Abstract
Background/Objectives:
The use of bisphenol A (BPA) in consumer products and food packaging has been associated under certain conditions with a risk of negative health outcomes. This prompted its removal from many products and replacement with structural analogs. Bisphenol S (BPS) is one such analog, but its metabolic effects have not been fully characterized. The objective of our study was to determine whether BPS functions similarly to BPA at inducing adipogenesis.
Methods:
Murine 3T3-L1 preadipocytes were used to evaluate and compare the adipogenic potential of BPS to BPA. Cells were treated with 0.01–50 μM BPS or 0.01–50 μM BPA and adipogenic effects were measured. Further, their ability to activate peroxisome proliferator-activated receptor gamma (PPARγ), an adipogenic transcription factor, was also determined.
Results:
Our results indicate that treatment of 3T3-L1 cells with BPS induced lipid accumulation and increased mRNA and protein expression of key adipogenic markers (1–50 μM; P<0.05). BPS treatment resulted in a higher expression of adipogenic markers as well as greater lipid accumulation when compared with BPA treatment. We showed that BPS can upregulate lipoprotein lipase, adipocyte protein 2, PPARγ, perilipin, adipsin and CCAAT/enhancer-binding protein alpha mRNA expression levels. Furthermore, using transcriptional assays, we showed that BPS and BPA can modestly activate PPARγ using a PPRE (PPARγ response element)-dependent luciferase construct by 1.5-fold (P<0.05). However, BPS but not BPA was able to competitively inhibit rosiglitazone (ROSI)-activated PPARγ, suggesting that BPS interacts with PPARγ distinctly from BPA. Co-treatment of cells with the selective PPARγ antagonist GW9662 inhibits BPS-, BPA-, ROSI- but not dexamethasone-dependent adipogenic differentiation.
Conclusions:
Both BPA and BPS can enhance 3T3-L1 adipocyte differentiation in a dose-dependent manner and require PPARγ to induce adipogenesis. Through direct comparison, we show that BPS is a more potent adipogen than BPA.
Similar content being viewed by others
Introduction
Bisphenol A (BPA) is used in many consumer products including: polycarbonate plastics, epoxy lining of food packaging, epoxy resins in dental sealants, and thermal paper receipts. In 2011, it was estimated that >5.5 million metric tons of BPA was produced.1 Epidemiological studies have shown that BPA is detectable in the nanogram range in both urine and serum samples of adults, children and infants, highlighting its ubiquitous nature and potential for continuous exposure.2, 3, 4 BPA exposure has been associated with obesity and metabolically linked diseases.5, 6, 7 Human studies have correlated BPA levels in urine and serum with obesity, cardiovascular disease and type 2 diabetes.5, 8, 9 This idea that environmental chemicals such as BPA could promote and induce adipogenesis has been supported by both in vitro and in vivo studies.10, 11 Its potential link to obesity and other human diseases has led scientists, regulators and the general public to raise concerns about the safety of BPA, prompting manufacturers to replace BPA with other structural analogs. One such analog is bisphenol S (BPS). BPS is now used in many industrial applications and in products marketed as BPA-free.3, 12, 13 In humans, BPS has been detected in urine at concentrations and frequencies similar to BPA.14, 15 Because of the structural similarity between BPS and BPA, it is unclear whether BPS is inert or at least less efficient at inducing various toxic end points previously associated with BPA exposure.
Understanding the mechanisms and potential role of environmental chemicals in adipose tissue formation in vitro is vital to evaluating their potential link to metabolic outcomes, including obesity. The murine 3T3-L1 preadipocyte cell line is currently accepted as an appropriate in vitro model to study adipocyte differentiation.16 The process of adipogenesis is tightly regulated by a network of transcription factors that coordinate the expression of genes leading to adipocyte maturation.17, 18, 19 Central to this pathway are two transcription factors; PPARγ (peroxisome proliferator-activated receptor gamma) and C/EBPα (C/CAAT enhancer-binding protein alpha).16 When PPARγ is knocked out, adipogenesis is abolished giving rise to PPARγ being considered the master regulator of adipogenesis.19 To date, in vitro studies have shown that BPA can induce adipocyte differentiation of 3T3-L1 preadipocytes in part owing to enhanced glucocorticoid receptor (GR)-mediated activity.10, 20, 21 The effects of BPS on adipogenesis and its mechanism of action have not yet been fully elucidated.
To determine whether BPS can induce adipogenesis and to determine whether cells treated with BPS achieve levels of differentiation comparable to BPA-treated cells, 3T3-L1 cells were exposed to both compounds at equivalent concentrations. Key transcription factors as well as their downstream targets were evaluated in both a dose- and time-dependent manner. Furthermore, to evaluate the role of PPARγ in mediating these effects, both transcriptional and differentiation assays were performed using the selective PPARγ antagonist GW9662. We are the first to show that both BPA and BPS weakly activate PPARγ and require PPARγ for their adipogenic potential.
A detailed understanding of the processes governing adipose tissue formation will be instrumental in combating the obesity epidemic. Much progress has been made in the past two decades in defining transcriptional events controlling the differentiation of mesenchymal stem cells into adipocytes. A complex network of transcription factors and cell-cycle regulators, in concert with specific transcriptional coactivators and corepressors, respond to extracellular stimuli to activate or repress adipocyte differentiation. This review summarizes advances in this field, which constitute a framework for potential antiobesity strategies.
