Abstract
CLC proteins transport chloride (Cl−) ions across cellular membranes to regulate muscle excitability, electrolyte movement across epithelia, and acidification of intracellular organelles. Some CLC proteins are channels that conduct Cl− ions passively, whereas others are secondary active transporters that exchange two Cl− ions for one H+. The structural basis underlying these distinctive transport mechanisms is puzzling because CLC channels and transporters are expected to share the same architecture on the basis of sequence homology. Here we determined the structure of a bovine CLC channel (CLC-K) using cryo-electron microscopy. A conserved loop in the Cl− transport pathway shows a structure markedly different from that of CLC transporters. Consequently, the cytosolic constriction for Cl− passage is widened in CLC-K such that the kinetic barrier previously postulated for Cl−/H+ transporter function would be reduced. Thus, reduction of a kinetic barrier in CLC channels enables fast flow of Cl− down its electrochemical gradient.
Similar content being viewed by others
Main
CLC proteins, which transport Cl− or other similar ions such as fluoride (F−) and nitrate (NO3−) across membranes, form a large family of membrane-spanning proteins that are ubiquitously found from bacteria to multicellular organisms (for reviews, see refs 1, 2, 3). In eukaryotes, CLC proteins control membrane potential in muscle, transepithelial electrolyte flow in the kidney, and regulation of pH and ion composition of intracellular organelles. Defects in CLC-encoding genes cause several hereditary diseases in humans1, including myotonia congenita4, Bartter syndrome5, Dent’s disease6 and osteopetrosis7.
Functionally, CLC proteins can be divided into two distinct subgroups: Cl− channels and Cl−/H+ transporters. Channel-type CLC proteins catalyse passive permeation of Cl− ions down a Cl− electrochemical gradient across the membrane8,9. By contrast, CLC transporters exchange Cl− ions and protons (H+) in opposite directions, with a stoichiometry of 2 Cl−:1 H+ (that is, Cl−/H+ antiporters)10,11,12. This coupled movement of the two ion species enables active transport of one species across an ‘uphill’ electrochemical gradient when the other moves across a ‘downhill’ gradient. Channel-type CLC proteins transport Cl− ions faster than transporter-type CLC proteins13,14. Despite such differences, they exhibit considerable homology in their amino acid sequences and therefore are expected to share a common structural architecture. This fact challenges the traditional view that channels and transporters have largely unrelated structures.
CLC proteins are homodimers with each subunit bearing a separate transport pathway8,15,16. Crystal structures of CLC Cl−/H+ transporters from bacteria and the eukaryotic algae Cyanidioschyzon merolae have identified three consecutive Cl−-binding sites referred to as the external, central and internal sites, the first two of which are fundamental to our current understanding of the transport mechanism16,17,18. The amino acids surrounding the external and central sites, including a glutamate residue (denoted ‘gating’ glutamate (Glugate)) and tyrosine and serine residues (denoted TyrC and SerC, respectively) are conserved in both transporters and channels and thus offer few clues as to the molecular distinction between CLC transporters and channels. While mutations of these residues convert transporters to passive, channel-like conductors by uncoupling H+ and Cl− transport, native CLC channels have somehow accomplished uncoupling in a different manner associated with higher conductance10,11,12,13,14. Despite conservation of primary sequences, the fundamental mechanistic distinction between CLC channels and transporters suggest that tertiary structural differences must exist between the two.
Characterization of bovine CLC-K
We screened several CLC channels and identified a bovine CLC-K channel as a promising candidate. CLC-K shows 84% amino acid sequence identity with the human CLC-K channel orthologues (CLC-Ka and CLC-Kb) and much less with Escherichia coli (EcCLC; 22% identity) and C. merolae (CmCLC; 32% identity) transporters. Expressed mainly in Henle’s loop, CLC-K channels play an important part in renal salt reabsorption5,19,20 and thus are potential targets of diuretic agents21.
We first examined CLC-K channel activity by two-electrode voltage clamp experiments (Extended Data Fig. 1a–e). Because previous studies have shown that many CLC-K channels require a β-subunit called barttin to detect their Cl− transport activity22,23, we performed experiments with and without co-expression of barttin. As previously reported for human CLC-K channels22, bovine CLC-K showed robust currents when expressed in Xenopus laevis oocytes together with barttin. Similar results were obtained when patch recordings were performed on mammalian cells (Extended Data Fig. 1f). The currents were Cl−-specific as the reversal potential approached the Nernst equilibrium potential of Cl− ions. Similar to other CLC-K channels studied22,23,24,25, bovine CLC-K displayed a linear current–voltage (I–V) relationship (Extended Data Fig. 1e–f). This contrasts with the other major CLC channel subgroups including CLC-0 and CLC-1, which show strong voltage-dependent gating15,26,27. This difference in gating properties is determined by the amino acid identity in the Glugate position. In CLC-K, the equivalent residue is a valine (Val166). Replacing Glugate with a neutral amino acid in CLC-0 and CLC-1 abolishes the voltage dependence and makes the I–V relationship linear, similar to CLC-K channels17,26,28; and conversely, mutation of Val166 to Glu reintroduces voltage dependence in CLC-K channels, while their activity (conductance) otherwise remains comparable to wild type23,24,25.
Previous data have shown that barttin has an essential role in trafficking channels to the plasma membrane22,25,29. We found that cell surface expression of bovine CLC-K requires coexpression of barttin (Extended Data Fig. 1g). To test further if barttin is required for activity, we purified bovine CLC-K and reconstituted it into lipid vesicles. Isolated bovine CLC-K showed robust channel activity in fluorescence-based flux assays18,30, indicating that channel function does not require barttin (Fig. 1a, b).
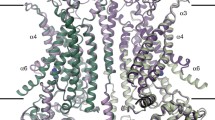
a, Fluorescence-based Cl− flux assay. Concentration-gradient-driven Cl− influx into vesicles induces H+ influx by the H+-ionophore carbonyl cyanide chlorophenylhydrazone (CCCP), which dissipates the electrical gradient built up by the Cl− transport. Intravesicular pH decrease is monitored by 9-amino-6-chloro-2-methoxyacridine (ACMA) fluorescence. b, Bovine CLC-K-incorporated vesicles (1:1,000 to 1:100 wt/wt protein-to-lipid ratios) containing <5 mM Cl− were diluted into buffer containing 150 mM NaCl (bottom). CCCP was added at ~100 s (arrowhead and grey dashed line), and ACMA fluorescence was monitored over time. As controls, empty (without CLC-K) vesicles were used, or vesicles were diluted into buffer lacking Cl− (replaced by gluconate; top). Means and standard errors of the mean (error bars) of five independent measurements are shown. c, d, Cryo-EM density map (c) and model (d) of the CLC-K channel (class 1). Blue and cyan, two subunits of the CLC-K homodimer. Grey, Fab variable domains. Black lines, membrane boundaries.
Cryo-electron microscopy
The small molecular weight (~150 kDa) of CLC-K poses considerable challenges for high-resolution structure determination by cryo-electron microscopy (cryo-EM). Therefore we raised monoclonal antibodies to increase the effective molecular mass. The complex of the channel and antigen-binding (Fab) fragments produced well-defined particle images, of which recognition was facilitated by the associated Fab molecules (Extended Data Fig. 2a). Three-dimensional classification of particles yielded two major groups with notably different dimer arrangements: while the main group (class 1) is similar to arrangements previously seen in CLC transporter structures, the second group (class 2) deviates from this (Extended Data Fig. 2c; see later). Individual three-dimensional reconstructions of the two classes using C2 symmetry yielded density maps with ~3.7 Å and ~3.9 Å overall resolutions, respectively (Extended Data Fig. 3). The maps, especially of class 1, were of sufficient quality to allow modelling of almost the entire channel and the variable domains of the Fab (Fig. 1c, d, and Extended Data Fig. 4). The local resolution estimation of the class 1 map indicates that a large portion of the transmembrane (TM) domain is beyond 3.5 Å resolution, whereas peripheral regions of the cytosolic domain and the Fab fragments are at lower resolution (Extended Data Fig. 3d).
