Abstract
Objective:
Sudden Infant Death Syndrome (SIDS) is defined as the sudden death of an infant <1 year of age that cannot be explained following a thorough investigation. Currently, no reliable clinical biomarkers are available for the prediction of infants who will die of SIDS.
Study Design:
This study aimed to profile the medulla oblongata from postmortem human brain from SIDS victims (n=16) and compare their profiles with that of age-matched controls (n=7).
Results:
Using LC-Orbitrap-MS, we detected 12 710 features in electrospray ionization positive (ESI+) mode and 8243 in ESI− mode from polar extracts of brain. Five features acquired in ESI+ mode produced a predictive model for SIDS with an area under the receiver operating characteristic curve (AUC) of 1 (confidence interval (CI): 0.995–1) and a predictive power of 97.4%. Three biomarkers acquired in ESI− mode produced a predictive model with an AUC of 0.866 (CI: 0.767–0.942) and a predictive power of 77.6%. We confidently identified 5 of these features (l-(+)-ergothioneine, nicotinic acid, succinic acid, adenosine monophosphate and azelaic acid) and putatively identify another 4 out of the 15 in total.
Conclusions:
This study underscores the potential value of metabolomics for studying SIDS. Further characterization of the metabolome of postmortem SIDS brains could lead to the identification of potential antemortem biomarkers for novel prevention strategies for SIDS.
Similar content being viewed by others
Introduction
Sudden Infant Death Syndrome (SIDS) is defined as the sudden death of an infant <1 year of age that remains unexplained following a thorough standardized investigation, including a complete autopsy, examination of the death scene and review of the clinical history.1 Each year in the United States, about 4000 infants die suddenly of no immediately obvious cause. About half of these sudden unexpected infant deaths are due to SIDS, the leading cause of sudden unexpected infant deaths and of all deaths among infants aged 1–12 months.1 According to the National Vital Statistics Report, published in December 2013, SIDS is the third leading cause of infant death in the United States of America, and rates for non-Hispanic black and American Indian or Alaskan native women are almost twice than those for non-Hispanic white women.2 There are numerous epidemiological and physiological factors that are associated with an increased risk for SIDS; however, these factors are still too generic to precisely predict high-risk infants.3 Intense efforts to understand the etiology of SIDS has led to the development of a triple-risk model, including: (i) a vulnerable infant; (ii) a critical period of development in homeostatic control, and (iii) an exogenous stressor.4 The triple-risk hypothesis emphasizes the interaction of multiple factors in pathogenesis and helps to understand why particular infants may die at particular ages when exposed to potential stressors.5 Despite many genetic alterations found in SIDS, the genetic contribution still remains unclear. For example there is a low rate of SIDS in siblings and lack of concordance in twins suggests a modest genetic impact and emphasizes the need for a multidisciplinary approach.6
Metabolomics, the systematic investigation of all metabolites present within a biological system, is used in biomarker development for many human diseases.7 This emerging member of the ‘omics’ field is not only concerned with the identification and quantification of metabolites but also concerned with relating metabolite data to genes,8 proteins,9 pathways,10 physiology and phenotypes.11 Metabolomic analysis of SIDS cases has the potential to elucidate pathogenesis and develop predictive biomarkers of this syndrome. Ultimately, this could assist in SIDS-prevention efforts.