Materials and methods
Murine adipocyte differentiation
3T3-L1 mouse embryonic fibroblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM) 1 g l−1 glucose (Hyclone, Mississauga, ON, Canada) containing 10% bovine calf serum (ATCC, Manassas, VA, USA) and grown to 70% confluence. Cells were then plated in six-well dishes using DMEM 1 g l−1 glucose supplemented with 10% fetal calf serum (Wisent, Montreal, QC, Canada) and 1% penicillin/streptomycin (Life Technologies, Burlington, ON, Canada) and left to reach confluence. Two days postconfluence (Day 0), cells were induced to differentiate using the cocktail consisting of 500 μM of the cAMP enhancer IBMX (3-isobutyl-1-methylxanthine; Sigma-Aldrich, Oakville, ON, Canada) and 100 nM of insulin (Roche Diagnostics, Laval, QC, Canada; MI), plus varying concentrations of BPS (ethanol, 0.01–50 μM) or BPA (ethanol, 0.01–25 μM). For positive control experiments, 3T3-L1 cells were supplemented with 250 nM of dexamethasone (DEX; Sigma-Aldrich) or 5 μM of rosiglitazone (ROSI; Sigma-Aldrich) along with IBMX and insulin (MID, MIR). Two days after differentiation was initiated, media was replaced to contain 100 nM of insulin and the test chemical or positive controls. The media was subsequently replaced every 2 days until the end of the experiment (2, 4, 6 or 8 days). For the PPARγ antagonist study, 5 μM of the irreversible antagonist GW9662 (Sigma-Aldrich) was added to the differentiation media and replaced daily owing to its short half-life.22
RNA extraction and reverse transcriptase-PCR
Total RNA was isolated from differentiating cells treated with varying concentrations of BPS or BPA as well as in the presence of the inhibitor GW9662 using the Qiagen RNeasy Kit (Qiagen, Toronto, ON, Canada). Five hundred nanograms of RNA was then reverse-transcribed using the iScript Reverse Transcription Kit (Bio-Rad, Mississauga, ON, Canada) following the manufacturer’s recommendations. Primers used to amplify markers of adipocyte differentiation are as follows: adipocyte protein 2 (aP2, also known as Fabp4) forward 5′-GGAAGCTTGTCTCCAGTGAA-3′ and reverse 5′-GCGGTGATTTCATCGAATTC-3′; Pparγ 5′-GCCTGCGGAAGCCCTTTGGT-3′ and reverse 5′-GCAGTTCCAGGGCCTGCAGC-3′; perilipin (Plin) forward 5′-TTGGGGATGGCCAAAGAGAC-3′ and reverse 5′-CTCACAAGGCTTGGTTTGGC-3′; lipoprotein lipase (Lpl) 5′-CAGGATGTGGCCCGGTTTAT-3′ and reverse 5′-CGGGGCTTCTGCATACTCAA-3′; adipsin (Adsn) forward 5′-CCTGAACCCTACAAGCGATG-3′ and reverse 5′-CAACGAGGCATTCTGGGATAG-3′; and CCAAT/enhancer-binding protein alpha forward 5′-TGCGCAAGAGCCGAGATAAA-3′ and reverse 5′-CCTTGACCAAGGAGCTCTCA-3′. All genes were amplified using Bio-Rad SsoFast SYBR Green 2X mix, normalized to β-actin levels and analyzed using the comparative CT method.
Lipid staining and quantification
Murine 3T3-L1 preadipocytes were differentiated as described above for 8 days with 250 nM DEX, 5 μM ROSI or increasing amount of BPS (0.01–50 μM) or with 25 μM BPA with media replenished every 2 days. Differentiated cells were then fixed using 4% paraformaldehyde and stained with Nile Red (stains cytoplasmic lipid droplets) and DAPI (4,6-diamidino-2-phenylindole; stains cell nuclei) as previously described.23 Nile Red fluorescence was quantified at 485/528 nm (excitation/emission) and normalized to DAPI staining measured at 360/460 nm (excitation/emission). All data were then normalized to MI control (data reported as fold change over MI). Fluorescence was measured using the Synergy 2 Microplate Reader (BioTek Instruments Inc., Winooski, VT, USA). Images of Nile Red and DAPI staining were taken using the Leica TCD SP8 confocal microscope (Leica Microsystems, Toronto, ON, Canada) at × 63 magnification. Images are representative of three independent experiments.
Western blotting analysis
For protein detection, cell extracts were prepared after 6 days in the presence of the differentiation media with increasing amounts of BPS (0.01–50 μM) or with the inhibitor GW9662. Whole-cell extracts were prepared using RIPA buffer in the presence of protease inhibitors (Roche Diagnostics). Twenty micrograms of total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. Membranes were incubated with anti-aP2 antibody (AF3150; R&D Systems, Minneapolis, MN, USA) and anti-LPL (AF7197; R&D Systems), after detection membranes were stripped and probed with anti-β-actin antibody (13E5; Cell Signalling, Danvers, MA, USA). Blots were then probed using the appropriate horseradish peroxidase-conjugated secondary antibodies and were visualized using Clarity Western ECL Substrate (Bio-Rad). Bands were detected using the ChemiDoc System (Bio-Rad) and then quantified using the Image Lab software (Bio-Rad) and normalized to β-actin levels.