Flexibility of the TM dimer interface
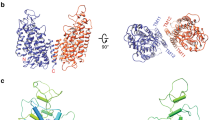
Comparison of the two structures of CLC-K (classes 1 and 2) indicates that conformational differences are mostly due to rigid-body movements of the two TM domains (Fig. 2). While the cytosolic domains of the two classes superimpose well, the TM domains show a ~6° tilt relative to each other. The static nature of the cytosolic domain is also suggested by the fact that a crystal structure of an isolated human CLC-Ka cytosolic domain dimer31 almost perfectly fits into our EM density maps.
The two subunits of the class 2 model (red and green) were superimposed on the class 1 model (grey) based on the superposition of the cytosolic domains. α-Helices are represented as cylinders. Left, a view from the extracellular side. Right, a side view. TMD, transmembrane domains. CTD, cytosolic domains.
The two distinct structural classes show that some flexibility seems to exist at the interface between the two TM domains (Fig. 2 and Extended Data Fig. 5a). This was unexpected considering the substantial interface area (~1,800 Å2), but in retrospect the finding seems compatible with the demonstration that mutations at the interface cause dissociation of EcCLC into monomers32. The extent of change on the dimer that allows this conformational difference is small. For example, a ~1.0 Å shift of TM helix αI and rearrangement of specific side chains (Extended Data Fig. 5a) appears sufficient. In class 2, TM helices across the dimer interface appear slightly less tightly packed than those of class 1 (Extended Data Fig. 5a). Corroborating this impression, the shape complementarity index of the interface drops from 0.63 in class 1 to 0.52 in class 2. An additional difference was observed at the N terminus of the channel, where the class 2 map shows extra density in which we could build 12 amino acids as an amphipathic helical extension lying parallel on the membrane surface (Extended Data Fig. 5b). This density is invisible in the class 1 map, implying disorder within this region. Careful comparison between the Cl− transport pathways of the two classes did not reveal marked differences.
The inter-subunit rearrangement of TM domains in the two CLC-K classes might be relevant to other CLC proteins: recent NMR and crosslinking data show that inter-subunit distances change during the transport cycle of EcCLC33. Such subunit rearrangements might also underlie a form of gating in CLC channels known as ‘slow’ gating, in which the protopores within each subunit open and close simultaneously15,27,34. This possibility is strengthened by the discovery of several dominant disease mutations in CLC-1 that are located on the dimer interface and influence slow gating35.
Architecture of the CLC-K channel
The overall structure of CLC-K is similar to crystal structures of CLC transporters16,17,18 (Fig. 3a and Extended Data Fig. 6a). Superposition of the class 1 CLC-K TM domains with the corresponding region of CmCLC (over 362 Cα atoms) or EcCLC (over 361 Cα atoms) shows a root mean square deviation (r.m.s.d.) of 1.5 Å and 1.8 Å, respectively. These values are comparable to an r.m.s.d. of 1.6 Å between CmCLC and EcCLC (over 356 residues).
a, Comparison of CLC-K (grey and magenta) and CmCLC (cyan). The TM domain of a CmCLC monomer was superimposed onto that of CLC-K. α-Helices are represented as cylinders. Dashed line, a boundary between the TM domain (TMD) and the cytosolic domain (CTD). b, The extracellular features of the CLC-K channel. A domain connecting αK and αM is shown in green. In the native channel, the domain becomes glycosylated through a surface-exposed Asn (N373; shown as sticks), which was mutated in our CLC-K construct to generate a binding site for antibodies. Blue mesh shows the EM density of the loop linking the ends of αI to αJ (indicated in cyan with amino acids positions). The entry to the ion transport pathway is marked in magenta. Shown is only one subunit with the dimer interface indicated as dark grey. c, Positions of disease-causing mutations in the extracellular domain (see also Extended Data Fig. 6e).
While absent in EcCLC, both CLC-K and CmCLC possess a cytosolic domain containing cystathionine-β-synthase (CBS) domains, a common feature of eukaryotic CLC proteins. The isolated monomeric cytosolic domains of CLC-K and CmCLC are superimposable (1.7 Å r.m.s.d.), but the two-fold axis between the cytosolic domains is skewed ~20° with respect to that of the TM domains (Fig. 3a). Conformational differences on the subunit interface and tilting of the cytosolic domains implies a degree of plasticity between structural components in CLC proteins.
On the extracellular side, the segment connecting TM helices αK and αM (residues 341 to 394) forms a novel structure in CLC-K (Fig. 3b). Tightly packed with four short α-helices, this segment is shaped into a globular domain that resides within the extracellular funnel. Sequence comparison suggests that a similar feature is present in other CLC channels but not in transporters (Extended Data Fig. 6e). Several disease-causing mutations of CLC-1 and CLC-Kb channels map to this region (Fig. 3c and Extended Data Fig. 6e). The CLC-K structure exhibits an elongated crevice between the extracellular and TM domains (Fig. 3b). This crevice houses an extracellular loop (αI–J) that bridges the ends of TM helices αI and αJ, which are separated by ~40 Å on the CLC dimer interface. Although the low resolution of this region precluded modelling, the density is continuous. The linker reaches deep into the extracellular funnel of CLC-K, with its tip reaching to within 8 Å of the Cl− selectivity filter (Extended Data Fig. 6b–d), suggesting a potential regulatory role in channel function. Perhaps relevant, the αI–J loop has been implicated in binding calcium (Ca2+) ions, which are known to enhance CLC-K channel currents29,36. Future studies in the presence of Ca2+ are needed to address such activation.
The Cl− transport pathway
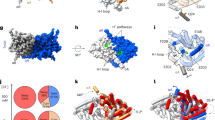
The TM domain of each subunit contains an hourglass-shaped penetration with extracellular and cytosolic ‘funnels’ tapering towards a constriction halfway across the membrane where the selectivity filter is located. The filter consists mainly of αN, αF and αD helices, whose N termini point towards the Cl− transport pathway in such a manner as to generate an electrostatically positive environment (Fig. 4a). In the constriction, six amide nitrogen atoms are positioned to coordinate completely or partially dehydrated Cl− ions in a cage-like manner in the external (Sext) and the central (Scen) sites (Extended Data Fig. 7a). These structural features are similar to those of EcCLC and CmCLC (Fig. 4c and Extended Data Fig. 7a). Likewise, a conserved TyrC (Tyr520), whose side chain is expected to interact with a Cl− ion bound in Scen, is positioned at essentially the same location (interposed between the selectivity filter and cytosolic funnel) as in EcCLC and CmCLC. An invariant conformation of TyrC observed across multiple different structures16,17,18,37,38 suggests that it is probably a stationary element of CLC proteins rather than a dynamic one, as proposed previously33,39,40.
a, Cl− selectivity filter of CLC-K. The side chains of Val166, Tyr520 and Ser121 are shown in ball-and-stick representation. The orientation of the Cl− transport pathway is indicated by grey arrows. b, EM density (grey mesh) and model of the αC–D loop and Tyr520 of CLC-K. Note that the rotamer of Glu125 is uncertain due to lack of clear side-chain density. c, As in a, but the CLC-K structure (magenta) was compared with the structures of EcCLC (yellow; Protein Data Bank (PDB) accession 1OTS) and CmCLC (cyan; PDB accession 3ORG). Grey spheres indicate the positions of Cl−-binding sites seen in a Glugate-mutant EcCLC structure (PDB accession 1OTT). d, As in c, but the same view as in b. The density of CLC-K (mesh) is shown.
One major structural difference exists between the CLC-K channel and CLC transporters: the cytosolic loop (αC–D) connecting αC to αD, which contains SerC, adopts a unique conformation (Fig. 4b, d). In the CLC transporter structures this loop reaches towards TyrC, with the side chain of SerC directed towards Scen, where its hydroxyl group interacts with a bound Cl− ion. In the CLC-K structures, SerC (Ser121) and the preceding Gly120 residues are flipped away from Scen and instead are directed towards the cytosol. In addition, the ‘elbow’ connecting αC to the loop is markedly shifted towards the cytosol. The same conformation was revealed in both class 1 and class 2 maps, which were reconstructed from completely independent particles (Extended Data Fig. 7b).
