Abstract
Currently, drug resistance of anti-cancer therapy has become the main cause of low survival rate and poor prognosis. Full understanding of drug resistance mechanisms is an urgent request for further development of anti-cancer therapy and improvement of prognosis. Here we present our N-glycoproteomics study of putative N-glycoprotein biomarkers of drug resistance in doxorubicin resistance breast cancer cell line michigan cancer foundation-7 (MCF-7/ADR) relative to parental michigan cancer foundation-7 (MCF-7) cells. Intact N-glycopeptides (IDs) from MCF-7/ADR and MCF-7 cells were enriched with zwitterionic hydrophilic interaction liquid chromatography (ZIC-HILIC), labeled with stable isotopic diethylation (SIDE), and analyzed with C18-RPLC-MS/MS (HCD with stepped normalized collision energies); these IDs were identified with database search engine GPSeeker, and the differentially expressed intact N-glycopeptides (DEGPs) were quantified with GPSeekerQuan. With target-decoy searches and control of spectrum-level FDR ≤ 1%, 322 intact N-glycopeptides were identified; these intact N-glycopeptides come from the combination of 249 unique peptide backbones (corresponding to 234 intact N-glycoproteins) and 90 monosaccharide compositions (corresponding to 248 putative N-glycosites). The sequence structures of 165 IDs were confirmed with structure-diagnostic fragment ions. With the criteria of observation at least twice among the three technical replicates, ≥ 1.5-fold change and p value < 0.05, 20 DEGPs were quantified, where five of them were up-regulated and 15 of them were down-regulated; the corresponding intact N-glycoproteins as putative markers of drug resistance were discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most malignant disease in women with high mortality rate. There have been many studies on anticancer treatment like photothermal therapy (Yang et al. 2018), while chemotherapy and surgery are still common threapies of breast cancer. However, drug resistance often occurs rapidly, resulting in the low survival rate.
Glycosylation is one of the most common protein post-translational modifications (PTMs), and aberrant glycosylation is closely related to many diseases including cancer (Xue et al. 2020). Biomarkers has been widely used in different phases of cancer including tumor formation, metastasis, diagnosis, drug treatment and prognosis (Yang et al. 2020). So far, most cancer biomarkers approved by Food and Drug Administration (FDA) for early diagnosis and treatment are glycoproteins. Thus, identification and discovery of aberrant glycoprotein biomarkers are essential for the studies of drug response mechanisms, early-stage diagnosis and ultimate precision medicine.
Multidrug resistance (MDR) to chemotherapy, a major hurdle in current cancer treatment, often leads to poor prognosis with metastasis as well as reoccurrence and progression; as such, World Health Organization (WHO) predicts that there will be 27 million new cancer cases and 17.1 million deaths per year by 2050 (Saraswathy and Gong 2013). Underlying molecular mechanisms of MDR have been widely studied, but it is not fully understood yet; the reported ones containing ATP-binding cassette (ABC) transporter family, anti-apoptosis, cancer stem cells regulation, microRNA (miRNA) regulation, and epithelial-mesenchymal transition (EMT). Many ABC transporters such as P-glycoprotein (P-gp /MRP1/ABCC1), ATP-binding cassette sub-family C member 2 (MRP2/ABCC2) and ATP-binding cassette sub-family G member 2 (BCRP/ABCG2) have been reported. P-gp is an MDR related N-glycosylated membrane protein. With the inhibitors of glycosylation, the activity of P-gp is affected, which results in the decrease of MDR (Wojtowicz et al. 2012). In addition, unusual glycosylation types cause by aberrant glycosyltransferases (GTs) expression are also reported as a hallmark of malignant tumor and associated with cancer formation, metastasis and drug resistance (Wu et al. 2019). To date, it is unclear that how glycosylation impacts the drug transport activity, and the identification of MDR-related differentially expressed N-glycoproteins is essential for understanding the underlying molecular mechanisms and developing more efficient drugs and therapies.
Breast cancer is the leading cause of cancer deaths among females, and chemotherapy is a first-choice therapy; unfortunately, most of the patients (up to 90%) demonstrated drug resistance within one year (Cobleigh et al. 1999; Slamon et al. 2001; Vogel et al. 2001). Knockdown of ABCG2 improved chemosensitivity in estrogen receptor- negative breast cancer (Arumugam et al. 2019). MCF-7 cell line is a very common breast cancer cell model for the study of MDR mechanisms. The MCF-7/ADR cell line is resistant to the chemotherapeutic agent doxorubicin, and some MDR-related proteins are also identified in this cell line (Bogush et al. 2018).In the study of immunofluorescent staining with antibodies, differential expression of mesenchymal protein vimentin in MCF-7/ADR cells relative to MCF-7 cells was found to be much higher than that of P-gp, which indicated that mesenchymal phenotype of epithelial cells contributed much more to MDR than the overexpression of ABC transporter (Bogush et al. 2018). Down‐regulation of proto-oncogene c-fos in MCF‐7/ADR cells led to decreased expression of P-gp and enhanced apoptosis with increased expression of B-cell lymphoma 2 (Bcl-2) as well as decreased expression of apoptosis regulator Bax, cellular tumor antigen p53 and Bcl-2-binding component 3, isoforms 1/2 (PUMA) (Shi et al. 2013).