Reporter gene assay
COS-7 cells were seeded in phenol red-free DMEM (Wisent) supplemented with 5% dextran-coated charcoal-stripped serum (Sigma-Aldrich). Twenty-four hours after plating, cells were transfected with plasmid DNA using Fugene HD (Promega, Madison, WI, USA) according to the manufacturer’s recommendations. For the PPARγ transcriptional assays, cells were transfected with 10 ng of pRL-CMV (renilla; internal control), 25 ng of pcDNA mPPARγ, 25 ng of pCMV6 mRXR and 125 ng of 3 × PPARγ response element (PPRE)-luciferase (PPRE-luc). All plasmids were generous gifts from Dr Jae Bum Kim.24 Six hours after transfection, cells were treated with vehicle control and the indicated concentrations of BPS or BPA, as well as increasing amount of ROSI (20 nM, 200 nM and 5 μM) in the presence of increasing amounts of BPS or BPA (1–50 μM). Twenty-four hours after treatment, cells were lysed using 1 × Passive Lysis Buffer (Promega). Luciferase activity was quantified with the Dual Luciferase Assay Kit (Promega) using the Glomax96 Luminometer (Promega). Luciferase activity was normalized to renilla levels and to vehicle control (dimethyl sulfoxide).
Statistical analysis
All data were presented as means and s.e.m. All analyses were carried out by a one-way analysis of variance followed by Tukey’s multiple comparison tests or using the GraphPad Prism Software (GraphPad Software, San Diego, CA, USA).
Results
Dose-dependent comparison of BPS- and BPA-induced gene expression levels of adipogenic markers in the mature adipocyte
We set out to investigate whether BPS, a BPA analog currently replacing BPA in many consumer products, could induce differentiation of murine 3T3-L1 preadipocytes equivalent to what was previously reported for BPA.20 Genes under investigation include transcription factors important for adipogenesis as well as genes expressed in the mature adipocyte. Our results demonstrate that both BPS and BPA increased the expression of all genes examined; however, compared with BPA, BPS treatment resulted in significantly higher levels of gene expression at certain concentrations (Figures 1a–f). Treatment with 0.01–1 μM of BPS or BPA did not cause significant increases in gene expression but treatment with 10 μM of BPS or BPA were able to induce equivalent expression levels of ap2, Lpl and Adsn, indicating that at lower concentrations both chemicals behave similarly (Figures 1a). However, treatment with 25 μM of BPS was significantly better than 25μM of BPA at inducing the expression of all genes examined (Figures 1a–f).
Dose-dependent comparison of BPS- and BPA-induced gene expression levels of adipogenic markers. mRNA expression levels were determined in murine 3T3-L1 preadipocytes 6 days posttreatment with the differentiation cocktail consisting of IBMX, insulin and increasing amounts of either BPS or BPA (0.01–25 μM) or vehicle control (MI). Six days after treatment, aP2 (a), Pparγ (b), Plin (c), Lpl (d), Adsn (e) and Cebpα (f) mRNA levels were determined and normalized to β-actin levels and are expressed as a fold change relative vehicle control. Data represent the mean±s.e.m. (n=4). *P<0.05 relative to vehicle control, #P<0.05 relative to dose-matched BPA treatment using a one-way analysis of variance followed by Tukey’s post-hoc analysis.
Evaluation of temporal differences in BPS- and BPA-mediated gene expression profiles
To further characterize the enhanced ability of 25 μM of BPS at inducing adipogenesis of murine 3T3-L1 cells compared with 25 μM of BPA, we completed a time-course experiment looking at markers of adipogenesis after 2, 4 and 6 days of treatment in the presence of the differentiation cocktail. Our results indicate that there were no significant differences between BPS and BPA after 2 days of treatment except that BPA was significantly better than BPS at increasing perilipin levels (2.5-fold vs. 3.8-fold; Figure 2c). However, this was not maintained over time as Plin expression was significantly higher for BPS at both days 4 and 6. Furthermore, Pparγ levels were upregulated after BPA treatment at day 2 that was not seen for BPS but that comparable expression levels were achieved by day 6 (Figure 2b). After 4 days in the presence of the differentiation cocktail, 25 μM of BPS was significantly better at inducing the expression of aP2 and Plin and its superiority was maintained until day 6 (Figures 2a and c). It is also important to note that, at day 4, BPS but not BPA was able to upregulate Lpl and Cebpα (Figures 2d and f), suggesting that BPS is able to promote adipogenesis at earlier time points and may account for the enhanced gene expression in mature adipocytes.
Evaluation of temporal differences in BPS- and BPA-mediated gene expression profiles. mRNA expression levels were determined in murine 3T3-L1 preadipocytes 2, 4 and 6 days posttreatment with the differentiation cocktail consisting of IBMX, insulin and 25μM BPS, 25μM BPA or vehicle control (MI). After the indicated time points, RNA was extracted and reverse transcribed, and we measured the levels of aP2 (a), Pparγ (b), Plin (c), Lpl (d), Adsn (e) and Cebpα (f), which were normalized to β-actin levels and relative to time-matched vehicle control. Data represent the mean±s.e.m. (n=4). *P<0.05 relative to time-matched vehicle control, #P<0.05 relative to time-matched BPS treatment using a one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc analysis. Murine 3T3-L1 preadipocytes were treated with control (MI), 25 μM BPS or 25 μM BPA for 8 days and lipid accumulation was visualized using Nile Red staining (g) and then quantified (h). *P<0.05 relative to vehicle control, #P<0.05 relative to BPS treatment using a one-way ANOVA followed by Tukey’s post-hoc analysis. Lipid accumulation was normalized to DAPI staining and relative to vehicle-treated cells. Data represent mean±s.e.m. of three independent experiments performed in triplicate.