Although density for a Cl− ion bound to Sext was absent in the density maps (determined in the presence of ~100 mM Cl−), we observed weak density in the Scen site, which probably corresponds to a Cl− ion (Extended Data Fig. 8). A Cl− ion at this position would be expected to make anion–dipole interactions with two amide nitrogen atoms and the phenyl group of TyrC, as in CLC transporters, but not with the hydroxyl group of Ser121 because of its displacement in the CLC-K structures. Instead, unique to the CLC-K channel, additional anion–dipole/quadrupole interactions41 are provided by Tyr425 and Phe519, whose aromatic side chains direct their edges towards Scen. The third Cl− binding site (Sint) in EcCLC and CmCLC, which we believe represents a vestibule ion and is not critical to the mechanism of Cl−/H+ exchange, is not seen in CLC-K. Instead, a density peak possibly due to a Cl− ion is present at a different, adjacent position surrounded by the side chains of Thr523, Tyr425 and Lys527 (Extended Data Fig. 8).
The displacement of the αC–D loop substantially reshapes the cytosolic vestibule of the Cl−-conduction pathway. In all transporter structures, SerC is located between Scen and Sint, forming a narrow (1–1.5 Å radius) constriction together with TyrC, which would prevent Cl− ions (~1.7–1.8 Å radius) from freely moving between Scen and Sint (or the cytosol) (Fig. 5). Thermal motions of polypeptides may transiently widen this constriction to allow Cl− transfer. On the other hand, our new EM structures indicate that such a constriction is largely removed in the CLC-K channel due to the displacement of SerC. Cytosolic Cl− ions would be able to access Scen through a continuous, although still relatively narrow, vestibule without much restriction (Fig. 5a). In addition to lowering the barrier for Cl− ion movement, the displacement of the αC–D loop also exposes a highly positive surface in the cytosolic vestibule (Extended Data Fig. 9). Such an electrostatic potential may enhance the Cl− transport activity of the channel by increasing the local concentration of Cl− ions in this region.
a, Regions in the ion transport pathway immediately accessible to Cl− ions (radius of 1.7 Å) from extracellular and cytosolic solutions were calculated (blue mesh). The side chains of Glugate (or V166), TyrC and SerC are shown in a stick representation. For EcCLC and CmCLC, Cl− ions bound in the indicated sites are shown as grey spheres. The selectivity filter formed by αN and αF is shown in orange. The area of the view is depicted on the side view of CLC-K (top left). b, Pore radii along the ion transport pathway from Scen towards the cytosol were calculated. Arrowheads indicate the approximate positions of SerC hydroxyl groups.
Summary
The structures of CLC-K demonstrate that this CLC channel exhibits a marked difference in the ion transport pathway from transporter-type CLCs. The displacement of the αC–D loop (Fig. 6) provides a structural explanation for the higher conductance of CLC channels by lowering a kinetic barrier. Although it remains to be demonstrated, we suspect that a similar mechanism may be present in other CLC channels to enable fast Cl− conduction.
a, Model for CLC transporters. A swinging motion of the protonatable Glugate side chain (purple) coordinates countertransport of two Cl− (green sphere) and one H+ (blue sphere) ions. Cl−-binding sites are shown with grey dashed circles. A kinetic barrier preventing undesirable leakage of Cl− during the cycle is depicted as an orange bump. The αC–D loop and SerC are shown in red, and TyrC (Y) in black. For simplicity, some intermediate steps and the internal Cl−-binding site (Sint) are omitted, and only one direction of the reversible reaction is shown18,30. b, Model for the CLC-K channel mechanism, depicting fast Cl− permeation due to removal of the kinetic barrier. Steady-state Cl− occupancy of Sext and Scen may be lower than in CLC transporters.
The structures offer a straightforward explanation for why some members of the CLC protein family are channels and others are transporters (Fig. 6). Previously, our group proposed a working model for CLC transporters based on crystal structures18,30. The model explains the coordinated countertransport of two Cl− ions and one H+ by invoking the swinging action of a protonatable Glugate side chain, which both competes with Cl− ions for Sext and Scen in its deprotonated state and transfers H+ in its protonated state42. However, to preclude a certain ‘forbidden’ state, where Cl− ions could leak through a channel-like pore, uncoupled from H+ transfer (for example, Fig. 6a, top left), it was necessary to hypothesize a kinetic barrier for Cl− ions in the cytosolic vestibule. The CLC-K structures now suggest that SerC and the αC–D loop are a main constituent of the kinetic barrier in CLC transporters and that reduction of such a kinetic barrier is a major mechanism by which CLC channels function. This idea is further supported by the experimental observation that mutation of SerC to Gly in EcCLC uncouples Cl− transport from H+ transport with increased throughput14.
In contrast to structures of CLC transporters in which prominent densities corresponding to Cl− ions were observed in the Sext and Scen sites16,17,18, densities for Cl− were absent or weak in the CLC-K structures. Low Cl− occupancy in CLC-K might be consistent with biophysical and crystallographic observations previously made with EcCLC, in which there was found a strong correlation between H+ coupling of anion transport and anion occupancy of the sites43,44—for example, polyatomic pseudohalide ions, such as SCN−, SeCN− and NO3−, are robustly transported by EcCLC uncoupled from H+ transport, and yet their steady-state occupancy at both Sext and Scen sites was found to be very low43, presumably due to low affinity for the sites45. These observations lead us to speculate that low Cl− affinity of Sext and Scen sites, in addition to a reduced kinetic barrier, may contribute to the channel nature of CLC-K.
SerC has been implicated in determining the anion preference of CLC transporters46,47. In the case of Arabidopsis thaliana CLC transporters, which prefer NO3− to Cl− (ref. 48), the amino acid corresponding to SerC is a Pro, and mutating it to Ser reverses the anion preference46 (that is, Cl− > NO3−). Similar observations were made with the human CLC-5 transporter, in which mutating SerC to Pro renders transporter preference for NO3− over Cl− (ref. 47). These effects can be explained if such mutations alter physical properties of the protein that in turn influence the relative kinetic barrier for different ionic species. The fact that Cl−/H+ CLC transporters are permeable to polyatomic pseudohalides largely uncoupled from H+ transport43,46 suggests that the kinetic barrier made by SerC is ineffective for these anions. Mutations of SerC in the CLC-0 channel also moderately affect conductance and anion preference15, which might be explained by the fact that SerC still lines the narrow cytosolic vestibule in CLC-K.
The structure of CLC-K provides an explanation for the distinction between CLC transporters and channels. Local conformations of the ion permeation pathway can account for the magnitude of an energy barrier, which distinguishes the Cl− conduction pathway of CLC-K from CLC transporters. Such a lowered kinetic barrier can also explain the channel properties of CLC-0 and CLC-1, which contain the Glugate, by creating a ‘forbidden’ high Cl− conductance state in the transport cycle18.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Expression constructs
Gene synthesis was used to obtain DNA segments coding bovine CLC-K (UniProt accession E1B792) and barttin (UniProt accession E1BEP9). We note that unlike several other mammalian species, which possess two highly homologous genes (CLCNKA and CLCNKB), only one annotated Bos taurus CLCNK gene was found in databases. The synthesized barttin construct encodes amino acids 1–145, excluding its C-terminal domain, which has been shown to be dispensable for function, and contains a point mutation (Y98A), which increases currents of CLC-K (ref. 22). For two-electrode voltage clamp experiments, a wild-type full-length CLC-K was used. In all other experiments, CLC-K contains the deletion of a predicted unstructured N-terminal segment (amino acids 2–26) and a mutation (N373Q) abolishing N-glycosylation. Expression in mammalian cells was carried out with a modified pFastBac vector (Invitrogen) expressing the CLC-K channel as a fusion to a green fluorescent protein (GFP). The vector contains a CMV promoter, a chimaeric intron, CLC-K, an HRV 3C protease cleavage sequence, GFP, a WPRE element, and an SV40 poly-A signal in this order. For co-expression of barttin, a bicistronic expression cassette was used, where a DNA segment encoding a fusion of barttin, a Flag-tag, and a porcine teschovirus 2A self-cleaving peptide49, was inserted between the CMV promoter and the start codon of CLC-K. All cell lines used were obtained from ATCC. The cell lines have not been further authenticated, nor tested for mycoplasma contamination.