The brain has long been known to have a critical role in the pathogenesis of SIDS.12 Although there are many regions throughout the brain that influence arousal or respiratory and autonomic function (for example, hypothalamus, amygdala), SIDS research has historically focused on the brainstem.13 Combating exogenous stressors that trigger asphyxia, hypoxia, hypercapnia, thermal imbalance and/or cardiovascular instability require intact brainstem defense mechanisms to protect against lethal consequences.12 Accumulating evidence points to increased gliosis, increased apoptotic neurons,14 activated caspase-3 immunoreactivity,15 caspase-3 immunoreactivity16 and expression of c-jun and c-fos17 in the brainstems of infants dying of SIDS, however, these findings are not unequivocal.18 Further, there is compelling evidence that shows medullary homeostatic network dysfunction as a key underlying abnormality in SIDS.19 In the medulla oblongata, the arcuate nucleus is formed from a group of neurons located on the anterior surface of the medullary pyramids that contributes to the regulation of respiration in humans. It has been proposed that the arcuate nucleus at the ventral medullary surface is a candidate region for central chemosensitivity in humans and structural underdevelopment of the arcuate nucleus was subsequently observed in SIDS cases.20 Moreover, neurotransmitter abnormalities, including 5-hydroxytryptamine (5-HT),21, 22, 23 γ-aminobutyric acid (GABA)24 and the family of 14-3-3 signal transduction proteins,24 were most consistently reported in the medulla oblongata of brainstem in cases with SIDS. Thus this study aims to evaluate the metabolomic profile of the medula oblongata of brainstem in SIDS cases compared with controls. In doing so, we are confident of identifying central biomarkers for SIDS and discovering novel biochemical pathways associated with the disease.
Materials and methods
Tissue samples and data acquisition
Only a limited number of specimens and tissue volume was available for this pilot study. Tissue samples (medulla) were obtained from postmortem SIDS cases with no co-sleeping (n=16) and age- and gender-matched control subjects (n=7). Tissues were obtained from the University of Maryland Brain and Tissue Bank, which is a Brain and Tissue Repository of the NIH NeuroBioBank. This study was approved by the Beaumont Health System’s Human Investigation Committee (HIC no.: 2014-210). The methods were carried out in accordance with the approved guidelines. Details such as age, gender, race and postmortem delay can be found in Table 1 (Supplementary Material). Samples were prepared and analyzed using an LTQ-Orbitrap Elite (Thermo Scientific, San Jose, CA, USA) in both positive (ESI+) and negative (ESI−) electrospray ionization modes as previously described by Graham et al.25, 26, 27 For ESI− acquired data, the gradient remained the same as for ESI+; however, the flow rate changed to 0.36 ml min−1.
Data analysis
The raw data form the spectral analysis of the tissue extracts was processed using the Progenesis QI Software v2.0 (Waters, Milford, MA, USA) and XCMS Online (http://xcmsonline.scripps.edu/), as previously described by Graham et al.26, 27. Subsequently data were exported to Simca v14 (Umetrics, Umea, Sweden) for multivariate analysis. Quality-control measures were employed to all data sets as previously described by Graham et al.26, 27 (see Figure 1, Supplementary Material) to ensure chromatographic reproducibility of retention times and peak intensities. All univariate analyses were conducted using the SAS System for Windows version 9.3 (Cary, NC, USA) as previously described by Graham et al.26
Metabolite identifications
Using the accurate mass of precursor and fragment ions, Progenesis QI 2.0 and XCMS Online28 searched the KEGG, Human Metabolome Database29, 30, 31 and Massbank32 spectral libraries (a total of 4928 were identified from 12 710 m/z values; 5 p.p.m. tolerance). Ions of interest were fragmented at various normalized collision energies in the high-collision dissociation cell as previously described by Graham et al.26, 27 Further, fragment intensities were also used to search m/z cloud (http://www.mzcloud.org/; HighChem, Bratislava, Slovakia) and MassBank for metabolite identification. The chromatographic peak, accurate mass of the precursor ion, fragmentation spectra for the identified metabolites and their matching fragmentation spectra from the library are shown in Supplementary Figure S3.