To determine whether the enhanced gene expression achieved after BPS treatment led to greater lipid accumulation in the mature adipocyte, Nile Red lipid staining was performed in cells treated with 25 μM of BPS or 25 μM of BPA. As expected, lipid accumulation was significantly higher in BPS-treated cells when compared with BPA-treated wells (Figures 2g and h)
BPS-induced lipid accumulation and increases in protein levels of adipogenic markers
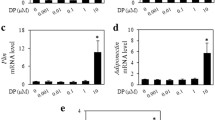
In order to quantify the ability of BPS to induce differentiation of 3T3-L1 preadipocytes, we visualized lipid accumulation on day 8 by Nile Red lipid staining after 0.01–50 μM treatment (Figures 3a and b). Although lipid accumulation was slightly increased at concentrations as low as 10 nM, statistically significant increases were not seen until after 10 μM treatment (Figure 3b). The positive controls consisting of 250 nM DEX or 5 μM ROSI treatment induced comparable levels of lipid accumulation, suggesting that either direct GR activation or direct PPARγ activation are equivalent at promoting lipid droplet formation despite having different mechanisms of action (Figure 3b).
BPS-induced lipid accumulation and increases in the protein expression of adipogenic markers. Murine 3T3-L1 preadipocytes were treated with vehicle (MI), the positive controls 250 nM DEX or 5 μM ROSI and increasing amounts of BPS (0.01–50 μM) for 8 days and lipid accumulation was visualized using Nile Red staining (a) and then quantified (b). Lipid accumulation was normalized to DAPI staining and relative to vehicle-treated cells. Data represent mean±s.e.m. for three independent experiments performed in triplicate. Statistical significance *P<0.05 was determined relative to vehicle control (MI) using a one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc analysis. Images were visualized using the Leica TCD SP8 confocal microscope at × 63 magnification and are representative of three independent experiments. (c) Immunoblot showing the ability of BPS (0.01–50 μM) to induce the protein expression of LPL and aP2 following 6 days of treatment. β-Actin was used as a loading control. (d) Quantification of aP2 and LPL protein levels (n=5) using the Image Lab software and β-actin as the loading control (Bio-Rad). Asterisks denoted protein levels significantly different (P<0.05) than vehicletreated cells (MI) using a one-way ANOVA. aDenotes statistical significance using a one-way ANOVA with the highest doses removed during analysis (P<0.05). mRNA expression of aP2 (e) and Lpl (f) after 2, 4 and 6 days of treatment using low doses of BPS (0.01–1 μM). Data represent the mean±s.e.m. (n=4). *P<0.05 relative to vehicle control, using a one-way ANOVA followed by Tukey’s post-hoc analysis.
We then determined the extent of differentiation achieved after BPS treatment by measuring the protein levels of select markers of adipogenesis after 6 days of treatment. There was a dose-dependent increase in LPL and aP2 protein levels after BPS treatment (Figure 3c). Despite seeing increases in protein levels visually at 10 nM–1 μM doses, we did not see significant increases in protein levels below 10 μM for aP2 and 25 μM for LPL using a one-way analysis of variance with all concentrations included (Figure 3d). Furthermore, looking at the temporal changes in mRNA expression of aP2 and Lpl at the low-dose treatment (10 nM–1 μM) indicate that aP2 levels were upregulated at days 4 and 6 of differentiation while Lpl levels were significantly upregulated on day 2 (Figures 3e and f). These changes in mRNA levels may account for the small increases in protein expression observed at the low doses examined. Taken together, our data suggest that BPS can induce lipid accumulation and mRNA and protein expression of key markers of adipogenesis in a dose-dependent manner.
Mechanistic insight into BPS- and BPA-dependent activation of PPARγ
It has been previously reported that BPA and BPS can induce estrogen response element-dependent luciferase activity. However, having estrogenic potential does not lead to enhanced adipogenesis.25, 26 We and others have shown that estrogen treatment alone does not induce differentiation of 3T3-LI preadipocytes.26, 27 Furthermore, all of our experimental procedures were carried out in the presence of non-stripped serum, which contains estrogen, indicating that any differentiation achieved was most likely owing to other pathways being activated and not estrogen receptor (ER) mediated. It has also been reported that BPA may have intrinsic GR activation capability determined by its ability to activate a GRE-dependent luciferase system.21 However, similar activity was not seen for BPS using both a GRE-luciferase and MMTV-luciferase reporter plasmids (Boucher et al., 2015, submitted).
In order to determine other possible mechanisms by which BPS and BPA treatment induces adipogenesis of murine 3T3-L1, we investigated whether BPS and BPA can activate PPARγ in a PPRE-dependent luciferase assay system. Activation of PPARγ has a critical function in adipocyte differentiation.18, 19 We show that, in COS-7 cells transfected with mPPARγ, mRXR, and a 3 × PPRE-luciferase reporter plasmid BPS treatment caused a significant increase of reporter gene activity at 25 and 50 μM (Figure 4a). These effects were approximately 10-fold lower than that achieved after 5 μM treatment with the full agonist ROSI (1.5-fold vs 12-fold) (Figure 4b). We also completed parallel experiments in the presence of increasing amounts of BPA that displayed similar increases in PPARγ activity, suggesting that both BPS and BPA can weakly activate PPARγ (Figure 4a).
Mechanistic insight into BPS- and BPA-dependent activation of PPARγ. (a) COS-7 cells were transfected as described in the Materials and methods section with pcDNA-mPPARγ, pcDNA mRXR, 3 × PPRE-luciferase and pCMV-RL and treated with increasing amounts of BPS or BPA (0.1-50 μM). Twenty-four hours after treatment, reporter gene activity was determined. Data represent the mean±s.e.m. of three independent experiments. Significantly different (*P<0.05) reporter gene activity was determined relative to transfected vehicle-treated cells using a one-way analysis of variance (ANOVA) followed by Tukeys post-hoc analysis. (b) COS-7 cells were transfected as described above and then treated with increasing amounts of ROSI (20 nM, 200 nM and 5 μM) as well as in the presence of increasing amounts of BPS (1, 25 and 50 μM). Similar experiments were performed in the presence of increasing amounts of (c) BPA (1, 25 and 50 μM). Data represent the mean±s.e.m. of three independent experiments. Significantly different (*P<0.05) reporter gene activity was determined relative to ROSI-treated cells using a one-way ANOVA followed by Tukeys post-hoc analysis.