Two-electrode voltage clamp
Bovine CLC-K and barttin constructs were separately cloned into a pGEM vector containing T7 and β-globin promoters. cRNA was synthesized in vitro using linearized pGEM templates and AmpliCap-Max T7 High Yield Message Maker Kit (CellScript) according to the manufacturer’s instructions. cRNA was then purified by phenol-chloroform extraction and ethanol precipitation, and resuspended in H2O. Xenopus laevis oocytes were injected with 2.4 ng of CLC-K cRNA and/or 2.2 ng of barttin cRNA (in a total volume of 50 nl), which were then kept for 1 day at 16 °C in ND96 solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES, pH 7.5) containing 50 μg/ml gentamycin before recordings. TEVC recordings were performed at 23 °C using an oocyte clamp amplifier (OC-725C, Warner Instrument) and pCLAMP10 software (Molecular Devices). Oocytes were held at the resting membrane potential (−50 to −20 mV), then clamped for 0.3 s to voltages between −60 to +60 mV in 10-mV steps, followed by repolarization to the initial holding potential for 0.24 s. Pipettes were filled with 3 M KCl solution (pipette resistance 0.3–0.4 MΩ), and ND96 buffer containing 0.3 mM CaCl2 was used as the extracellular solution for recordings.
Immunofluorescence and patch clamp
For immunofluorescence imaging, Chinese hamster ovary (CHO) cells (ATCC CLL-61) were cultured on 18-mm poly-l-lysine-coated glass coverslips placed in a 12-well plate containing DMEM/F12 medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Invitrogen). At ~30–50% confluency, cells were transiently transfected using 1.5 μg of plasmids and 4.5 μl of FuGENE HD transfection reagent (Promega) 1 day before staining. CHO cells for patch clamp experiments were prepared similarly except that they were grown on 12-mm Thermanox plastic coverslips (Nunc) placed in a 35-mm dish and transfected with using 3 μg of plasmids and 9 μl of FuGENE HD 1–2 days before recording.
To perform non-permeabilized immunofluorescence staining, 600 μl of 1 ml medium was removed from the well, and 40 μl of anti-bovine CLC-K hybridoma supernatant (clone 16E3 at ~10 μg/ml; see later) was added to the well. After 30-min incubation in a 5% CO2-incubator, cells were washed three times with fresh medium and once with phosphate-buffered saline (PBS) at 37 °C, and then fixed with 4% paraformaldehyde (in PBS) at 23 °C for 20 min (hereafter, all procedures were done at 23 °C). Fixed cells were washed twice with PBS and blocked with 2% FBS for 30 min. Coverslips were incubated for 1 h with secondary anti-mouse IgG antibody conjugated with Dylight 594 (Thermo Fisher #35510; diluted 1:300 with 5% FBS in PBS), and then washed with PBS three times. Coverslips were mounted on glass slides using FluorMount-G mounting medium (SouthernBiotech) for imaging. Immunofluorescence images were acquired using a Nikon Eclipse Ti widefield microscopes equipped with a ×100 oil immersion objective (Plan Apo VC; Nikon) and a CoolSNAP HQ2 CCD camera (Photometrics). Images were adjusted for levels and combined using Adobe Photoshop.
Whole-cell patch clamp recordings were performed at 23 °C with an Axopatch 200B amplifier (Molecular Devices). The current signal was low-pass filtered at 1 kHz (Bessel) and digitized at 20 kHz with a Digidata 1440A digitizer (Molecular Devices), and recorded on a computer using pCLAMP10 software. Patch pipettes (2–4 MΩ) were pulled on a P-97 puller (Sutter Instrument) using filamented borosilicate glass capillaries (1.5-mm outer diameter). The extracellular solution contained 104 mM Na-gluconate, 42 mM NaCl, 4 mM CsCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, pH 7.5 (~300 mOsM). The pipette solution contained 140 mM CsCl, 2 mM MgCl2, 5 mM ethylene glycol tetraacetic acid (EGTA), 10 mM HEPES, pH 7.0 (~290 mOsM). Membrane potential was initially held at a voltage producing near-zero currents (for example, near +25 mV for CLC-K/barttin), then clamped for 0.25 s at a voltage ranged from −80 mV to +80 mV (10-mV steps), followed by repolarization for 0.1 s at −80 mV.
Expression and purification of bovine CLC-K
Bovine CLC-K was expressed in HEK293 GnTI− cells (ATCC CRL-3022) by transduction using a modified baculovirus50,51. Baculovirus bacmid expressing a CLC-K–GFP fusion was generated by transformation of DH10Bac E. coli cells with pFastBac plasmid (Bac-to-Bac; Invitrogen). Baculoviruses were generated by transfecting Spodoptera frugiperda (Sf9; ATCC CRL-1711) cells with the bacmid using Cellfectin-II (Invitrogen). Viruses were then amplified twice for large-scale transduction. HEK293 GnTI− cells were grown at 37 °C in suspension in Freestyle 293 medium (Invitrogen) supplemented 2% FBS in the presence of 8% CO2. At a cell density of ~2.5 × 106 ml−1, baculovirus was added to the culture (6–8% v/v). After 12 h, the culture was supplemented with 10 mM sodium butyrate, then further incubated for 1.5 days before harvest.
All protein purification steps were carried out at 4 °C. Approximately 15 g of HEK293 cells (from 2–3 l of culture) were suspended in 75 ml buffer (WB) containing 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM dithiothreitol (DTT), 1 mM EDTA (EDTA) and 10% v/v glycerol, and supplemented with a cocktail of protease inhibitors (1 mM benzamidine, 50 μM leupeptin, 1 μg/ml aprotinin, 1 μM pepstatin and 1 mM phenylmethylsulfonyl fluoride). Cells were lysed by brief sonication, then membranes were extracted by addition of 1% dodecyl-β-maltoside (DDM) and 0.1% cholesteryl semisuccinate (CHS). After extraction for 2 h, the lysate was clarified by centrifugation (Beckman Ti70 rotor, 40,000 RPM, 1.5 h). The clarified lysate was then mixed with 5 ml of CNBr-sepharose beads (GE Healthcare) coupled with anti-GFP nanobody (GFP enhancer; ref. 52) for 2.5 h. Beads were washed on a column with 60 ml WB containing 0.04% DDM and 0.004% CHS. Bound protein was released from beads by overnight incubation with 5 ml WB containing 0.04% DDM, 0.004% CHS and 0.2 mg HRV 3C protease. The retrieved protein was concentrated to 0.5 ml using Amicon Ultra (100-kDa cut-off; Millipore) and applied to a Superose-6 column (GE Healthcare) equilibrated with 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM DTT, 10% glycerol, 0.04% DDM, and 0.004% CHS. The peak fractions were pooled and concentrated with Amicon Ultra. Purified CLC-K was mixed with Fab fragments (clone 25E7; see later) at a molar ratio of 1.5 Fab fragments per CLC-K monomer for 1–2 h. The CLC-K/Fab complex was separated using a Superdex-200 column (GE Healthcare) equilibrated with 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM DTT, 0.04% DDM, and 0.004% CHS. The peak fractions were collected, concentrated to ~3 mg/ml, and immediately used for cryo-EM grid preparation.