Results
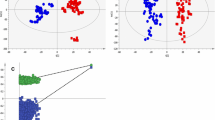
Raw data acquired in ESI+ and ESI− modes and analyzed using Progenesis QI revealed 12 710 and 8243 features, respectively; following background filtration to remove noise (>20% zero values), the number of detected features were reduced to 4952 for ESI+ and to 2310 for ESI− acquired data. Figures 1a and b display the chromatographs of the postmortem brain extracts in both ESI+ and ESI−, respectively. The filtered data sets were used to produce the multivariate models. Using principal component analysis (PCA) all acquired data were found to be highly reproducible as evident from the tight clustering of the quality controls (at the center of the score plot) and each individual extract (n=6) over the duration of the data acquisition (Figure 1, Supplementary Material).25, 26 Figure 2 displays the OPLS-DA scores plots for the data acquired in ESI+ (Figure 2a) and ESI− (Figure 2c) modes. The OPLS-DA plot in Figure 2a was produced with one orthogonal and three latent components; R2=92.9 % and Q2=89.3 %. Complete separation between controls and SIDS data was visualized from the score plots, and as the R2 and Q2 values would suggest, excellent clustering of the data was observed. Figure 2c displays the OPLS-DA plot for data acquired in ESI− mode. As previously, complete separation was observed between the two groups producing a model with R2=96.4 % and Q2=92.4 %, denoting excellent clustering of the data. The model was developed using one orthogonal component and three latent components. Figures 2b and d display the results of the permutation testing (n=999) for both sets of data acquired in ESI+ and ESI−, respectively. The results of these permutation tests demonstrate the accuracy of both models. The criteria to which the validity of each model is critiqued include: all Q2 values to the left must be lower than the original points to the right and the Q2 regression line must intersect the y axis below zero.26, 33
(a) The scores plot for data acquired in electrospray ionization positive (ESI+) mode displaying the separation between the two sample groups (blue squares=controls; red circles=Sudden Infant Death Syndrome (SIDS)). (b) The results of the permutation analysis for the model build using ESI+ acquired data (green circles=R2; blue squares=Q2). (c) The scores plot for data acquired in ESI− mode displaying the separation between the two sample groups (blue squares=controls; red circles=SIDS). (d) The results of the permutation analysis for the model build using ESI+ acquired data (green circles=R2; blue squares=Q2). A full color version of this figure is available at the Journal of Perinatology journal online.
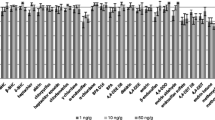
Following the development of the multivariate models, the filtered data were analyzed to determine whether a small panel of predictive biomarkers was capable of discriminating SIDS brains from controls. All the data from both modes of acquisition were uploaded to the Metaboanalyst (http://www.metaboanalyst.ca)34, 35, 36 and analyzed using the Biomarker option. This function developed a variety of models based on the top 5, 10, 15, 25, 50 and 100 features. For both ESI+ and ESI− data, we found that the model’s predictive ability did not increase with >15 features being used to develop the models. These 15 features were analyzed to determine whether they were indeed statistically significantly different between SIDS and control samples. Tables 1 and 2 display the results of the univariate analyses for both ESI+ and ESI− data, respectively. Each individual feature was inspected manually to determine the validity of their chromatographic peak in a number of pooled samples. Some peaks were deemed to be noise and removed. This reduced the number of peaks used to build the predictive models from 15 to 9 (one removed by the Metaboanalyst software, n=8) for the ESI+ data and from 15 to 5 for the ESI− data (one removed by the Metaboanalyst software, n=4). The results of the receiver operating characteristic (ROC) analysis for all the models using a variety of features is displayed as Figures 2a and b (Supplementary Material) for ESI+ and ESI− data, respectively.
Using the top nine features from the ESI+ acquired data, it was shown following cross validation (model developed using 2/3 of the data and blindly tested using the remaining 1/3) that five features produced the best model. Following an ROC analysis, it produced an AUC of 1 CI: 0.9995–1) and a predictive accuracy of 97.4 % when differentiating between SIDS and control postmortem brain extracts (Figures 3a and b, respectively). Of the top five features for the ESI− acquired data, three variables produced the best model following an ROC analysis with an AUC of 0.866 (CI: 0.767–0.942) and a predictive accuracy of 77.6 % following crossvalidation (Figures 3c and d, respectively). Permutation testing using 1000 repeat analyses were performed and yielded a P-value of 1.21E−05 and 2.10E−04 for both ESI+ and ESI−, respectively. These significant P-values indicate the observed differentiation achieved by both panels of features was not coincidental.