To further characterize the ability of BPS and BPA to activate PPARγ, we measured luciferase activity in the presence of increasing amounts of the full PPARγ agonist ROSI. Our data confirms that there is a dose-dependent increase in ROSI-mediated activation of PPARγ and that this activity was inhibited by BPS co-treatment (Figure 4b). This data suggest that BPS was able to bind to PPARγ and displace ROSI and may function as a partial agonist or interact with the receptor similarly to ROSI. Furthermore, BPA did not inhibit ROSI-mediated PPRE-luciferase activity, suggesting that its interaction with PPARγ is not similar to ROSI (Figure 4c). Taken together, our findings indicate that although BPS and BPA activate a PPARγ-dependent luciferase their interaction with the nuclear receptor may be different.
BPS- and BPA-mediated adipogenesis requires PPARγ activation
To determine the importance of PPARγ in BPS- and BPA-dependent adipogenesis, we completed differentiation in the presence of the selective PPARγ antagonist GW9662. We compared the differentiation achieved after treatment with 250 nM DEX, 5 μM ROSI, 25 μM BPS or 25 μM BPA in the presence or absence of 5 μM of the PPARγ antagonist GW9662. We measured the mRNA and protein levels of aP2 and LPL after 6 days of treatment with the differentiation cocktail. As expected, treatment with DEX caused a significant increase in aP2 and Lpl mRNA levels that were unaffected by co-treatment with GW9662, suggesting that direct PPARγ activation is not involved in meditating DEX-dependent adipogenesis (Figures 5a, b and i). Similar experiments could not be performed with the selective GR antagonist RU486 as it is a potent inducer of differentiation in murine 3T3-L1 (data not shown28, 29) to confirm the importance of direct GR activation in DEX-mediated differentiation. Treatment of ROSI enhanced the differentiation of murine 3T3-L1 cells that was significantly inhibited by co-treatment with 5 μM GW9662, leading to approximately 50% reduction in the mRNA and protein levels of aP2 and LPL (Figures 5c). These results reinforce the importance of PPARγ in ROSI-mediated differentiation as it is known to be a potent activator of PPARγ. Interestingly, similar results were observed after BPS and BPA co-treatment with GW9662 (Figures 5e–i). A significant decrease in mRNA expression and protein levels were achieved, confirming the role of PPARγ in BPS- and BPA-mediated adipogenesis and suggests that both chemicals may be able to directly activate the receptor (Figures 5e–i). Taken together, our data suggest that PPARγ activation has an important role in mediating BPS- and BPA-dependent differentiation similar to the full PPARγ agonist ROSI but not the GR agonist DEX.
BPS- and BPA-mediated adipogenesis requires PPARγ. 3T3-L1 cells were treated with ethanol (MI), 5μM GW9662 alone, 250 nM DEX, 5 μM ROSI, 25 μM BPS or 25 μM BPA as well as co-treatment with the PPARγ inhibitor GW9662 for 6 days. mRNA expression levels for aP2 (a, c, e, g) and Lpl (b, d, f, h) were determined after DEX, ROSI, BPS and BPA treatment in the presence or absence of the inhibitor GW9662. Data represent the mean±s.e.m. of three independent experiments. Significantly different (*P<0.05) gene expression was determined relative to MI-control cells as well as to relative to chemical-matched cells (#P<0.05) using a one-way analysis of variance followed by Tukeys post-hoc analysis. (i) Representative immunoblot showing the effects of the inhibitor GW9662 on DEX-, ROSI- and BPS- and BPA-mediated expression of adipogenic markers.
Discussion
Concerns raised by scientists, regulators and the general public over the endocrine-disruptive effects of BPA have prompted the industry to seek alternatives to BPA, which include structural analogs of BPA. Our study focused on one such analog: BPS and understanding its metabolic effects in vitro compared with BPA. To our knowledge, we are the first to show that BPS is more potent than BPA at inducing adipogenesis and that PPARγ activity is required for these effects. Previous studies have tested the ability of BPS to induce adipogenesis using the murine 3T3-L1 cell model but their experimental procedures involved treatment with DEX, limiting the impact of their results.10, 30 In contrast, our study was conducted in the absence of DEX or ROSI, suggesting that BPS can mimic one of these chemicals to promote adipogenesis. Using the selective PPARγ antagonist GW9662, we show that PPARγ is required for BPS and BPA adipogenic potential. However, their interaction with PPARγ is distinct giving a possible mechanism for their differential ability to promote differentiation.
The molecular mechanisms controlling adipogenesis in the 3T3-L1 cells involves two well-defined phases: clonal expansion and the timely expression of key adipogenic transcription factors.16 The expression of the transcription factor PPARγ is sufficient and required for adipocyte formation and maturation.19, 31 The large and promiscuous ligand-binding pocket of PPARγ has led to the identification of numerous compounds that have unique interactions within the ligand-binding domain that may promote and facilitate adipogenesis.32 Our transcriptional and differentiation assays suggest that the interactions of BPS and BPA with PPARγ were unique and sufficient to promote differentiation. The ability of BPS but not BPA to competitively displace ROSI from the ligand-binding pocket of PPARγ indicates that BPS interaction sites are similar to ROSI. ROSI directly interacts with the ligand-binding domain and stabilizes helix H12 and creates five hydrogen bonds with PPARγ unlike weak or partial agonists, which tend to interact with H3 and the β-sheet S1/S2.33, 34 Furthermore, the inability of BPA to displace ROSI may account for why we see lower mRNA expression levels of adipogenic transcription factors and their downstream effectors. It may be that BPA-bound receptor conformation may not facilitate the binding of co-activators similar to the receptor conformation achieved after BPS binding.