Here, we identified 322 intact N-glycopeptides with FDR ≤ 1% and quantified 20 DEGPs with the criteria of observation at least twice, ≥ 1.5-fold change and p value < 0.05 in MCF-7/ADR cells relative to MCF-7 cells with the characterization of quantitative site- and structure-specific N-glycoproteomics. The identified intact N-glycoproteins related to drug resistance were discussed in this study.
Materials and Methods
Chemicals and Reagents
Ammonium hydroxide solution (NH4OH, 28–30% NH3 basis, 1336–21-6), aldehyde (CH3CH2O, 99 atom% 13C, 1632–98-0), dithiothreitol (DTT, 3483-12-3), formic acid (FA, 64-18-6), iodoacetamide (IAA, 144-48-9), sodium cyanoborohydride (NaBH3CN, 25895–60-7), 2,2,2-trifluoroethanol (TFE, ≥ 99%, 75-89-8), trifluoroacetic acid (TFA, 99%, 76-05-1), trypsin (T6567, proteomics grade) and all HPLC solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetone (67-64-1), acetaldehyde solution (40% in H2O, 75-07-0) and hydrochloric acid (HCl, 7647-01-0) were obtained from General Reagent (Shanghai). Ammonium bicarbonate (NH4HCO3, 1066-33-7), bicinchoninic acid (BCA) protein assay kit, urea (57-13-6), phosphate buffer saline (20 × PBS), sodium dodecyl sulfate (SDS, 151-21-3) and tris (hydroxymethyl) aminomethane (Tris, 77-86-1) were purchased from Sangon Biotech (Shanghai). Ultrapure water was produced on site by Millipore Simplicity System (Billerica, MA, USA).
Cell Culture of MCF-7 and MCF-7/ADR Cells
The breast cancer cell line MCF-7 (ATTC, Manassas, VA) was cultured with DMEM (Thermo Scientific Hyclone, MA, USA), 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C with 5% CO2. MCF-7/ADR cell line was maintained in the same culture medium plus 1000 ng/mL adriamycin to induce the doxorubicin resistance. With periodically re-selection, a highly drug-resistant cell population of MCF-7/ADR cells was obtained. Both MCF-7 cells and MCF-7/ADR cells were centrifuged and then washed with pre-cooled PBS thrice for further use.
Sample Preparation of Intact N-Glycopeptides
The protocol of stable isotopic diethylated intact N-glycopeptides from whole cell lysate was reported in our previous study (Wang et al. 2019). Briefly, MCF-7 and MCF-7/ADR cell pellets (from one 10-cm dish each) were sonicated in 1 mL lysis buffer (0.1 M Tris/HCl, 4% SDS, pH 8.0) on ice with the cycle of 2 s on and 3 s off for 15 min. After centrifugation at 13,500 rpm and 4 °C for 15 min, the supernatant was collected and precipitated with ice-cold acetone (1:6, v/v). After 3-h precipitation at −20 °C, the protein mixture was centrifuged at 10,000 rpm for 10 min. Acetone was discarded, and the protein pellets were subsequently washed with ice-cold acetone (about 1 mL) for three times. Then the protein pellets were dissolved in 200 μL 8 M urea and diluted with 800 μL 50 mM NH4HCO3. The protein concentration was measured with BCA assay.
The protein (1 mg) was reduced with 10 mM DTT at 55 °C for 30 min, alkylated with 20 mM IAA at RT in the dark for 20 min and quenched by additional 10 mM DTT. Then the protein was disolved in 5 mL 50 mM NH4HCO3 with 20 μg trypsin for overnight digestion at 37 ºC. A home-made C18 Solid-Phase Extraction (SPE) tip (Jupiter C18, particle size 15 μm, pore size 300 Å) was used for desalting. The C18 tip was pre-treated sequentially by 80% ACN with 5% TFA and then by 0.1% TFA. The digestion solution was loaded, washed with 0.1% TFA, and peptides were eluted by 100 μL 50% ACN with 0.1% TFA for four times and 100 μL 80% ACN with 5% TFA for four times, respectively. The eluted peptides were dried in a SpeedVac and re-suspended in 100 μL 80% ACN with 5% TFA.
Intact N-glycopeptides were enriched with home-made SPE columns packed with ZIC-HILIC particles (Merk Millipore, particle size 5 μm, pore size 200 Å). The column was first equilibrated with 0.1% TFA and then followed by 80% ACN with 5% TFA. The peptide mixtures were loaded on the ZIC-HILIC SPE column. After washing with the washing buffer [80% ACN with 5% TFA (Xue et al. 2018)], intact N-glycopeptides were eluted by 100 μL 0.1% TFA for three times and 50 μL 50 mM NH4HCO3 twice. The eluants were combined and dried in the SpeedVac. The concentration of obtained intact N-glycopeptides was measured by BCA assay.
Equal amount of intact N-glycopeptides either from MCF-7 or MCF-7/ADR cells was isotopically diethylated with 100 μL TFE, 10 μL 20% acetaldehyde or acetaldehyde-13C and 10 μL 600 mM NaBH3CN; the reaction was carried out at 37 °C for 1 h with gentle shaking and stopped with 10 μL 4% (v/v) NH4OH. The reaction mixture was neutralized with 9.6 μL 5% (v/v) FA. The labelled intact N-glycopeptides from the two types of cells were mixed, concentrated and desalted with C18 SPE tips as described above. Intact N-glycopeptides were eluted with 50 μL of 50% ACN with 0.1% TFA for four times and 50 μL of 80% ACN with 0.1% TFA for four times. After dried in the SpeedVac, the labelled intact glycopeptides was resuspended in 45 μL ultrapure water for further LC–MS analysis.