(a) ROC curve for the data acquired in ESI+ mode using the top five features (AUC=1 (CI: 0.995–1)). (b) The predicative ability of all the models created using a varying number of features with ESI+ data, with five features providing the best predictive model following cross validation (97.4%). (c) The ROC curve for the data acquired in ESI− mode using the top five features (AUC=0.866 (CI: 0.767–0.942)). (d) The predicative ability of all the models created using a varying number of features with ESI− data, with four features providing the best predictive model following crossvalidation (77.6%).
Tables 1 and 2 list the putative identifications (level 2 reporting according to the Metabolomics Standards Initiative37) for the features of interest acquired in ESI+ and ESI− modes, respectively. Of the 15 variables, 5 were confidently identified (2 for ESI+ and 3 for ESI−) matching library fragmentation patterns with experimental data. Figure 3 (Supplementary Material) displays the chromatographic peak, precursor ion accurate mass, fragmentation spectra for the identified metabolites and their matching fragmentation spectra from the library. An additional four features (3 for ESI+ and 1 for ESI−) were putatively identified using their accurate masses to search online libraries. The remaining six potential biomarkers remain unidentified following a comprehensive process of identification. The fragmentation pattern for all the features is available as Table 1 (Supplementary Material). An additional model was created using a combination of the identified metabolites from both models. Supplementary Figure S4a displays the ROC analyses using the relative levels of L-(+)-ergothioneine, nicotinic acid, succinic acid, adenosine monophosphate (AMP) and azelaic acid. The ROC analysis produced an AUC=0.798 (CI: 0.71–0.885) following 100 independent cross validations and had a predictive power of 70.6 %. However, when we further tested the model using permutation analyses we found it to be not statistically significant (P-value=0.078; Supplementary Figure S4b).
Discussion
To our knowledge, this is the first study applying metabolomics and using high resolution mass spectrometry (HRMS) to biochemically fingerprint postmortem SIDS brain tissue versus matched controls. Using ESI+ and ESI− acquired data, two predictive metabolomic models were developed that differentiated SIDS subjects from controls. The fit and clustering of the models were excellent, with Q2 and R2 values for both models ⩾90 %. Furthermore, the predictive accuracies of the models using the top 5 and top 3 features for both the ESI+ and ESI− acquired data were exceptional, with 97.4% and 77.6% predictive accuracy, respectively.
The biomarker panel identified using ESI+ acquired data performed much better when differentiating between the two sample groups than the data acquired in ESI− mode. Of the five features, only one (230.0958 m/z) did not reach statistical significance (P<0.05; false discovery rate (FDR)<0.05) when analyzed using univariate techniques. However, as is the case with the predictive models, the issue of statistical significance is not the most important one (Supplementary Figure S5a). A feature can be important to the model but yet not reach statistical significance. In an instance such as this study, it can be difficult to detect differences between groups with small cohorts. In addition, the adjusted P-values are the minimum of two quantities, one of which depends on the number of features in the set. Altman and Bland38 famously noted that absence of evidence is not evidence of absence. In other words, a non-small P-value does not ‘prove’ that there is no difference; rather the data does not allow you to reasonably exclude the possibility of no difference.38 This holds for the ESI− recorded data; one of the top ranked variables (199.0370 m/z; no. 6 VIP) did not prove to be statistically significantly different (P<0.05; FDR<0.05) between the two groups, yet it was found to be important for the development of the statistical model (Supplementary Figure S5b).
Of the 15 features to be identified as important for each of the respective models, only 5 were confidently characterized by matching the fragmentation patterns with those freely available online (http://www.mzcloud.org/; HighCHem; Figure 3, Supplementary Material). Of these 5 confidently identified metabolites, nicotinic acid, also known as niacin or vitamin B3, is a water-soluble vitamin whose derivatives such as NADH, NAD, NAD+ and NADP have essential roles in energy metabolism, biosynthesis, DNA repair and respiratory burst (http://www.HMDB.ca). However, in this study the relative levels of nicotinic acid were found not to be statistically different between case and control postmortem brain tissue specimens. L-(+)-ergothioneine is a sulfur-containing amino acid that is widely distributed throughout both the animal and plant kingdom, yet its physiological function has yet to be clearly characterized.39 Ergothioneine is believed to be a strong neuroprotectant. Aruoma et al.40 demonstrated its neuroprotective effect in an in vivo excitotoxicity system where cell loss was reduced from 81% in ganglion cells and 43% of non-ganglion cells to 44% and 31%, respectively, when treated with ergothioneine as opposed to N-methyl-D-aspartate. However, as the levels of ergothioneine were upregulated in the postmortem brain extract from SIDS infants, we cannot hypothesize as to its role in SIDS.