Fluorescence-based flux assay
E. coli polar lipids (Avanti Polar Lipids) and egg l-α-phosphotidylcholine (60%; Avanti Polar Lipids) were mixed in chloroform at a 3:1 wt/wt ratio and dried with an argon stream, followed by overnight incubation in a vacuum chamber. Dried lipids were suspended by sonication in buffer containing 10 mM HEPES, 100 mM Na2SO4, 2 mM tris(2-carboxyethyl)phosphine-HCl (pH 7.2). An aliquot of 10 mg/ml lipid suspension was mixed with 0.7% DDM (from a 10% stock) for 3 h at 23 °C. Purified CLC-K protein was then added to the lipid/DDM mixture in a protein-to-lipid ratios of 1:1,000 to 1:100 (wt:wt). Following 1.5-h incubation at 23 °C, DDM was removed by incubating the mixture with SM-2 bio-beads (Bio-rad) overnight at 4 °C. Generated proteoliposome vesicles were collected and briefly sonicated. Three microlitres of vesicles were mixed with 40 μl of flux assay buffer containing 10 mM HEPES pH 7.2, 150 mM NaCl, 2 μM ACMA in a well of a 384-well fluorescence assay plate. After measuring initial AMCA fluorescence intensity (λEx = 410 nm, λEm = 490 nm), Cl−-flux was initiated by addition of 1 μM CCCP, followed by monitoring fluorescence over time (10-s intervals) using a plate reader (Tecan Infinite M1000) at 27 °C. Note that there is a dead time for measurement between t = 80 s to t = 120 s due to handling of the plate during CCCP addition. CCCP was added to the reactions at t = ~100 s.
Generation of monoclonal antibodies and Fab fragments
Mouse monoclonal antibodies against bovine CLC-K were raised using standard procedures. Hybridomas were generated by immunization of mice (female BALB/c) with CLC-K protein purified in DDM, followed by fusion of splenocytes with mouse myeloma cells and selection of clones by enzyme-linked immunosorbent assay (ELISA). The positive clones from ELISA (performed using CLC-K purified in DDM as the antigen) were further tested for high-affinity binding to the channel based on ability to completely shift the peak of DDM-extracted, GFP-tagged CLC-K channel on gel-filtration chromatography (injected samples contained the channel at ~5 nM and antibodies in excess) and lack of binding to a denatured form of the channel (denaturation ELISA; ref. 53). Antibody sequencing was done by 5′-rapid amplification of cDNA ends (5′-RACE) of messenger RNA isolated from hybridomas.
Monoclonal antibodies were produced by culturing hybridoma cells in hybridoma serum-free medium (Invitrogen) supplemented with 1% ultra-low IgG FBS (Invitogen) using CELLine disposable bioreactors (Wheaton). Medium supernatant (~50 ml) from cultures was dialysed (50-kDa cut-off) against 10 mM Tris-HCl pH 7.5, 10 mM NaCl. Dialysed samples were spun at 15,000 r.p.m. for 30 min, and the supernatant was applied to a 5-ml Q-Sepharose column (GE Healthcare) equilibrated with 10 mM Tris-HCl pH 7.5. After washing the column with 20 ml of 10 mM Tris-HCl pH 7.5, antibodies were eluted by a gradient to 10 mM Tris-HCl pH 7.5, 0.3 M NaCl. Eluted antibodies were digested for 3 h at 37 °C in a reaction mixture containing 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 mM cysteine-HCl, 10 mM β-mercaptoethanol, 10 mM EDTA and papain (1:50 wt/wt ratio with respect to IgG). Digestion was stopped by addition of 23 mM iodoacetamide for 20 min. Cleaved antibodies were dialysed (15-kDa cut-off) against 10 mM Tris-HCl pH 8.0, 10 mM NaCl, and then applied to a 5-ml Q-Sepharose column equilibrated with 10 mM Tris-HCl pH 7.5, 10 mM NaCl. Fab fragments were recovered from the flow-through and a low-salt fractions collected during a gradient to 10 mM Tris-HCl pH 8.0, 0.2 M NaCl, and concentrated 10–20 mg/ml using Amicon Ultra (25-kDa cut-off).
Cryo-EM analysis
Three microlitres of purified CLC-K–Fab complex was applied to a glow-discharged copper Quantifoil R 1.2/1.3 holey carbon grids (Quantifoil) and incubated for 20 s. Grids were then blotted for 4 s at ~90% humidity and plunge-frozen in liquid-nitrogen-cooled liquid using Vitrobot Mark IV (FEI).
The data sets were collected on a Titan Krios electron microscope (FEI) operated at an acceleration voltage of 300 kV. Dose-fractionated images were recorded on a K2 Summit direct electron detector (Gatan) operated in super-resolution counting mode (a super-resolution pixel size of 0.65 Å) using SerialEM software54. The dose rate was 10 e− per pixel per s, and total exposure time was 15 s with 0.3 s for each frame (total cumulative dose of ~89 e− per Å2 over 50 frames). Microscope set defocus values in the final data set ranged from −1.0 μm to −3 μm.
Dose-fractionated images (movies) were first gain-corrected and 2× pixel-binned (resulting in a pixel size of 1.3 Å). Frame-based motion-corrected, summed micrographs were generated by the unblur program55 using all 50 frames and unblur’s exposure filter (radiation damage filter) option. Defocus values were estimated using CTFFIND4 (ref. 56). All subsequent image processing, except for per-particle motion correction, was carried out with RELION 1.4 (ref. 57), using a particle box size of 240 pixels and a mask dimeter of 180–220 Å (Extended Data Fig. 3). Initially, ~5,000 particles were manually picked and subjected to reference-free two-dimensional classification. Six to ten representative two-dimensional class averages were selected as templates for automated particle picking. A total of 442,331 auto-picked particles from 3,392 micrographs, which were divided into 4 subsets, were subjected to 1–3 rounds of reference-free two-dimensional classification to clean up false positives and defective particles. Particles were further manually cleaned up by visual inspection of micrographs. An initial three-dimensional reference was obtained with EMAN2 (ref. 58) from a subset of particles. To obtain ‘polished’ particle images corrected for individual particle movements, frame-based motion-corrected movies, generated by unblur without using its exposure filter option, were 2× frame-binned (resulting in a total of 25 frames per movie), and then applied to the alignparts-lmbfgs program59 together with coordinates of cleaned-up particles. Alignparts-lmbfgs’s exposure filter option was used to filter radiation damage. All ‘polished’ particle images were then subjected to one more round of two-dimensional classification for clean-up, and then to three-dimensional classification, sorting into 10 classes. By visual inspection in UCSF Chimera60, particles from seven classes were combined to two groups (117,208 particles for class 1 and 76,272 particles for class 2) based on similarity, which were separately refined. For each group, low-quality particles were removed based on rlnMaxValueProbDistribution values generated by the first-round three-dimensional refinement (<0.09 for class 1 and <0.07 for class 2), and the remaining particles (82,167 particles for class1 and 67,475 particles for class 2) were subjected to the second-round three-dimensional refinement, yielding the final reconstructions. For both three-dimensional classification and refinement, C2 rotational symmetry was imposed. Three-dimensional refinement was performed using a soft mask including the channel, detergent micelle, the variable (VH/VL) domains of Fab fragments, but excluding the constant (CH1/CL) domains of Fab fragments. Gold-standard Fourier shell correlation (FSC) of independently refined half maps and overall resolutions (based on the 0.143 cut-off criterion) were calculated in RELION’s post-processing procedure using user-provided soft masks (Extended Data Fig. 3). Local resolution was estimated using blocres61 with a box size of 15 pixels (Extended Data Fig. 3d, i). We note that local resolution calculated using Resmap62 was significantly higher than resolution calculated with blocres (for example, most parts of the channel being at ~3.0 Å resolution in the case of the class 1 map), but this is likely to be an overestimation given the observed structural details and the overall resolution estimates based on FSC. Maps shown in figures are combined maps, which were sharpened and low-pass filtered in RELION. The map in Fig. 1c was sharpened with a B-factor of −100 Å2 and low-pass filtered at 3.7 Å. The density map in Fig. 4b (and Fig. 4d) was sharpened with a B-factor of −100 Å2 and low-pass filtered at 3.9 Å.