C18-RPLC-MS/MS (HCD) Analysis
The 1:1 mixtures of the labelled intact N-glycopeptides (about 15 μg) of MCF-7 and MCF-7/ADR cells were subjected to the RPLC-MS/MS analysis, which was performed on a Dionex Ultimate 3000 RSLC nano-HPLC system (Thermo Fisher Scientific) equipped with nano-ESI-Q Exactive tandem MS (Thermo Fisher Scientific, San Jose, CA, USA).
Briefly, the separation of intact N-glycopeptides was carried out with a 5 cm long C18 trap column (360 μm o.d. × 200 μm i.d., pore size 300 Å, particle size 5 μm) and a 70 cm long C18 analysis column (360 μm o.d. × 75 μm i.d., pore size 300 Å, particle size 5 μm). The gradient time was set to 240 min, and the flow rate was 300 nL/min. Mobile phase A was 0.2% FA, and mobile phase B consisted of 95% ACN, 4.8% H2O and 0.2% FA. Mobile phase B was held at 2% for 10 min for sample loading, ramped to 40% from 10 to 200 min, then increased to 95% in 10 min and kept for another 5 min, then went down to 2%, maintained it until the run completed. ESI–MS spectra were acquired with the following settings: ion transfer capillary temperature of 250 °C, spray voltage of 2.8 kV, m/z range 700 to 2,000, mass resolution 70 k. MS/MS spectra were acquired in the data-dependent acquisition manner (Top 20) as follows: mass resolution 17.5 k, HCD with stepped NCEs of 20.0%, 30.0% and 30.0% (Wang and Tian 2020). With the aforementioned MS and MS/MS settings, three RPLC-MS/MS technical replicates were acquired.
Database Search and Identification of Intact N-Glycopeptides
The raw datasets from RPLC-MS/MS analysis were processed with the database search engine GPSeeker, which was reported in detail elsewhere (Xiao and Tian 2019b), and only a brief description is given here. Before searching, four customized databases including light diethylation in forward direction (LF), light diethylation in decoy direction (LR), heavy diethylation in forward direction (HF) and heavy diethylation in decoy direction (HR) were created, and each raw data file was searched against all the four databases independently. Isotopic peak abundance cutoff (IPACO, %), isotopic peak abundance deviation (IPAD, %) and isotopic peak m/z deviation (IPMD, ppm) were set as 40%, 50% and 20 ppm, respectively, for the search of both the precursor and fragment ions. For output of intact N-glycopeptide spectrum matches (GPSMs), additional parameters were: percentage of matched fragment ions of the peptide backbone ≥ 10%, matched fragment ions of the N-glycan moiety ≥ 1, TopN hits N = 2 (Top 1 hits have the lowest p score), Y1 ions Top4. After searching, GPSMs from each diethyl/diethyl-13C modified forward/decoy database (LF, LR, HF or HR) were filtered with various criteria and ranked in the increasing order of -lg (p score); spectrum index, m/z and z were used to remove duplicates. GPSMs from each pair of forward and decoy databases (LF vs. LR, or HF vs. HR) were combined and ranked in the increasing order of -lg (p score), and a threshold score was chosen to obtain IDs with spectrum-level FDR ≤ 1%.
The filtering criteria of GPSMs were as followed. For each MS2 spectrum, only the peptide backbone with the highest confidence (often highest percentage of matchhed fragment ions and p score value) was kept. If multiple putative N-glycosites existed for the selected peptide backbones, only the site with the most site-determining fragment ions was kept. The N-glycan composition was identified with the mass difference between the intact N-glycopeptide and the peptide backbone, where only the composition with the most matched fragment ions and abundance coverage was kept.
For the GPSMs with the control spectrum-level FDR ≤ 1%, the combination of peptide sequence, peptide PTMs, N-glycosite and N-glycan linkage were utilized to distinguish a unique intact N-glycopeptide, and it was defined as a qualitative ID. Every qualitative ID had at least one matched structure-diagnostic fragment ion, which was defined as a N-glycan fragment ion that could individually distinguish a sequence structure from its isomers with the same molecular formula (the same or different monosaccharide compositon). IDs without any structure-diagnostic fragment ion were interpreted and quanified at the level of monosaccharide compotion.
Relative Quantitation of Differentially Expressed Intact N-Glycopeptides
For relative quantitation of DEGPs in MCF-7/ADR relative to MCF-7 cells, GPSeekerQuan (Xiao and Tian 2019b) was utilized with the IPMD and delta mass set as 20 ppm and 4.01344 Da, respectively. The paired isotopic envelopes of the precursor ions for each ID were searched, and the summed abundance of Top 3 isotopic peaks was used for relative quantitation. For each ID, all six isotopic peaks were required to be observed for the calculation of relative ratios, and at least two ratios were required to be observed among three technical replicates. Quantitative IDs with a fold change of no less than 1.5 and p value < 0.05 were classified as DEGPs.