Succinic acid and its anion, succinate, are components of the citric acid cycle capable of donating electrons to the electron transfer chain (http://www.HMDB.ca). Succinic acid is produced from succinic semialdehyde by succinic semialdehyde dehydrogenase (SSADH) in the mitochondrial matrix; it has been reported to affect the central nervous system degradation of the neurotransmitter GABA.41 SSADH deficiency is an autosomal recessively inherited disease and is considered to be an under-recognized and potentially manageable neurological disorder.42, 43 Children who have been diagnosed with SSADH deficiency have been shown to display ataxia, seizures, psychomotor retardation, language delay and hypotonia.44 From our analyses, we report significant differences in the levels of succinic acid between those infants who died from SIDS and control cases. Postmortem brain from SIDS patients were identified as having significantly lower levels of succinic acid as opposed to controls (P=0.0268; FDR=0.016). One hypothesis could be energy metabolism, and in particular, the TCA cycle is directly related to SIDS or another hypothesis could be that the failure to degrade secondary metabolites of GABAergic and 5-HT systems in the brain has contributed to the syndrome. The idea that GABAergic and 5-HT systems are associated with SIDS is not a new theory, Broadbelt et al.45 report that GABAA are abnormal in SIDS infant and that SIDS is a complex malady in the medulla that involves deficits in the GABAergic and 5-HT systems. Moreover, succinic acid is known to be the end product of the methylmalonic acid pathway, which is also known to be essential for myelin synthesis. Perturbations in myelin synthesis could lead the underdevelopment of conduction velocity in neuron cells that could possibly cause vulnerability to such aforementioned defense mechanisms.12 In addition to succinic acid, we also found and confidently identified serotonin to be significantly lower (P<5E−06; FDR=5E−05; data not presented) in SIDS postmortem brain extracts. This evidence would support our hypothesis that the potential difficulty to degrade secondary metabolites of GABAergic system and serotonin itself may be directly linked with SIDS.
AMP is a nucleotide that is found as a monomer in RNA. Nucleotides such as AMP are directly involved in a number of major metabolic functions, such as energy metabolism and allosteric regulation. They perform functions such as activated intermediates, physiological mediators and cellular agonists and are considered important structural components of coenzymes.46 AMP have also been reported to be involved in the reversal of malnutrition and starvation-induced immunosuppression47 and the enhancement of T-cell maturation and function.48 Moreover, AMP is a metabolite of cyclic AMP, a well-known secondary messenger in many excitatory neurotransmitter signal transduction pathways, including glutamic acid (secreted in response to chemoreceptor stimulation during hypoxia).49 In this study, we found levels of AMP to be at significantly lower levels in SIDS postmortem brain extracts as opposed to controls. Increased need for cyclic AMP triggered by hypoxia could explain the lower levels of AMP in infants with SIDS. The mechanism of the immune-enhancing activity of nucleic acids/nucleotides is still not clear; however, based on its role in energy production one could hypothesis, as for succinic acid, that energy production and mitochondrial malfunction could be directly related to SIDS.
Azelaic acid is a naturally occurring dicarboxylic acid that possesses antibacterial, keratolytic, anti-inflammatory, comedolytic and antioxidant activity.50 In this study, we found axelaic acid to be at lower concentrations in SIDS postmortem brain extracts as opposed to controls. The reason for this remains unclear, but it could be directly related to its antibacterial, anti-inflammatory and strong antioxidant capabilities, which have been reported to be linked with SIDS previously.51, 52 However, the link between infection and SIDS remains unclear.