Model building
An initial homology model of the CLC-K monomer was generated with the SWISS-MODEL online server using the CmCLC model (PDB accession 3ORG) as a template. The resulting model was fit into the class 1 density map using Chimera. Further model building was done with Coot63. The density map was of sufficient quality for de novo building of several regions including the extracellular domain between residues 340 and 398. A starting model of the Fab fragment (VH and VL domains) was obtained by combining VH and VL models from PDB accessions 1ORS and 1NGX, respectively, which were selected based on sequence homology to the 25E7 VH/VL sequences using BLAST. This model was then docked into the density map using UCSF Chimera and further rebuilt in Coot. A dimer model of the CLC-K–Fab complex was obtained by a symmetry operation of the monomer model. Reciprocal space refinement was done with Refmac 5.8 using a script developed for cryo-EM model refinements64,65 (Extended Data Table 1). Two-fold symmetry was imposed during refinement using strong non-crystallographic symmetry (NCS) restraints. However, the segment between residues 228 and 234 of CLC-K was excluded from NCS restraints during refinement of the class 1 model due to steric clash of two F231 side chains at twofold symmetric axis. As a result, this segment in the final model shows asymmetry between two subunits, mainly in the rotamer of F231. Secondary structure restraints were generated using ProSMART (ref. 66). To prevent overfitting during refinement, the weight in Refmac between the geometric restraints and the fit to the experimental density map was adjusted using a method previously described67. Briefly, atoms in the model built with the combined map were randomly displaced up to 0.2 or 0.3 Å, and then subjected to fully restrained refinement against one (‘work’) of two half maps obtained from independent half data sets. An FSC curve (FSCWork) was calculated between the refined model and the half map used for refinement. As cross-validation, a second FSC curve (FSCFree) was calculated between the refined model and the other half map (‘free’) not used in refinement. The weight in Refmac was reduced until it did not produce any significant difference between FSCWork and FSCFree (Extended Data Fig. 3e, j). To this end, a weight of 0.0004 was chosen and used for refinement against the combined map reconstructed from all particles. To build the model of the class 2 structure, the TM and cytosolic domains and Fab of the refined class 1 model were separately fit into the density map in Chimera. The fit was further improved in Coot by rigid body fitting of smaller fragments and minor rebuilding. A dimer model was generated by applying symmetry operators and then subjected to refinement with Refmac as described earlier. The following segments of CLC-K were not modelled due to invisible or poor density features in the corresponding regions: in class 1, N–47, 258–275, 454–456, 606–617, 684–C; in class 2, N–35, 186–192, 258–275, 454–458, 606–617, 684–C.
To evaluate Cl− ion accessibility in the cytosolic and extracellular vestibules of CLC-K (Fig. 5a), internal cavities were probed using HOLLOW (ref. 68) using a probe radius of 1.7 Å. After removing small isolated cavities, the resulting regions were represented as surface using PyMOL (Schrödinger). Pore radii along the Cl− ion transport pathway from the central Cl−-binding site (Scen) towards the cytosol was measured using Caver (ref. 69). In the case of CmCLC, Glu210, whose side chain is occupying Scen, was mutated to Val before estimation. Three-dimensional coordinates of probe positions were converted to one-dimensional distances by projection onto a straight line between Scen and the entrance to the cytosolic funnel. Protein electrostatics were calculated using the Adaptive Poisson-Boltzmann Solver (ref. 70). A monovalent salt concentration of 150 mM was given as a parameter. The dimer interface shape complementarity was calculated by Sc (ref. 71). UCSF Chimera and PyMOL were used to prepare structure figures.
Data availability
Atomic coordinates of CLC-K have been deposited in the PDB under accessions 5TQQ (class 1) and 5TR1 (class 2). The three-dimensional cryo-EM density maps (combined, unfiltered maps and sharpened, low-pass filtered maps) have been deposited in Electron Microscopy Data Bank under accessions EMD-8435 (class 1) and EMD-8454 (class 2).
References
Jentsch, T. J. Discovery of CLC transport proteins: cloning, structure, function and pathophysiology. J. Physiol. 593, 4091–4109 (2015)
Maduke, M., Miller, C. & Mindell, J. A. A decade of CLC chloride channels: structure, mechanism, and many unsettled questions. Annu. Rev. Biophys. Biomol. Struct. 29, 411–438 (2000)
Miller, C. ClC chloride channels viewed through a transporter lens. Nature 440, 484–489 (2006)
Koch, M. C. et al. The skeletal muscle chloride channel in dominant and recessive human myotonia. Science 257, 797–800 (1992)
Simon, D. B. et al. Mutations in the chloride channel gene, CLCNKB, cause Bartter’s syndrome type III. Nature Genet. 17, 171–178 (1997)
Fisher, S. E. et al. Isolation and partial characterization of a chloride channel gene which is expressed in kidney and is a candidate for Dent’s disease (an X-linked hereditary nephrolithiasis). Hum. Mol. Genet. 3, 2053–2059 (1994)
Kornak, U. et al. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 104, 205–215 (2001)
Miller, C. & White, M. M. Dimeric structure of single chloride channels from Torpedo electroplax. Proc. Natl Acad. Sci. USA 81, 2772–2775 (1984)
Jentsch, T. J., Steinmeyer, K. & Schwarz, G. Primary structure of Torpedo marmorata chloride channel isolated by expression cloning in Xenopus oocytes. Nature 348, 510–514 (1990)
Accardi, A. & Miller, C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature 427, 803–807 (2004)
Scheel, O., Zdebik, A. A., Lourdel, S. & Jentsch, T. J. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature 436, 424–427 (2005)
Picollo, A. & Pusch, M. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature 436, 420–423 (2005)
Walden, M. et al. Uncoupling and turnover in a Cl−/H+ exchange transporter. J. Gen. Physiol. 129, 317–329 (2007)
Jayaram, H., Accardi, A., Wu, F., Williams, C. & Miller, C. Ion permeation through a Cl−-selective channel designed from a CLC Cl−/H+ exchanger. Proc. Natl Acad. Sci. USA 105, 11194–11199 (2008)
Ludewig, U., Pusch, M. & Jentsch, T. J. Two physically distinct pores in the dimeric ClC-0 chloride channel. Nature 383, 340–343 (1996)
Dutzler, R., Campbell, E. B., Cadene, M., Chait, B. T. & MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002)
Dutzler, R., Campbell, E. B. & MacKinnon, R. Gating the selectivity filter in ClC chloride channels. Science 300, 108–112 (2003)
Feng, L., Campbell, E. B., Hsiung, Y. & MacKinnon, R. Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle. Science 330, 635–641 (2010)
Adachi, S. et al. Two isoforms of a chloride channel predominantly expressed in thick ascending limb of Henle’s loop and collecting ducts of rat kidney. J. Biol. Chem. 269, 17677–17683 (1994)
Kieferle, S., Fong, P., Bens, M., Vandewalle, A. & Jentsch, T. J. Two highly homologous members of the ClC chloride channel family in both rat and human kidney. Proc. Natl Acad. Sci. USA 91, 6943–6947 (1994)
Denton, J. S., Pao, A. C. & Maduke, M. Novel diuretic targets. Am. J. Physiol. Renal Physiol. 305, F931–F942 (2013)
Estévez, R. et al. Barttin is a Cl− channel β-subunit crucial for renal Cl− reabsorption and inner ear K+ secretion. Nature 414, 558–561 (2001)
Scholl, U. et al. Barttin modulates trafficking and function of ClC-K channels. Proc. Natl Acad. Sci. USA 103, 11411–11416 (2006)
Waldegger, S. & Jentsch, T. J. Functional and structural analysis of ClC-K chloride channels involved in renal disease. J. Biol. Chem. 275, 24527–24533 (2000)
L’Hoste, S. et al. Characterization of the mouse ClC-K1/Barttin chloride channel. Biochim. Biophys. Acta 1828, 2399–2409 (2013)