Results
Intact N-Glycopeptides Identification from Both MCF-7/ADR and MCF-7 Cell Lines
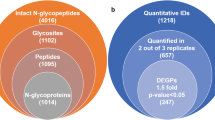
With ZIC-HILIC enrichment, stable isotopic diethyl labelling (SIDE) (Wang and Tian 2020) and GPSeeker DB search, intact N-glycopeptide mixtures from MCF-7/ADR and MCF-7 cell lines were analyzed using C18-RPLC-nanoESI-MS/MS (HCD with stepped NCEs) (Wang et al. 2020a); three technical replicates (TR1, TR2, TR3) were acquired. The venn diagram of identified intact N-glycopeptides was displayed in Fig. S1, and the MS-only base peak chromatograms from each technical replicate were displayed in Fig. S2. To improve the identification confidence, we used homemade ZIC-HILIC SPE columns to improve enrichment efficiency (Mysling et al. 2010; Neue et al. 2011; Alagesan et al. 2017) of intact N-glycopeptides from both MCF-7/ADR and MCF-7 cell lines. In this study, approximately 50 µg intact N-glycopeptides were enriched from 1 mg protein extracted from either MCF-7 or MCF-7/ADR cell pellets (from one 10-cm dish each). After target and decoy database searches by GPSeeker search engine (Xiao and Tian 2019a) with spectrum-level FDR control (≤ 1%) and duplicates removal, 322 intact N-glycopeptides were identified from TR1, TR2 and TR3; these IDs corresponded to 248 N-glycosites, 249 unique peptide backbones and 234 intact N-glycoproteins (Fig. 1a), 90 monosaccharide compositions and 58 putative N-glycan linkages. Among the 165 structure-specific IDs, each of them has no less than one structure-diagnostic fragment ion; in regard to N-glycan types, 89 were high-mannose (53.9%), 46 were complex (27.9%) and 30 were hybrid (18.2%) (Fig. 1c). The detailed information for each of the 322 intact N-glycopeptide IDs was listed in Table S1 including ID type, dataset number, spectrum index, retention time (RT, min), precursor ion (experimental and theoretical m/z, charge, z, IPMD), intact N-glycoprotein (peptide sequence, peptide PTMs, accession number, N-glycosite, glycan linkage, monosaccharide composition), structure-diagnostic fragment ions, -lg (p score), G-bracket and GF score. G-bracket was defined as the number of site-determining peptide backbone b/y fragment ion pairs, each of which could independently localized the N-glycosite; GF score was defined as the number of structure-diagnostic fragment ions of the N-glycan moiety, each of which could independently and unambiguously dinstinguish the given N-glycan structure with all the other isomers.
Identification and quantification of intact N-glycopeptide IDs. a Intact N-glycopeptide IDs identified (FDR ≤ 1%) and b differentially expressed N-glycopeptides quantified (two out of three technical replicates, ≥ 1.5-fold, p < 0.05) from C18-RPLC-ESI–MS/MS (stepped HCD) analysis of 1:1 mixture of stable isotopically diethylated intact N-glycopeptides from MCF-7 and MCF-7/ADR cells; c bar chart for relative abundances of N-glycosylation type from MCF-7 and MCF-7/ADR cells
Among the 248 putative N-glycosites, 247 were identified with single N-X-S/T/C (X ≠ P) motif in peptide amino acid sequence (G-bracket > 0). Take protein nuclear pore membrane glycoprotein 210 (Q8TEM1, PO210_HUMAN, N405) as an example, the intact N-glycopeptide IETVLPAEFFEVLSSSQNGSYHR_N2H9F0S0 was found with G-brackets = 5 of singly charged y8*, y9*, y11*, y12* and y18* (* = GlcNAc) (Fig. S3). Furthermore, 95 of them were newly discovered N-glycosites with G-brackets > 0, which have not been annotated in UniProt as of July 1, 2021 (Table S2).
For micro-heterogeneity (i.e., more than one glycan compositions at the same glycosite of the peptide backbone), two out of 165 structure-specific IDs were modified with two different N-glycans; for macro-heterogeneity (i.e., more than one glycosite in one protein), 11 out of 234 intact N-glycoproteins were identified with no less than two N-glycosylation sites with G-brackets > 0. For hypoxia up-regulated protein 1 (Q9Y4L1, HYOU1_HUMAN), three compositons (N2H6F0S0, N2H7F0S0 and N2H8F0S0) were identified on the peptide backbone LSALDNLL830NHSSMFLK, and two compostions (N2H5F0S0 and N2H6F0S0) on AEPPL931NASASDQGEK (Table 1).
Intact N-Glycopeptides Quantification from Both MCF-7/ADR and MCF-7 Cell Lines
To obtain the quantitative results from MCF-7 cells and MCF-7/ADR cells, the Top 3 isotopic peaks of the paired precursor ions with 4.01344 Da mass delta in MS spectrum (as shown in Fig. 2a–c) were searched by GPSeekerQuan. With the criteria of all the six isotopic peaks observed, 100 IDs were quantified with the relative abundance ratio of the summed isotopic abundance, and 39 IDs were quantified with no less than two ratios out of the three technical replicates. Moreover, for DEGPs, the p value calculated with a t test must be less than 0.05, and the fold change should be bigger than 1.5. Twenty DEGPs were obtained (Fig. 1b) including 15 down-regulated and five up-regulated (Table S3). For example, down-regulated (0.45 ± 0.06) intact N-glycopeptide YHNQTLR_N2H5F0S0 was found on N-glycosite N400 of cation-independent mannose-6-phosphate receptor (P11717, MPRI_HUMAN) (Fig. 2); up-regulated (2.00 ± 0.28) intact N-glycopeptide QVVENMTR_N2H3F0S0 was found on N-glycosite N390 of N-glycoprotein lysosomal alpha-glucosidase (P10253, LYAG_HUMAN) in MCF-7/ADR cells (Fig. 3).