An additional four ions of interest were identified based on their accurate mass; however, the online spectral libraries do not contain these compounds to enable us to confidently assign their identities other than putatively. The remaining six continue to be unidentified following a comprehensive search of the all the available libraries. To this end, we have made all the fragmentation patterns acquired for each of the individual ions available in Supplementary Material (Table 2). This is one of the inherent problems with untargeted metabolomics; although it remains the strongest method of identifying novel metabolomic biomarkers of interest, identification will continue to be difficult owing to limited spectral libraries.
Conclusion
This study applies a previously reported method for the metabolomic profiling of postmortem brain25, 26, 27 capable of differentiating SIDS from controls. To the best of our knowledge, this is the first reported study to apply HRMS to the biochemically profile postmortem human brain from infants who have died from SIDS, further emphasizing the uniqueness of this current report. Employing a panel of five and three ions (ESI+ and ESI− acquired data, respectively), we produced statistical models with high predictive accuracy underscoring the power of HRMS and metabolomics for studying SIDS. As with most untargeted metabolomic experiments, one of the major drawbacks is that we were not able to positively identify all the features of interest. However, we have identified 60% of the features in which we are interested and proposed some previously unreported mechanisms for their biochemical involvement in SIDS. Additional studies encompassing larger sample cohorts will facilitate the development of statistical models with increased predictive power while enabling our group to continue our ongoing efforts to identify the remaining biomarkers in our panels. Increasing the heterogeneity of the sample population by selecting from a variety of different sources will also allow us to validate our findings as reported herein. This metabolomics approach has the potential to contribute to the generation of new hypotheses related to SIDS, which at present is poorly understood. Further, it could help uncover timely innovative therapeutic targets to help treat those at greatest risk of dying as a direct result of this severe syndrome.
References
Centers for Disease Control and Prevention (CDC). Sudden infant death syndrome—United States, 1983-1994. MMWR Morb Mortal Wkly Rep 1996; 45 (40): 859–863.
Mathews TJ, Marian FM . Infant mortality statistics from the 2010 period linked birth/infant death data set. Natl Vital Stat Rep 2010; 58 (17): 1–31.
Creery D, Mikrogianakis A . Sudden infant death syndrome. Clin Evid 2005; 13: 434–443.
Filiano JJ, Kinney HC . A perspective on neuropathologic findings in victims of the sudden infant death syndrome: the triple-risk model. Biol Neonate 1994; 65 (3-4): 194–197.
Willinger M, Hoffman HJ, Hartford RB . Infant sleep position and risk for sudden infant death syndrome: report of meeting held January 13 and 14, 1994, National Institutes of Health, Bethesda, MD. Pediatrics 1994; 93 (5): 814–819.
Malloy MH, Freeman DH Jr . Sudden infant death syndrome among twins. Arch Pediatr Adolesc Med 1999; 153 (7): 736–740.
Wishart DS . Advances in metabolite identification. Bioanalysis 2011; 3 (15): 1769–1782.
Filipp FV . A gateway between omics data and systems biology. J Metabolomics Syst Biol 2013; 1 (1): 1.
Matsuda R, Bi C, Anguizola J, Sobansky M, Rodriguez E, Vargas Badilla J et al. Studies of metabolite-protein interactions: a review. J Chromatogr B Anal Technol Biomed Life Sci 2013; 966: 48–58.
Graham S, Holscher C, Green B . Metabolic signatures of human Alzheimer’s disease (AD): 1H NMR analysis of the polar metabolome of post-mortem brain tissue. Metabolomics 2013; 1–10.
Ramautar R, Berger R, van der Greef J, Hankemeier T . Human metabolomics: strategies to understand biology. Curr Opin Chem Biol 2013; 17 (5): 841–846.
Kinney HC . Neuropathology provides new insight in the pathogenesis of the sudden infant death syndrome. Acta Neuropathol 2009; 117 (3): 247–255.
Hunt CE, Brouillette RT . Sudden infant death syndrome: 1987 perspective. J Pediatr 1987; 110 (5): 669–678.
Sawaguchi T, Patricia F, Kadhim H, Groswasser J, Sottiaux M, Nishida H et al. Pathological data on apoptosis in the brainstem and physiological data on sleep apnea in SIDS victims. Early Hum Dev 2003; 75 (Suppl): S13–S20.
