in addition to the orthosteric binding pocket, extra-helical binding sites accessed from the membrane or within the cell may provide alternative strategies to modulate receptor function.
Change history
12 May 2016
The MK-0893 structure in Fig. 1b was corrected.
References
Ahrén, B. Glucagon—early breakthroughs and recent discoveries. Peptides 67, 74–81 (2015)
Bagger, J. I., Knop, F. K., Holst, J. J. & Vilsbøll, T. Glucagon antagonism as a potential therapeutic target in type 2 diabetes. Diabetes Obes. Metab. 13, 965–971 (2011)
Siu, F. Y et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature 499, 444–449 (2013)
Xiong, Y. et al. Discovery of a novel glucagon receptor antagonist N-[(4-(1S)-1-[3-(3, 5-dichlorophenyl)-5-(6-methoxynaphthalen-2-yl)-1H-pyrazol-1-yl]ethylphenyl)carbonyl]-β-alanine (MK-0893) for the treatment of type II diabetes. J. Med. Chem. 55, 6137–6148 (2012)
Hollenstein, K. et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499, 438–443 (2013)
Serrano-Vega, M. J., Magnani, F., Shibata, Y. & Tate, C. G. Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proc. Natl Acad. Sci. USA 105, 877–882 (2008)
Shibata, Y. et al. Thermostabilization of the neurotensin receptor NTS1. J. Mol. Biol. 390, 262–277 (2009)
Robertson, N. et al. The properties of thermostabilised G protein-coupled receptors (StaRs) and their use in drug discovery. Neuropharmacology 60, 36–44 (2011)
Ballesteros, J. A. & Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995)
Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl Acad. Sci. USA 110, 5211–5216 (2013)
Hollenstein, K. et al. Insights into the structure of class B GPCRs. Trends Pharmacol. Sci. 35, 12–22 (2014)
Deupi, X. & Standfuss, J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 21, 541–551 (2011)
Zhang, D. et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 520, 317–321 (2015)
Tehan, B. G., Bortolato, A., Blaney, F. E., Weir, M. P. & Mason, J. S. Unifying family A GPCR theories of activation. Pharmacol. Ther. 143, 51–60 (2014)
Acknowledgements
We thank D. Hall and J. Waterman at I04, Diamond Light Source, Oxford, UK for technical support. We thank colleagues at Heptares Therapeutics Ltd for suggestions and comments, specifically R. K. Y. Cheng.
Author information
Authors and Affiliations
Contributions
D.L. and A.J. devised the strategy and carried out the conformational thermostabilization of the receptor and construct engineering. H.K. established procedures for, and H.K. and S.M.S. carried out expression and optimized purification of the final construct. H.K., S.M.S. and A.S.D. established the platform/protocols for LCP crystallization, harvested crystals, collected and processed X-ray diffraction data, and solved and refined the structure. N.J.R. supported thermostabilization. M.K. and J.C.E. supported expression and purification of the final StaR. A.H.B., I.T. and A.J.H.B. carried out and analysed the pharmacology data. Computational analysis of the structure and modelling was carried out by A.B. S.P.A. identified and sourced the chemical compounds used in the study. Project management was carried out by A.J., R.M.C., M.W. and F.H.M. The manuscript was prepared by A.S.D., A.J. and F.H.M. All authors contributed to the final editing and approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The work described here was carried out by employees of Heptares Therapeutics Limited.
Extended data figures and tables
Extended Data Figure 1 Packing interactions in the GCGR-StaR(136–417)–T4L primitive orthorhombic crystal system.
a, View along b-axis, GCGR-StaR TMD in blue ribbon representation, T4L in magenta ribbon representation, MK-0893 in stick representation with carbon, nitrogen, oxygen and chlorine atoms coloured yellow, blue, red and green, respectively. b, View as in a rotated 90° to view along c-axis. c, Close-up of the hydrophobic/shape complementarity interaction of the dichlorophenyl ‘head’ group of MK-0893 with residues on TM4 of a symmetry-related copy.