It has been previously shown that halogenated analogs of BPA can directly interact with PPARγ.35 Rui et al.35 have shown through functional and structural studies that the halogenated BPA analogs TBBPA (tetrabromobisphenol A) and TCBPA (tetrachlorobisphenol A) act as partial agonists of PPARγ. Our data for BPS indicate that its interactions with PPARγ are similar to the halogenated BPA compounds. We were able to show competitive inhibition of ROSI-bound PPARγ similar to TBBPA and TCBPA and both the sulfur and oxygen atoms of BPS might engage in more hydrogen bonds than BPA, which lacks the halogen functional groups. The inability of BPA co-treatment to inhibit ROSI-mediated PPARγ transcriptional activity may be due to its weak and distinct interaction with the ligand-binding domain. The rather weak PPARγ activation achieved after BPA and BPS treatment can be explained by their smaller size and fewer direct atomic contacts with the transcription factor unlike the full agonist ROSI, which is a much bulkier ligand with five hydrogen bonds. Furthermore, the ability of the selective PPARγ antagonist GW9662 to inhibit BPA-, BPS- and ROSI-mediated differentiation of the 3T3-L1 preadipocytes supports our findings that both chemicals may be working through PPARγ to induce adipogenesis. This is in contrast to DEX-mediated differentiation, which was not affected by direct PPARγ inhibition. We postulate that the GR-mediated upregulation of CEBPδ and CEBPα, whose transcriptional activity has been shown to overlap with many PPARγ targets, is sufficient to overcome the direct inhibition caused by GW9662 treatment.16, 36
BPS binding to other nuclear receptors involved in adipogenesis may have a role in the enhanced adipocyte differentiation achieved after BPS treatment when compared with BPA. ER alpha (ERα), which is expressed in murine and human adipocytes, is activated by both BPS and BPA and has been previously shown to be involved in adipogenesis by increasing adipocyte number.37, 38, 39 However, the ability of BPS when compared with BPA to bind and activate ERα are in the same order of magnitude, demonstrating that ERα is most likely not involved in the enhanced differentiation we observe.13, 40 Recently, the membrane-bound ER, GPR30 (G-protein coupled receptor 30), has been implicated in obesity. GPR30 knockout mice exhibit increased adiposity highlighting its role in metabolic regulation in vivo.41, 42 However, several studies have shown that the GPR30 expression in mouse adipose tissue is quite low,43, 44, 45 which implies that in our murine cell model this receptor may not be responsible for the differential BPS activation. Furthermore, the ability of BPS to bind to GPR30 has not been evaluated yet. In a recent study that evaluated the role of the estrogen-related receptor alpha (ERRα) in adipogenesis, they determined ERRα as a novel adipogenic marker involved in the expression of genes involved in differentiation through its interactions with the coactivator PGC1α (PPARγ coactivator 1α).46 Currently, there are no studies showing that BPS and BPA can bind and activate ERRα directly. However, using human peripheral blood isolated from adult male men, there was a positive association between high blood BPA concentrations and high expression of ERRα but, to date, no studies have evaluated BPS levels and ERRα expression.47 Therefore, the involvement of ERRα expression or binding in BPS-mediated adipogenesis cannot be ruled out.
The human health effects of BPA have been intensely studied.48 Body mass index and obesity are two of the most studied end points assessed for human BPA exposure. However, a causal relationship between BPA exposure and obesity cannot be drawn owing to the cross-sectional nature of these studies. Using in vitro cell models, we and others have shown that BPA can promote adipogenesis of both murine and human preadipocytes.20, 21, 27 This and the overwhelming evidence regarding the endocrine-disrupting capabilities of BPA have led to replacement chemicals, such as BPS. However, the in vitro data we have generated suggest that such replacement chemicals may not be safer when evaluating metabolic outcomes. These results add to the increasing evidence showing that BPA replacements, such as BPS, may have adverse human health effects similar to the chemical they are replacing. This study suggests that replacement chemicals need to be evaluated for potential risks before they are incorporated into consumer products. Further, end points such as obesogenicity may need to be taken into account by policy makers in the future. Nevertheless, epidemiological studies assessing the impact of BPS are warranted. Epidemiological studies assessing the impact of BPS are warranted.
In this report, we directly compared the metabolic effects of BPA and BPS in vitro. We show that BPS is better than BPA in both a dose- and time-dependent manner at promoting adipocyte differentiation and lipid accumulation. Furthermore, we are the first to show that both BPA and BPS activate PPARγ albeit distinctly and that their ability to induce adipogenesis was inhibited by a selective PPARγ antagonist, providing evidence that PPARγ is required for mediating their effects.
References
Rubin B, Bisphenol A . An endocrine disruptor with widespread exposure and multiple effects. J Steroid Biochem Mol Biol 2011; 127: 27–34.
Calafat AM, Weuve J, Ye X, Jia LT, Hu H, Ringer S et al. Exposure to bisphenol A and other phenols in neonatal intensive care unit premature infants. Environ Health Perspect 2009; 117: 639–644.
Liao C, Liu F, Guo Y, Moon HB, Nakata H, Wu Q et al. Occurrence of eight bisphenol analogues in indoor dust from the United States and several Asian countries: implications for human exposure. Environ Sci Technol 2012; 46: 9138–9145.