Quantification of down-regulated (0.45 ± 0.06) intact N-glycopeptide YHNQTLR_N2H5F0S0 from cation-independent mannose-6-phosphate receptor (P11717, MPRI_HUMAN) on N-glycosite N400 in MCF-7/ADR relative to MCF-7 cells. a–c the isotopic envelope fingerprinting maps of the precursor ions from three technical replicates; d selective fragmentation and the graphical fragmentation map of N-glycan moiety with the peptide backbone, e fragmentation and the graphical fragmentation map of the peptide backbone with one core GlucNAc, and f the annotated MS/MS spectrum with the matched fragment ions in representative spectrum 9,487 of TR3
Quantification of up-regulated (2.00 ± 0.28) intact N-glycopeptide QVVENMTR_N2H3F0S0 from N-glycoprotein lysosomal alpha-glucosidase on N-glycosite N390 (P10253, LYAG_HUMAN) in MCF-7/ADR relative to MCF-7 cells. a–c the isotopic envelope fingerprinting maps of the precursor ions in the three technical replicates; d selective fragmentation and the graphical fragmentation map of N-glycan moiety with the peptide backbone, e fragmentation and the graphical fragmentation map of the peptide backbone with one core GlucNAc, and f the annotated MS/MS spectrum with the matched fragment ions in representative spectrum 13,312 of TR2
Simultaneous up- and down-regulations were quantified in intact N-glycoprotein N-acetylglucosamine-6-sulfatase (P15586, GNS_HUMAN, N362); down-regulated (0.46) GPGIKPNQTSK_N2H6F0S0 and up-regulated (2.50 ± 0.24) GPGIKPNQTSK_N2H7F0S0 were quantified in MCF-7/ADR relative to MCF-7 cells (Fig. 4).
In our present system of MCF-7/ADR cells vs. MCF-7 cells, the down expression level of SLSNSTAR_N2Hx (x = 5–9) was gradually levelled off; from N2H5 to N2H9, fold changes were 0.27 ± 0.03, 0.49 ± 0.08, 0.76 ± 0.08, 0.92 ± 0.08 and 1.09 ± 0.29, respectively (Fig. 5). This intact N-glycopeptide SLSNSTAR_N2H5F0S0 (P50454, SERPH_HUMAN, N120) was also quantified in our previous studies (Wang et al. 2019, 2020b). For lysosome-associated membrane glycoprotein 1 (P11279, LAMP1_HUMAN), intact N-glycopeptides GHTLTLNFTR with N2H6F0S0 and N2H7F0S0 on N-glycosite N103 were quantified with fold changes of 1.79 ± 0.38 and 0.89 (only quantified once in spectrum 20,345 of TR2), respectively. Wheras, AANGSLR with N2H8F0S0 and N2H9F0S0 on N-glycosite N322 were down-regulated with fold changes of 0.55 (only quantified once in spectrum 8,053 of TR1) and 0.65 ± 0.08 (Fig. 6). Different from above-mentioned fold changes, GHTLTLNFTR with N2H6F0S0 and N2H7F0S0 on N-glycosite N103 and AANGSLR with N2H9F0S0 on N-glycosite N322 were all down-regulated in MCF-7 cells vs. MCF-10A cells (0.23 ± 0.03, 0.20 ± 0.02, 0.29 ± 0.02, respectively) (Xue et al. 2020) and all up-regulated in MCF-7 CSCs vs. MCF-7 cells (7.41 ± 1.43, 3.28 ± 0.35, 1.10 ± 0.42, 3.05 ± 0.30) (Wang et al. 2019).
On N-glycosite N120 of serpin H1 (P50454), down-regulations of intact N-glycopeptides SLSNSTAR_N2HxF0S0 (x = 5–9) were gradually levelled off. a from N2H5F0S0 to N2H9F0S0, fold changes were 0.27 ± 0.03, 0.49 ± 0.08, 0.76 ± 0.08, 0.92 ± 0.08 and 1.09 ± 0.29, respectively; b the corresponding isotopic envelope fingerprinting maps of the precursor ions in this study; SLSNSTAR_N2H5F0S0 was identified as GPSMs and quantified with Top2 isotopic peaks of three technical replicates; SLSNSTAR_N2H9F0S0 was quantified at the monosaccharide composition level in two technical replicates
Different fold changes of the intact N-glycopeptides GHTLTLNFTR with N2H6F0S0 and N2H7F0S0 (LAMP1_HUMAN, N103) and AANGSLR_N2H8F0S0 and N2H9F0S0 (LAMP1_HUMAN, N322) from MCF-7 relative to MCF-10A cells (orange) (Xue et al. 2020), MCF-7/ADR relative to MCF-7 cells (green) and MCF-7 CSCs relative to MCF-7 (violet) cells (Wang et al. 2019) were showed with bar chart. AANGSLR_N2H8F0S0 was only quantified once in spectrum 8,053 of TR1. GHTLTLNFTR_N2H7F0S0 was only quantified once in spectrum 20,345 of TR2
Gene Ontology (GO) Analysis and Protein–Protein Interaction (PPI) Network
Gene ontology (GO) analysis of 20 DEGPs in MCF-7/ADR relative to MCF-7 cells were carried out using PANTHER (protein annotation through evolutionary relationship) classification system (http://pantherdb.org/). Most intact N-glycoproteins corresponding to DEGPs came from intracecellular and cellular anatomical entity (Fig. S4), which were mainly involved in biological processes of metabolic process, celluar process and biological regulation; whereas molecular functions were concentrated in binding and catalytic activitity.