Fahlke, C., Yu, H. T., Beck, C. L., Rhodes, T. H. & George, A. L. Jr. Pore-forming segments in voltage-gated chloride channels. Nature 390, 529–532 (1997)
Accardi, A. & Pusch, M. Fast and slow gating relaxations in the muscle chloride channel CLC-1. J. Gen. Physiol. 116, 433–444 (2000)
Estévez, R., Schroeder, B. C., Accardi, A., Jentsch, T. J. & Pusch, M. Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. Neuron 38, 47–59 (2003)
Waldegger, S. et al. Barttin increases surface expression and changes current properties of ClC-K channels. Pflugers Arch. 444, 411–418 (2002)
Feng, L., Campbell, E. B. & MacKinnon, R. Molecular mechanism of proton transport in CLC Cl−/H+ exchange transporters. Proc. Natl Acad. Sci. USA 109, 11699–11704 (2012)
Markovic, S. & Dutzler, R. The structure of the cytoplasmic domain of the chloride channel ClC-Ka reveals a conserved interaction interface. Structure 15, 715–725 (2007)
Robertson, J. L., Kolmakova-Partensky, L. & Miller, C. Design, function and structure of a monomeric ClC transporter. Nature 468, 844–847 (2010)
Khantwal, C. M. et al. Revealing an outward-facing open conformational state in a CLC Cl−/H+ exchange transporter. eLife 5, e11189 (2016)
Miller, C. Open-state substructure of single chloride channels from Torpedo electroplax. Phil. Trans. R. Soc. Lond. B 299, 401–411 (1982)
Duffield, M., Rychkov, G., Bretag, A. & Roberts, M. Involvement of helices at the dimer interface in ClC-1 common gating. J. Gen. Physiol. 121, 149–161 (2003)
Gradogna, A., Fenollar-Ferrer, C., Forrest, L. R. & Pusch, M. Dissecting a regulatory calcium-binding site of CLC-K kidney chloride channels. J. Gen. Physiol. 140, 681–696 (2012)
Jayaram, H., Robertson, J. L., Wu, F., Williams, C. & Miller, C. Structure of a slow CLC Cl−/H+ antiporter from a cyanobacterium. Biochemistry 50, 788–794 (2011)
Lim, H. H., Stockbridge, R. B. & Miller, C. Fluoride-dependent interruption of the transport cycle of a CLC Cl−/H+ antiporter. Nature Chem. Biol. 9, 721–725 (2013)
Bell, S. P., Curran, P. K., Choi, S. & Mindell, J. A. Site-directed fluorescence studies of a prokaryotic ClC antiporter. Biochemistry 45, 6773–6782 (2006)
Basilio, D., Noack, K., Picollo, A. & Accardi, A. Conformational changes required for H+/Cl− exchange mediated by a CLC transporter. Nature Struct. Mol. Biol . 21, 456–463 (2014)
Jackson, M. R. et al. A preference for edgewise interactions between aromatic rings and carboxylate anions: the biological relevance of anion-quadrupole interactions. J. Phys. Chem. B 111, 8242–8249 (2007)
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966)
Nguitragool, W. & Miller, C. Uncoupling of a CLC Cl−/H+ exchange transporter by polyatomic anions. J. Mol. Biol. 362, 682–690 (2006)
Accardi, A., Lobet, S., Williams, C., Miller, C. & Dutzler, R. Synergism between halide binding and proton transport in a CLC-type exchanger. J. Mol. Biol. 362, 691–699 (2006)
Picollo, A., Malvezzi, M., Houtman, J. C. & Accardi, A. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nature Struct. Mol. Biol . 16, 1294–1301 (2009)
Bergsdorf, E. Y., Zdebik, A. A. & Jentsch, T. J. Residues important for nitrate/proton coupling in plant and mammalian CLC transporters. J. Biol. Chem. 284, 11184–11193 (2009)
Zifarelli, G. & Pusch, M. Conversion of the 2 Cl−/1 H+ antiporter ClC-5 in a NO3−/H+ antiporter by a single point mutation. EMBO J. 28, 175–182 (2009)
De Angeli, A. et al. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442, 939–942 (2006)
Kim, J. H. et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6, e18556 (2011)
Hofmann, C. et al. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl Acad. Sci. USA 92, 10099–10103 (1995)
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014)
Kirchhofer, A. et al. Modulation of protein properties in living cells using nanobodies. Nature Struct. Mol. Biol . 17, 133–138 (2010)
Lim, H. H., Fang, Y. & Williams, C. High-efficiency screening of monoclonal antibodies for membrane protein crystallography. PLoS One 6, e24653 (2011)
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005)
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015)
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 184, 226–236 (2013)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015)
Nicholls, R. A., Long, F. & Murshudov, G. N. Low-resolution refinement tools in REFMAC5. Acta Crystallogr. D 68, 404–417 (2012)
Fernández, I. S., Bai, X. C., Murshudov, G., Scheres, S. H. & Ramakrishnan, V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157, 823–831 (2014)
Ho, B. K. & Gruswitz, F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008)
Chovancova, E. et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput. Biol. 8, e1002708 (2012)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Lawrence, M. C. & Colman, P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 (1993)
Fialho, D. et al. Chloride channel myotonia: exon 8 hot-spot for dominant-negative interactions. Brain 130, 3265–3274 (2007)
Vindas-Smith, R. et al. Identification and functional characterization of CLCN1 mutations found in nondystrophic myotonia patients. Hum. Mutat. 37, 74–83 (2016)
Mailänder, V., Heine, R., Deymeer, F. & Lehmann-Horn, F. Novel muscle chloride channel mutations and their effects on heterozygous carriers. Am. J. Hum. Genet. 58, 317–324 (1996)
Trip, J. et al. In tandem analysis of CLCN1 and SCN4A greatly enhances mutation detection in families with non-dystrophic myotonia. Eur. J. Hum. Genet. 16, 921–929 (2008)
Dupré, N. et al. Clinical, electrophysiologic, and genetic study of non-dystrophic myotonia in French-Canadians. Neuromuscul. Disord. 19, 330–334 (2009)
Lorenz, C., Meyer-Kleine, C., Steinmeyer, K., Koch, M. C. & Jentsch, T. J. Genomic organization of the human muscle chloride channel CIC-1 and analysis of novel mutations leading to Becker-type myotonia. Hum. Mol. Genet. 3, 941–946 (1994)
Acknowledgements
We thank M. Ebrahim at the Rockefeller University Cryo-EM Resource Center for help with microscope operation, staff at the Memorial Sloan Kettering Cancer Center Antibody & Bioresource Core Facility for hybridoma generation, Y. C. Hsiung for help with large-scale cell culture, members of the MacKinnon laboratory for helpful discussions, and J. Chen for critical reading of the manuscript. E.P. is supported by the Jane Coffin Childs Memorial Fund fellowship (#61-1513). R.M. is a Howard Hughes Medical Institute investigator.
Author information
Authors and Affiliations
Contributions
E.P. performed experiments. E.B.C. assisted in development of monoclonal antibodies. E.P. and R.M. designed experiments, analysed and interpreted results, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks C. Miller, M. Pusch, S. Scheres and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Functional characterization of the bovine CLC-K channel.
a, Representative two-electrode voltage clamp (TEVC) recordings of bovine CLC-K channel in X. laevis oocytes. Clamping voltages were from −60 to +60 mV (10-mV steps). b–e, I–V curves of recordings in a. Shown are means and standard deviations (s.d.; error bars) of 5, 9, 10 and 11 independent oocyte recordings, respectively. f, I–V curves of whole-cell patch recordings on Chinese hamster ovary (CHO) cells expressing CLC-K and barttin. The pipette and bath solutions contain 144 and 52 mM Cl−, respectively. Shown are means and s.d. (error bars) of 3 or 5 independent recordings using different cells. g, Immunofluorescence staining of the bovine CLC-K channel expressed on the plasma membrane. CHO cells were transiently transfected with a green fluorescent protein (GFP)-tagged channel construct alone or together with barttin, and then cell-surface-targeted channels were probed by non-permeabilized immunofluorescence (IF) staining using monoclonal anti-CLC-K antibodies (clone 16E3), which specifically recognize an extracellular epitope of the CLC-K channel. Hoechst 33342 was used to stain nuclei. The same exposure parameters were used for the left and right panels. Shown are representative images of reproducible results. Scale bar, 10 μm.