Vandenberg LN, Chahoud I, Padmanabhan V, Paumgartten FJ, Schoenfelder G . Biomonitoring studies should be used by regulatory agencies to assess human exposure levels and safety of bisphenol A. Environ Health Perspect 2010; 118: 1051–1054.
Takeuchi T, Tsutsumi O, Ikezuki Y, Takai Y, Taketani Y . Positive relationship between androgen and the endocrine disruptor, bisphenol A, in normal women and women with ovarian dysfunction. Endocr J 2004; 51: 165–169.
Ranciere F, Lyons JG, Loh VH, Botton J, Galloway T, Wang T et al. Bisphenol A and the risk of cardiometabolic disorders: a systematic review with meta-analysis of the epidemiological evidence. Environ Health 2015; 14: 46.
Watkins DJ, Peterson KE, Ferguson KK, Mercado-Garcia A, Tamayo YOM, Cantoral A et al. Relating phthalate and BPA exposure to metabolism in peripubescence: the role of exposure timing, sex, and puberty. J Clin Endocrinol Metab 2016; 101: 79–88.
Lang IA, Galloway TS, Scarlett A, Henley WE, Depledge M, Wallace RB et al. Association of urinary bisphenol A concentration with medical disorders and laboratory abnormalities in adults. JAMA 2008; 300: 1303–1310.
Shankar A, Teppala S, Sabanayagam C . Urinary bisphenol a levels and measures of obesity: results from the national health and nutrition examination survey 2003-2008. ISRN Endocrinol 2012; 2012: 965243.
Masuno H, Iwanami J, Kidani T, Sakayama K, Honda K . Bisphenol a accelerates terminal differentiation of 3T3-L1 cells into adipocytes through the phosphatidylinositol 3-kinase pathway. Toxicol Sci 2005; 84: 319–327.
Somm E, Schwitzgebel VM, Toulotte A, Cederroth CR, Combescure C, Nef S et al. Perinatal exposure to bisphenol a alters early adipogenesis in the rat. Environ Health Perspect 2009; 117: 1549–1555.
Liao C, Kannan K . A survey of bisphenol A and other bisphenol analogues in foodstuffs from nine cities in China. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 2014; 31: 319–329.
Rochester JR, Bolden AL . Bisphenol S and F: a systematic review and comparison of the hormonal activity of bisphenol A substitutes. Environ Health Perspect 2015; 123: 643–650.
Liao C, Liu F, Alomirah H, Loi VD, Mohd MA, Moon HB et al. Bisphenol S in urine from the United States and seven Asian countries: occurrence and human exposures. Environ Sci Technol 2012; 46: 6860–6866.
Zhou X, Kramer JP, Calafat AM, Ye X . Automated on-line column-switching high performance liquid chromatography isotope dilution tandem mass spectrometry method for the quantification of bisphenol A, bisphenol F, bisphenol S, and 11 other phenols in urine. J Chromatogr B Analyt Technol Biomed Life Sci 2014; 944: 152–156.
Farmer SR . Transcriptional control of adipocyte formation. Cell Metab 2006; 4: 263–273.
Koutnikova H, Cock TA, Watanabe M, Houten SM, Champy MF, Dierich A et al. Compensation by the muscle limits the metabolic consequences of lipodystrophy in PPAR gamma hypomorphic mice. Proc Natl Acad Sci USA 2003; 100: 14457–14462.
Rosen ED . Molecular mechanisms of adipocyte differentiation. Ann Endocrinol (Paris) 2002; 63 (2 Pt 1): 79–82.
Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 2002; 16: 22–26.
Atlas E, Pope L, Wade MG, Kawata A, Boudreau A, Boucher JG . Bisphenol A increases aP2 expression in 3T3L1 by enhancing the transcriptional activity of nuclear receptors at the promoter. Adipocyte 2014; 3: 170–179.
Sargis RM, Johnson DN, Choudhury RA, Brady MJ . Environmental endocrine disruptors promote adipogenesis in the 3T3-L1 cell line through glucocorticoid receptor activation. Obesity (Silver Spring) 2010; 18: 1283–1288.
Li X, Ycaza J, Blumberg B . The environmental obesogen tributyltin chloride acts via peroxisome proliferator activated receptor gamma to induce adipogenesis in murine 3T3-L1 preadipocytes. J Steroid Biochem Mol Biol 2011; 127: 9–15.
Greenspan P, Mayer EP, Fowler SD . Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 1985; 100: 965–973.
Kim JB, Wright HM, Wright M, Spiegelman BM . ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc Natl Acad Sci USA 1998; 95: 4333–4337.
Heim M, Frank O, Kampmann G, Sochocky N, Pennimpede T, Fuchs P et al. The phytoestrogen genistein enhances osteogenesis and represses adipogenic differentiation of human primary bone marrow stromal cells. Endocrinology 2004; 145: 848–859.
Lea-Currie YR, Wen P, McIntosh MK . Dehydroepiandrosterone reduces proliferation and differentiation of 3T3-L1 preadipocytes. Biochem Biophys Res Commun 1998; 248: 497–504.
Boucher JG, Boudreau A, Atlas E . Bisphenol A induces differentiation of human preadipocytes in the absence of glucocorticoid and is inhibited by an estrogen-receptor antagonist. Nutr Diabetes 2014; 4: e102.
Caprio M, Feve B, Claes A, Viengchareun S, Lombes M, Zennaro MC . Pivotal role of the mineralocorticoid receptor in corticosteroid-induced adipogenesis. FASEB J 2007; 21: 2185–2194.