Intact N-glycoproteins corresponding to the 100 quantified intact glycopeptides in MCF-7/ADR relative MCF-7 cells were subject to STRING (https://string-db.org/) analysis, and 32 intact N-glycoproteins were correlated on the PPI network (Fig. 7). The correlation among ITGAE, ITGA3, ITGAV, LAMA1 and ILK were discussed as below.
Discussion
Resistance to chemotherapy can be caused by many factors like drug efflux and uptake transporters (e.g.,ABCC9, ABCA9 and SLC22A4), positive DNA damage repair (e.g.,GPAT), cancer stem cells (CSCs) and tumor microenvironment (e.g., ITGAE, ITGA3, ITGAV and LAMA1), etc. Hence, it is still urgent to figure out the mechanisms of drug resistance. In this regard, intact N-glycoproteins about the development of drug resistance, either identified or quantified in current and previous studies (Wang et al. 2019, 2020b; Xu et al. 2020; Xue et al. 2020), were discussed.
Intact N-Glycoproteins Associated with Drug Transporters
In this study, we focused on the differential N-glycosylation in breast cancer cell lines MCF-7 and MCF-7/ADR to find putative drug resistance N-glycoprotein biomarkers. The ATP-binding cassette (ABC) transporters family is the most common cause of drug resistance. As typical efflux proteins, ABC transporters have the ability to transport drug from outside the cell, and increased ABC transporters can promote the drug efflux, which reduces the accumulation of the drug in cells (Imperio et al. 2019). Among the ABC transport protein family, ATP-binding cassette sub-family A member 9 (Q8IUA7, ABCA9_HUMAN) plays a key role in the ATPase-coupled transmembrane transporter activity and lipid transporter activity. Intact N-glycopeptide SMDELDLNYSIDAVR_N4H5F0S2 on N-glycosite N120 of ABCA9 was up-regulated with a fold change of 9.38 from spectrum 29,356 of TR2. High-level expression of ABCA9 in ovarian cancer (OC) was related to poor cancer prognosis with the validation of RT-qPCR, expression microarray data and immunohistochemistry, which illustrated that lipid trafficking might be associated with serous epithelial ovarian cancer (EOC) (Hedditch et al. 2014). ABCC9, a member of ATP-binding cassette sub-family C, is another multidrug efflux transporter. With the assessment of RT-qPCR, ABCA2/8/9/10, ABCC9 and SLC16A14 genes showed high expression level in EOC and high correlation with longer progression-free survival of EOC patients (Elsnerova et al. 2017). Different from increased drug efflux, decreased drug uptake could also cause resistance to anti-cancer agents, which was induced by uptake transporters like solute carriers (Okabe et al. 2008). The collaboration of uptake and efflux transporters resulted in drug resistance. We identified intact N-glycopeptide NKSEIR_N2H6F0S0 from solute carrier family 13 member 3(Q8WWT9, S13A3_HUMAN, N312) and intact N-glycopeptide DGWVYNR_N7H4F0S0 (A6NK97, S22AK_HUMAN, N114) from solute carrier family 22 member 20 (encoded by SLC22A20P). Down-regulation of solute carrier family 22 member 4 (SLC22A4 or OCTN1) was related to poor prognosis of acute myeloid leukemia patients after treated by cytarabine (Ara-C) (Buelow et al. 2021).
Intact N-glycoproteins Associated with DNA Damage Repair
The majority of anticancer drugs can cause DNA damage of malignant cells and subsequent cell death. However, when DNA damage becomes repairable, malignant cells would survive and be resistant to drugs together with cancer recurrence (Li et al. 2020). Bcl-2 functioned as an anti-apoptotic protein, which was associated with drug resistance (Allen et al. 2015). Bcl-2-associated transcription factor 1 (BCLAF1) showed higher expression level in cisplatin resistance non‑small cell lung cancer A549 (A549/DDP) cell line than in A549 cell line as characterized by western blotting and RT-qPCR (Jiang et al. 2020). Furthermore, BCLAF1 facilitated DNA damage repair by mediating the forming process of γH2A histone family member X foci formation via immunofluorescence and western blot (Jiang et al. 2020). Glycerol-3-phosphate acyltransferase (GPAT) is responsible for the catalysis of de novo synthesis of glycerolipids in cells (Pellon-Maison et al. 2014). Upregulated intact N-glycopeptide GMFATNVTENVLNSSR_N7H9F0S0 was quantified once with a fold change of 2.95 on N-glycosite N135 of glycerol-3-phosphate acyltransferase 1, mitochondrial (Q9HCL2, GPAT1_HUMAN). GPAT2, a mitochondrial isoform, was primarily expressed and upregulated in lung and breast cancer, and its expression was further validated in breast cancer MDA-MB-231 cells with immunohistochemistry. Increased expression of GPAT2 promoted cell growth, migration, proliferation and resistance to drug-induced apoptosis (Pellon-Maison et al. 2014).
Intact N-glycoproteins Associated with Cancer Stem Cells (CSCs)
CSCs have the capacity of self-proliferation and differentiation. Treated with chemotherapy drugs or radiation, a large number of CSCs die, and only a small part survive, resulting in the relapse of cancer and tumor cells metastasis. CSCs may be the source of primary malignant cell, the reason of drug resistance, or the root of tumor cell metastasis (Clarke 2019).