Extended Data Figure 2 Molecular dynamics analysis of the GCGR–MK-0893 complex.
a, Structural alignment of the wild-type GCGR–MK-0893 complex at 0, 4, 8, 12, 16 and 20 ns molecular dynamics, colour-coded from blue to red as indicated by the arrow. Protein is shown as ribbon, MK-0893 as sticks. b, Extracellular view of the ligand conformations during the molecular dynamics simulation, colour-coded as in a. c, Root-mean-squared deviation (r.m.s.d.) in Å for protein Cα (blue) and ligand heavy atom (pink) during the simulation, after structural alignment to the initial model. d, Number of protein–ligand contacts during the simulation: top, fluctuation of the total number of contacts over time; bottom, individual residues interacting with the ligand at a particular time are shown as rectangles colour-coded based on the number of contacts, from white (no contacts) to dark red (four contacts). e, Two-dimensional representation of the ligand–protein contacts. Green circles represent hydrophobic, cyan represents polar and purple are charged residues. Interactions that occur more than 20% of the simulation time in the selected trajectory are shown and the percentage frequency is marked. Hydrogen bonds are shown as pink arrows. A π–cation interaction is shown as a red line. The part of the ligand buried within the membrane is surrounded by a continuous line, while the water-exposed atoms (corresponding to the propionic acid moiety) are indicated by grey circles. f, GRID (Molecular Discovery) analysis of the ligand-binding site. The shape of the pocket is shown as grey mesh, hydrophilic and hydrophobic hotspots are shown using green and yellow transparent surfaces, respectively. This panel was prepared in Vida (OpenEye).
Extended Data Figure 4 Structural superposition of GCGR structures.
a, Structural superposition of the GCGR-StaR(136–417)–MK-0893 structure and GCGR–NNC0640 structure (PDB accession 4L6R) in blue and orange ribbon representation, respectively. b, Close-up view of the intracellular ends of TM5, TM6 and TM7 in 4L6R, as viewed from within the membrane. A PEG molecule modelled in the 4L6R coordinates is shown in stick representation with carbon coloured grey and oxygen coloured red. m|Fo| − Δ|Fc| map calculated using the 4L6R structure factors and 4L6R coordinates with the PEG molecule omitted, difference density is rendered at 2.5σ. c, View as in b, with the GCGR-StaR(136–417)–MK-0893 structure overlaid with MK-0893 in stick representation with carbon, nitrogen, oxygen and chlorine atoms coloured yellow, blue, red and green, respectively. d, Representation as in c, close-up and tilted towards the view from the cytoplasm.
Extended Data Figure 5 Conservation of residues in the MK-0893 bipartite allosteric pocket.
a, Sequence alignment of 15 human GPCR class B members across residues constituting the MK-0893 bipartite binding site. UniProt accession numbers are given. b, The GCGR MK-0893 allosteric binding site is shown in surface representation with residues in a coloured in rainbow spectrum according to conservation level (red = 100%; blue = 0%). MK-0893 is in stick representation with carbon, nitrogen, oxygen and chlorine atoms coloured purple, blue, red and green, respectively.
Extended Data Figure 6 Expression of GCGR mutants.
Cell-surface expression of GCGR mutants determined using FACS. Data are expressed as the ratio of APC (cell surface expression) to GFP (total expression) and calculated as percentage of wild type. Experiments were carried out in triplicate and error bars indicate s.e.m. P values are derived from an unpaired two-tailed t-test.
Extended Data Figure 7 Saturation binding analysis of mutants with [3H]MK-0893.
a–g, Saturation binding of [3H]MK-0893 to membrane containing the indicated variants of GCGR. Data are representative of three independent experiments. pKd values are average of three independent experiments and error bars represent s.e.m. P values are calculated from a two-tailed t-test. The data set for R346E and N404A did not fit the one-site binding unambiguously due to near complete loss of specific binding.
Extended Data Figure 8 Effect of binding-site mutations in functional assay.
Cells expressing either wild type or the indicated mutants were stimulated with a concentration range of glucagon in the presence of increasing concentrations of MK-0893. After determination of levels of cAMP generated, the data were analysed by global fitting to the half-maximum effective concentration (EC50) allosteric shift equation. pKB is the negative log of the antagonist affinity that mediates the allosteric inhibition of the glucagon response. Data are an average of three independent experiments and error bars represent s.e.m. P values are derived from an unpaired two-tailed t-test.
Supplementary information
Supplementary Information
This file contains Supplementary Methods and additional references. (PDF 527 kb)
Rights and permissions
About this article
Cite this article
Jazayeri, A., Doré, A., Lamb, D. et al. Extra-helical binding site of a glucagon receptor antagonist. Nature 533, 274–277 (2016). https://doi.org/10.1038/nature17414
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17414
- Springer Nature Limited
This article is cited by
-
Structural basis for recognition of antihistamine drug by human histamine receptor
Nature Communications (2022)
-
G protein-coupled receptors: structure- and function-based drug discovery
Signal Transduction and Targeted Therapy (2021)
-
Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist
Nature Communications (2021)
-
Ligand binding at the protein–lipid interface: strategic considerations for drug design
Nature Reviews Drug Discovery (2021)
-
Biologically active compounds and drugs in the tetrazole series
Chemistry of Heterocyclic Compounds (2021)