Extended Data Figure 2 Cryo-EM image processing procedure.
a, A representative micrograph of the CLC-K–Fab complex (scale bar, 50 nm). Boxed regions (white squares) are magnified on the right to show representative particle images. Note that in the top/bottom views, Fab signals are evident while the channel part is barely visible because of low contrast (also see b). b, Images of selected two-dimensional classes from reference-free two-dimensional classification by RELION. Note that the distal half of the Fab fragment shows much flexibility, resulting in blurred features. Scale bar, 10 nm. c, Summary of the image processing procedure (see Methods).
Extended Data Figure 3 Three-dimensional reconstruction of CLC-K.
a, Angular distribution histogram of class 1 particle projections. b, Fourier shell correlation (FSC) of class 1 half maps before (black) and after (blue and red) masking. Two soft masks were used (see also c): one (red) including only the channel portion and the other (blue) including the channel and the variable domain (VH/VL) of the Fab fragments but excluding the constant domain (CH1/CL) of Fab. When a mask was used, the FSC curve was corrected for masking effects during the RELION postprocessing procedure (phase randomization above 7.8 Å). c, Masks used in FSC calculations (red and blue) in b are shown with the unsharpened, unfiltered map (grey) superimposed. A contour level of 0.7 was used for the surface representation of the masks. d, Local resolution map of the class 1 reconstruction estimated by the blocres program. Shown is the combined map, which is not sharpened or filtered. e, FSC between model and map of the class 1 particles. The black curve shows FSC between the final refined atomic model and the combined map that the model was refined against. The blue and red curves show FSC between the atomic model and the half map it was refined against (half 1) and FSC between the atomic model and the other half map it was not refined against (half 2), respectively (see also Methods). f–j, As in a–e, but with the class 2 structure. For the FSC calculations with the masks in g, the FSC curves were corrected for masking effects (phase randomization above 8.7 Å).
Extended Data Figure 4 EM density of the CLC-K channel.
EM density segments (mesh) of the class-1 CLC-K reconstruction are superimposed with the model in a ribbon representation. Numbers indicate amino acid positions of segments. The density map was sharpened with a B-factor of −100 Å2 and low-pass filtered at 3.7 Å. CTD, cytosolic domain. VL, variable domain of the IgG light chain. VH, variable domain of the IgG heavy chain. Note that a model for the distal half of the Fab fragment was not built because of poor density features in this region caused by its high flexibility (see also Extended Data Fig. 2b).
Extended Data Figure 5 Comparison between the class 1 and 2 structures of CLC-K.
a, The dimer interfaces (dashed boxes) of the class 1 and 2 structures were compared. Views from the extracellular side were aligned based on the upper (red) subunit. In addition, the right panel shows superposition of a class 1 monomer (grey) onto a class 2 monomer (red). b, N-terminal amphipathic helix of class 2 structure. A side view of the class 2 structure is shown in a ribbon representation superimposed with density (mesh) of the N-terminal helix. The two subunits are shown in pale green and blue. The TM helix αB, which is visible in both class 1 and 2 structures is shown in dark green. A 12-amino-acid amphipathic helix preceding αB, visible only in class 2, is shown in yellow. The density map was sharpened with a B-factor of −120 Å2 and low-pass filtered at 4.1 Å. The approximate membrane boundaries are indicated by grey lines.
Extended Data Figure 6 Structural features of the CLC-K channel.
a, Comparison between the CLC-K (class 1; grey and magenta) and EcCLC (yellow) structures. The TM domain of an EcCLC monomer was superposed onto that of CLC-K. α-Helices are represented as cylinders. b, The density of the extracellular loops αI–J of the CLC-K channel (class 1) is shown in blue and orange. The density of the remaining parts is shown in pale blue and yellow. In the top right panel, the model of one subunit is shown in the ribbon representation. αN and αF are highlighted, and Val166 and Tyr520 are shown in a ball-and-stick representation (green). The two ends of the αI–J link are indicated as cyan spheres with amino acid positions. The bottom panels show side views of one subunit with the αI–J link in blue density. The density map (5σ cut-off) was sharpened with a B-factor of −100 Å2 and low-pass filtered at 4.0 Å. c, As in the top left panel of b, but additionally showing a model of the αI–J link of EcCLC in a ball-and-stick representation (yellow). d, The same view as in the bottom right panel of b, but with superposition of an EcCLC model (pink; PDB accession 1OTS) onto the CLC-K model. A model for the αI–J linker of EcCLC is shown in yellow balls and sticks. e, Sequence alignment of the extracellular segments between TM helices αK and αM from various CLC proteins. The segments forming α-helices in the extracellular domain of bovine CLC-K are indicated by red coils. Red and blue dots indicate positions of mutations causing Bartter syndrome (CLC-Kb) and myotonia congenita (CLC-1), respectively. The A349D (ref. 5) mutation in CLC-Kb causes Bartter syndrome. The P408A (ref. 72), Q412P (ref. 73), F413C (ref. 4), A415V (ref. 74), E417G (ref. 75) or W433R (ref. 76) mutations in CLC-1 (corresponding to positions 342, 346, 347, 349, 351 and 367 of CLC-K, respectively) cause myotonia congenita. In addition, mutations at R438 (R438C in CLC-Kb or R496S in CLC-1), of which the side chain directly interacts with W367 (W433 in CLC-1) cause Bartter syndrome5 or myotonia congenita77 (see Fig. 4c).
Extended Data Figure 7 Structure of the ion transport pathway.
a, Stereo images of the atomic model of the CLC-K channel’s Cl−-selective filter (class 1; light magenta) and comparisons with that of CmCLC (cyan) and EcCLC (yellow) are shown in a stick representation. In the cases of CmCLC and EcCLC, bound Cl− ions are shown as spheres. Predicted polar interactions are indicated by grey dashed lines. b, EM density and model of the αC–D loop are shown for the class-2 CLC-K structure. Also, density and model were shown for the Tyr520 side chain. The density map (grey mesh) was sharpened with a B-factor of −120 Å2 and low-pass filtered at 4.1 Å.
Extended Data Figure 8 Density in the Scen Cl−-binding site.
A view into Scen of CLC-K (from Sext towards the cytosol) is shown in a stereo diagram. Predicted polar interactions between bound Cl− ions and the protein, including anion–quadrupole interactions, are indicated by green dashed lines. The shown EM density map is of the class 1 structure, sharpened with a B-factor of −100 Å2 and low-pass filtered at 3.7 Å. Density for protein (grey mesh) and possible ions (magenta mesh) are shown at contour levels of 5.8σ and 3.2σ, respectively.
Extended Data Figure 9 Surface electrostatic potential.
Surface electrostatics of the CLC-K channel (class 1) compared with the EcCLC and CmCLC transporters. Left panels show side views with one subunit in a surface representation and the other in a ribbon representation. Electrostatic potential is indicated by the colour scale (bottom left). Views from the extracellular side (blue arrow) and intracellular side (red arrow) are shown in the middle and right panels, respectively. The entrances to the Cl− ion pathway are indicated by dashed circles. Mobile monovalent cation and anion species at 0.15 M each were included in the electrostatic potential calculations.
Rights and permissions
About this article
Cite this article
Park, E., Campbell, E. & MacKinnon, R. Structure of a CLC chloride ion channel by cryo-electron microscopy. Nature 541, 500–505 (2017). https://doi.org/10.1038/nature20812
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20812
- Springer Nature Limited
This article is cited by
-
Structural basis of pH-dependent activation in a CLC transporter
Nature Structural & Molecular Biology (2024)
-
Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger
Nature Communications (2024)
-
Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nature Communications (2023)
-
Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids
Nature Communications (2023)
-
Long-read sequencing identifies a common transposition haplotype predisposing for CLCNKB deletions
Genome Medicine (2023)