Drug‑resistance N‑glycosylation markers (ABCC5, ABCA4 and ABCB9 etc.) and CSC N‑glycosylation markers (centrosome-associated protein 350 CE350, zinc finger protein GLI1, CD63 antigen and CD13 etc.) were reported in our previous studies (Wang et al. 2020b; Xu et al. 2020).
In comparison of DEGPs in MCF-7 CSCs (relative to MCF-7 cells) (Wang et al. 2019) with those in MCF-7/ADR CSCs (relative to MCF-7/ADR cells) (Xu et al. 2020), intact N-glycopeptide SLSNSTAR_N2H5F0S0 (P50454, SERPH_HUMAN, N120) showed the same up-regulation with ratios of 4.23 ± 0.71 and 3.80 ± 0.55, respectively. We found that the expression level of intact N-glycopeptides SLSNSTAR_N2Hx (x = 5–9) was decreased progressively from up-regulation to down-regulation in MCF-7/ADR CSCs (relative to MCF-7/ADR cells) (Xu et al. 2020). Interestingly, the decreased expression level of SLSNSTAR_N2Hx (x = 5–9) was gradually levelled off in our present system of MCF-7/ADR cells vs. MCF-7 cells; from N2H5 to N2H9, fold changes were 0.27 ± 0.03, 0.49 ± 0.08, 0.76 ± 0.08, 0.92 ± 0.08 and 1.09 ± 0.29, respectively (Fig. 5). The result also inllustrated that high mannose N-glycan type was identified from N2H5 to N2H8, and hybrid N-glycan type was identified from N2H9.
Serpin H1 (also known as heat shock protein 47, Hsp 47) is a 47 kDa endoplasmic reticulum (ER) protein which can specifically bind to collagen and works as a chaperone in collagen synthesis (Ito and Nagata 2017). Hsp 47 (encoded by SERPINH1), a member from serpin family, participated in progression of cancer such as hepatocellular carcinoma (HCC) (Naboulsi et al. 2016) and cholangiocellular carcinoma (Padden et al. 2014). Overexpression of Hsp 47 was also reported in micro-vessels from invasive ductal carcinoma (IDC) of breast cancer (vs. para-carcinoma tissue), and Hsp 47 was involved in tumor formation and invasion. Down-regulated Hsp 47 was responsible for poor prognosis via western blot and immunohistochemistry analysis in laryngeal squamous cell carcinoma (LSCC) tissues relative to adjacent nonmalignant tissues (Hill et al. 2011). Hsp 47-upregulated LSCC cell line (amplified by plasmid vectors) showed the decreased ability of proliferation and invasion and increased sensitivity to cisplatin chemotherapy. Hence, down-regulation of Hsp 47 was associated with optimal LSCC prognosis and chemotherapy resistance (Song et al. 2017).
Intact N-glycoproteins Associated with Tumor Microenvironment
Up to now, many studies show that extracellular microenvironment (EMT) plays a key role in cancer progression and drug assistance. The extracellular matrix (ECM) is one of the most important parts of EMT which participates in many cellular physiological processes like cell proliferation and components attachment of ECM (fibronectin, collagen I, collagen IV, fibrinogen, etc.) (Hoshiba and Tanaka 2013). Integrins mediate signaling transduction between cells and ECM during cancer development and progression, which can affect the efficacy of chemotherapy (Wu et al. 2020). According to the PPI analysis (Fig. 7), ITGAE, ITGA3, ITGAV, LAMA1 and ILK show high correlation with each other.
The intact N-glycopeptide SSAHVSVVWQLEENAFPNRTADITVTVTNSNER_N4H5F0S2 of integrin alpha-E (ITGAE, P38570, ITAE_HUMAN, N934) was quantified once (3.00) from spectrum 36,036 of TR3. The intact N-glycopeptide SLDAYPILNQAQALENHTEVQFQK_N2H8F0S0 of integrin alpha-3 (ITGA3, P26006, ITA3_HUMAN, N605) was quantified to be up-regulated with a fold change of 2.15 ± 0.67. The intact N-glycopeptide TAADTTGLQPILNQFTPANISR_N2H5F0S0 of integrin alpha-V (ITGAV, P06756, ITAV_HUMAN, N615) was quantified once (0.39) from spectrum 28,503 of TR1. Upregulated intact N-glycopeptide TRPRNGTLNK_N2H7F0S0 of integrin-linked protein kinase (Q13418, ILK_HUMAN, N179) was quantified once (7.31) from spectrum 10,412 of TR2. ITGA3/B4 was an important prognostic marker in OC, and ITGAE acted as a residing memory CD8 + T cells marker which showed the correlation with early OC patients prognosis (Wu et al. 2020). ITGB5, one part from integrin beta subtype, worked as the receptor of fibronectin. Integrins were responsible for the mechanical link and signal communication between cells and microenvironment, which contributed to tumor formation and progression (Moon et al. 2019). ITGA5 and ITGB1/3/5 were involved in cell formation, cancer metastasis and chemotherapy resistance in lung cancer (Aksorn and Chanvorachote 2019). In etoposide-resistant human retinoblastoma (RB) cell line of WERI-ETOR cells (compared with WERI-RB1 cells), the mRNA expression of ITGA5 was confirmed to be down-regulated with a fold change of 0.198 (p < 0.001) via RT-qPCR. The same trend was also observed in etoposide resistant Y79 RB cells vs. sensitive Y79 RB cells with a fold change of 0.029 (p = 0.003) (Reinhard et al. 2020).