Machaalani R, Rodriguez M, Waters KA . Active caspase-3 in the sudden infant death syndrome (SIDS) brainstem. Acta Neuropathol 2007; 113 (5): 577–584.
Machaalani R, Waters KA . Neuronal cell death in the Sudden Infant Death Syndrome brainstem and associations with risk factors. Brain 2008; 131 (Pt 1): 218–228.
Lavezzi AM, Ottaviani G, Matturri L . Identification of neurons responding to hypoxia in sudden infant death syndrome. Pathol Int 2003; 53 (11): 769–774.
Paine SM, Jacques TS, Sebire NJ . Neuropathological features of unexplained sudden unexpected death in infancy: current evidence and controversies. Neuropathol Appl Neurobiol 2013; 40 (4): 364–384.
Kadhim H, Kahn A, Sebire G . Distinct cytokine profile in SIDS brain: a common denominator in a multifactorial syndrome? Neurology 2003; 61 (9): 1256–1259.
Matturri L, Biondo B, Mercurio P, Rossi L . Severe hypoplasia of medullary arcuate nucleus: quantitative analysis in sudden infant death syndrome. Acta Neuropathol 2000; 99 (4): 371–375.
Panigrahy A, Filiano J, Sleeper LA, Mandell F, Valdes-Dapena M, Krous HF et al. Decreased serotonergic receptor binding in rhombic lip-derived regions of the medulla oblongata in the sudden infant death syndrome. J Neuropathol Exp Neurol 2000; 59 (5): 377–384.
Kinney HC, Randall LL, Sleeper LA, Willinger M, Belliveau RA, Zec N et al. Serotonergic brainstem abnormalities in Northern Plains Indians with the sudden infant death syndrome. J Neuropathol Exp Neurol 2003; 62 (11): 1178–1191.
Paterson DS, Trachtenberg FL, Thompson EG, Belliveau RA, Beggs AH, Darnall R et al. Multiple serotonergic brainstem abnormalities in sudden infant death syndrome. JAMA 2006; 296 (17): 2124–2132.
Randall BB, Paterson DS, Haas EA, Broadbelt KG, Duncan JR, Mena OJ et al. Potential asphyxia and brainstem abnormalities in sudden and unexpected death in infants. Pediatrics 2013; 132 (6): e1616–e1625.
Graham SF, Chevallier OP, Roberts D, Holscher C, Elliott CT, Green BD . Investigation of the human brain metabolome to identify potential markers for early diagnosis and therapeutic targets of Alzheimer's disease. Anal Chem 2013; 85 (3): 1803–1811.
Graham SF, Chevallier OP, Kumar P, Türkoğlu O, Bahado-Singh RO . High resolution metabolomic analysis of ASD human brain uncovers novel biomarkers of disease. Metabolomics 2016; 12 (4): 1–10.
Graham SF, Kumar P, Bahado-Singh RO, Robinson A, Mann D, Green BD . Novel metabolite biomarkers of Huntington’s disease (HD) as detected by high resolution mass spectrometry. J Proteome Res 2016; 15: 1592–1601.
Gowda H, Ivanisevic J, Johnson CH, Kurczy ME, Benton HP, Rinehart D et al. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem 2014; 86 (14): 6931–6939.
Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y et al. HMDB 3.0—The Human Metabolome Database in. 2013 Nucleic Acids Res 2013; 41 (Database issue): D801–D807.
Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 2009; 37 (Database issue): D603–D610.
Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N et al. HMDB: the Human Metabolome Database. Nucleic Acids Res 2007; 35 (Database issue): D521–D526.
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 2010; 45 (7): 703–714.
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstrom C, Wold S . Multi- and Megavariate Data Analysis: Part I Basic principles and Applications. Umetrics: Umea, 2006.
Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS . MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res 2012; 40 (Web Server issue): W127–W133.
Xia J, Psychogios N, Young N, Wishart DS . MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009; 37 (Web Server issue): W652–W660.