Feve B, Antras J, Lasnier F, Hilliou F, Pairault J . The antiglucocorticoid RU38486 is a potent accelerator of adipose conversion of 3T3-F442A cells. Mol Cell Endocrinol 1989; 67: 17–27.
Helies-Toussaint C, Peyre L, Costanzo C, Chagnon MC, Rahmani R . Is bisphenol S a safe substitute for bisphenol A in terms of metabolic function? An in vitro study. Toxicol Appl Pharmacol 2014; 280: 224–235.
Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 1999; 4: 611–617.
Kroker AJ, Bruning JB . Review of the structural and dynamic mechanisms of PPARgamma partial agonism. PPAR Res 2015; 2015: 816856.
Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y et al. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure 2007; 15: 1258–1271.
Kallenberger BC, Love JD, Chatterjee VK, Schwabe JW . A dynamic mechanism of nuclear receptor activation and its perturbation in a human disease. Nat Struct Biol 2003; 10: 136–140.
Riu A, Grimaldi M, le Maire A, Bey G, Phillips K, Boulahtouf A et al. Peroxisome proliferator-activated receptor gamma is a target for halogenated analogs of bisphenol A. Environ Health Perspect 2011; 119: 1227–1232.
Cao Z, Umek RM, McKnight SL . Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 1991; 5: 1538–1552.
Heine PA, Taylor JA, Iwamoto GA, Lubahn DB, Cooke PS . Increased adipose tissue in male and female estrogen receptor-alpha knockout mice. Proc Natl Acad Sci USA 2000; 97: 12729–12734.
Joyner JM, Hutley LJ, Cameron DP . Glucocorticoid receptors in human preadipocytes: regional and gender differences. J Endocrinol 2000; 166: 145–152.
Yi KW, Shin JH, Seo HS, Lee JK, Oh MJ, Kim T et al. Role of estrogen receptor-alpha and -beta in regulating leptin expression in 3T3-L1 adipocytes. Obesity (Silver Spring) 2008; 16: 2393–2399.
Rosenmai AK, Dybdahl M, Pedersen M, Alice van Vugt-Lussenburg BM, Wedebye EB, Taxvig C et al. Are structural analogues to bisphenol a safe alternatives? Toxicol Sci 2014; 139: 35–47.
Sharma G, Hu C, Brigman JL, Zhu G, Hathaway HJ, Prossnitz ER . GPER deficiency in male mice results in insulin resistance, dyslipidemia, and a proinflammatory state. Endocrinology 2013; 154: 4136–4145.
Sharma G, Prossnitz ER . Mechanisms of estradiol-induced insulin secretion by the G protein-coupled estrogen receptor GPR30/GPER in pancreatic beta-cells. Endocrinology 2011; 152: 3030–3039.
Hugo ER, Brandebourg TD, Woo JG, Loftus J, Alexander JW, Ben-Jonathan N . Bisphenol A at environmentally relevant doses inhibits adiponectin release from human adipose tissue explants and adipocytes. Environ Health Perspect 2008; 116: 1642–1647.
Isensee J, Meoli L, Zazzu V, Nabzdyk C, Witt H, Soewarto D et al. Expression pattern of G protein-coupled receptor 30 in LacZ reporter mice. Endocrinology 2009; 150: 1722–1730.
Martensson UE, Salehi SA, Windahl S, Gomez MF, Sward K, Daszkiewicz-Nilsson J et al. Deletion of the G protein-coupled receptor 30 impairs glucose tolerance, reduces bone growth, increases blood pressure, and eliminates estradiol-stimulated insulin release in female mice. Endocrinology 2009; 150: 687–698.
Ijichi N, Ikeda K, Horie-Inoue K, Yagi K, Okazaki Y, Inoue S . Estrogen-related receptor alpha modulates the expression of adipogenesis-related genes during adipocyte differentiation. Biochem Biophys Res Commun 2007; 358: 813–818.
Cipelli R, Harries L, Okuda K, Yoshihara S, Melzer D, Galloway T . Bisphenol A modulates the metabolic regulator oestrogen-related receptor-alpha in T-cells. Reproduction 2014; 147: 419–426.
Rochester JR . Bisphenol A and human health: a review of the literature. Reprod Toxicol 2013; 42: 132–155.
Acknowledgements
We thank Dr M Wade and Dr E Tung for reviewing this manuscript. Grant funding was provided by the Health Canada Chemical Management Plan. SA is funded by the Natural Sciences and Engineering Research Council of Canada Visiting Scientist Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Ahmed, S., Atlas, E. Bisphenol S- and bisphenol A-induced adipogenesis of murine preadipocytes occurs through direct peroxisome proliferator-activated receptor gamma activation. Int J Obes 40, 1566–1573 (2016). https://doi.org/10.1038/ijo.2016.95
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2016.95
- Springer Nature Limited
This article is cited by
-
Tetra methyl bisphenol F: another potential obesogen
International Journal of Obesity (2024)
-
Bisphenol A substitutes and childhood obesity at 7 years: a cross-sectional study in Shandong, China
Environmental Science and Pollution Research (2023)
-
Toxicity of bisphenol A (BPA) and its derivatives in divers biological models with the assessment of molecular mechanisms of toxicity
Environmental Science and Pollution Research (2023)
-
Bisphenol A enhances adipogenic signaling pathways in human mesenchymal stem cells
Genes and Environment (2020)
-
Dechlorane Plus increases adipogenesis in 3T3-L1 and human primary preadipocytes independent of peroxisome proliferator-activated receptor γ transcriptional activity
International Journal of Obesity (2019)