Xia J, Sinelnikov IV, Han B, Wishart DS . MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res 2015; 43 (W1): W251–W257.
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007; 3 (3): 211–221.
Altman DG, Bland JM . Absence of evidence is not evidence of absence BMJ 1995; 311: 485 .
Paul BD, Snyder SH . The unusual amino acid L-ergothioneine is a physiologic cytoprotectant. Cell Death Differ 2010; 17 (7): 1134–1140.
Aruoma OI, Spencer JPE, Mahmood N . Protection against oxidative damage and cell death by the natural antioxidant ergothioneine. Food Chem Toxicol 1999; 37 (11): 1043–1053.
Tamazian G, Chang JH, Knyazev S, Stepanov E, Kim KJ, Porozov Y . Modeling conformational redox-switch modulation of human succinic semialdehyde dehydrogenase. Proteins 2015; 83 (12): 2217–2229.
Pearl PL, Gibson KM, Acosta MT, Vezina LG, Theodore WH, Rogawski MA et al. Clinical spectrum of succinic semialdehyde dehydrogenase deficiency. Neurology 2003; 60 (9): 1413–1417.
Ting Wong CG, Bottiglieri T, Snead OC . GABA, γ-hydroxybutyric acid, and neurological disease. Ann Neurol 2003; 54 (S6): S3–S12.
Gibson KM, Christensen E, Jakobs C, Fowler B, Clarke MA, Hammersen G et al. The clinical phenotype of succinic semialdehyde dehydrogenase deficiency (4-hydroxybutyric aciduria): case reports of 23 new patients. Pediatrics 1997; 99 (4): 567–574.
Broadbelt KG, Paterson DS, Belliveau RA, Trachtenberg FL, Haas EA, Stanley C et al. Decreased GABA(A) receptor binding in the medullary serotonergic system in the sudden infant death syndrome. J Neuropathol Exp Neurol 2011; 70 (9): 799–810.
Hess JR, Greenberg NA . The role of nucleotides in the immune and gastrointestinal systems: potential clinical applications. Nutr Clin Pract 2012; 27 (2): 281–294.
Pizzini RP, Kumar S, Kulkarni AD, Rudolph FB, Van Buren CT . Dietary nucleotides reverse malnutrition and starvation-induced immunosuppression. Arch Surg 1990; 125 (1): 86–89 discussion 90.
Li K, Anderson KJ, Peng Q, Noble A, Lu B, Kelly AP et al. Cyclic AMP plays a critical role in C3a-receptor–mediated regulation of dendritic cells in antigen uptake and T-cell stimulation. Blood 2008; 112 (13): 5084–5094.
Hoop B, Masjedi MR, Shih VE, Kazemi H . Brain glutamate metabolism during hypoxia and peripheral chemodenervation. J Appl Physiol 1990; 69 (1): 147–154.
Graupe K, Cunliffe WJ, Gollnick HP, Zaumseil RP . Efficacy and safety of topical azelaic acid (20 percent cream): an overview of results from European clinical trials and experimental reports. Cutis 1996; 57 (1 Suppl): 20–35.
Blood-Siegfried J . The role of infection and inflammation in sudden infant death syndrome. Immunopharmacol Immunotoxicol 2009; 31 (4): 516–523.
Prandota J . Possible pathomechanisms of sudden infant death syndrome: key role of chronic hypoxia, infection/inflammation states, cytokine irregularities, and metabolic trauma in genetically predisposed infants. Am J Ther 2004; 11 (6): 517–546.
Acknowledgements
We thank the University of Maryland Brain and Tissue Bank, which is a Brain and Tissue Repository of the NIH NeuroBioBank, for graciously providing the tissue samples for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Journal of Perinatology website
Supplementary information
Rights and permissions
About this article
Cite this article
Graham, S., Chevallier, O., Kumar, P. et al. Metabolomic profiling of brain from infants who died from Sudden Infant Death Syndrome reveals novel predictive biomarkers. J Perinatol 37, 91–97 (2017). https://doi.org/10.1038/jp.2016.139
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jp.2016.139
- Springer Nature America, Inc.