For laminin subunit alpha-1 (P25391, LAMA1_HUMAN), intact N-glycopeptide VNTTLR_N2H6F0S0 with glycan linkage 01Y41Y41M(31M21M)61M(31M)61M on N2047 was down-regulated from spectrum 16,457 in TR1 (0.18), and VNTTLR_N2H8F0S0 was down-regulated with a fold change of 0.27 ± 0.02. COL1A2, COL6A1, ITGB4, ITGA2 and LAMA3 were associated with the PI3K-AKT signaling pathway where tumor-resistance gene RAC1 served as an important role (Wei et al. 2019). Five hub genes (COL1A2, ITGB4, ITGA2, LAMA3 and RAC1) except COL6A1 were reported to be related to pancreatic cancer (Wei et al. 2019). LAMA3 was down-regulated in lung adenocarcinoma with over expression of long non-coding RNA LINC00628. Xu et al. (2019) illustrated the mechanism that highly expressed LINC00628 bound to promoter region of LAMA3 gene, resulting in the increase of methylase. With the promotion of LAMA3 promoter methylation, LAMA3 expression was inhibited, then tumor invasion, migration and drug resistance might occur (Xu et al. 2019).
Other Intact N-glycoproteins Associated with Drug Resistance
Besides those discussed above, we also found other putative drug resistant N-glycoprotein biomarkers. Some lysosomal proteins were identified in this work, including sulfatase (GNS), lysosomal thioesterase (PPT2) and prosaposin (PSAP), and some of them were reported to be upregulated in OC (Hu et al. 2020).
Up-regulation of intact N-glycopeptide CPMAGISHTAWHSNR_N2H6F0S0 was quantified with a fold change of 2.04 ± 0.09 from lysosomal thioesterase PPT2 (Q9UMR5, PPT2_HUMAN, N289). PPT2, also called palmitoyl protein thioesterase 2, is a lysosomal thioesterase, and it shows significant correlation with cell metabolism. Down-regulated PPT2 was identified in clear cell renal cell carcinoma (ccRCC) tissues paired with adjacent normal tissues, and it was regarded as a putative diagnostic marker for ccRCC (Yuan et al. 2020).
Up-regulation of intact N-glycopeptide TNSTFVQALVEHVKEECDR_N2H3F1S0 was identified from prosaposin (P07602, SAP_HUMAN, N215). Prosaposin (PSAP) was also a kind of lysosomal protein, which was located in lysosomal and secreted proteins (Choi et al. 2020). High expression of PSAP showed a high correlation with poor prognosis in breast cancer. PSAP worked as an androgen receptor (AR) activator in invasive phenotype (Ali et al. 2015). For N-acetylglucosamine-6-sulfatase (P15586, GNS_HUMAN), up-regulated intact N-glycopeptide GPGIKPNQTSK_N2H7F0S0 on N-glycosite N362 and down-regulated intact N-glycopeptide GPGIKPNQTSK_N2H6F0S0 on the same N-glycosite were quantified with fold changes of 2.50 ± 0.24 and 0.46, respectively. GNS had the function of metal binding with calcium. It was also identified in breast cancer cell line Hs578T and confirmed in plasma by western blot and LC–MS/MS (Ahn et al. 2010).
LAMP1 is also related to lysosomal protein, which belongs to the lysosome-associated membrane glycoprotein family (Dominguez-Bautista et al. 2015). It was found that LAMP1 was highly expressed in cisplatin-resistant gastric cancer SGC7901/CDDP cell line by western blot experiment (Qi and Zhang 2020), and increased LAMP1 expression in breast cancer cells was associated with poor prognosis (Wang et al. 2017). In this study, intact N-glycopeptides GHTLTLNFTR with compositions of N2H6F0S0 and N2H7F0S0 on N-glycosite N103 were quantified with fold changes of 1.79 ± 0.38 and 0.89 (only quantified once in spectrum 20,345 of TR2), respectively. However, intact N-glycopeptides AANGSLR with compositions of N2H8F0S0 and N2H9F0S0 on N-glycosite N322 were quantified to be down-regulated by fold changes of 0.55 (only quantified once in spectrum 8,053 of TR1) and 0.65 ± 0.08, respectively (Fig. 6). In our previous study, intact N-glycopeptides AANGSLR with compositions of N2H8F0S0 and N2H9F0S0 on N-glycosite N322 were also up-regulated by fold changes of 3.28 ± 0.35 and 7.41 ± 1.43, respectively (Wang et al. 2019), whereas intact N-glycopeptides SSCGKENTSDPSLVIAFGR_N2H3F1S0 on N-glycosite N84 and GHTLTLNFTR_N2H5F0S0 on N-glycosite N103 were quantified to be down-regulated by fold changes of 0.55 ± 0.02 and 0.32 ± 0.04, respectively (Xue et al. 2020).
For translocon-associated protein subunit beta encoded by SSR2 (P43308, SSRB_HUMAN), up-regulation (1.57 ± 0.26) of intact N-glycopeptide IAPASNVSHTVVLRPLK_N2H9F0S0 was quantified on N-glycosite N88. SSR2 was responsible for ER transport across membranes, and it was identified as a candidate gene in drug resistanct human osteosarcoma cell line U-2 OS relative to parental U-2 OS cells as characterized by differential display reverse transcription-polymerase chain reaction (DDRT-PCR) (Chano et al. 2004).
For lysophosphatidic acid receptor 5 (Q9H1C0, LPAR5_HUMAN, N323), intact N-glycopeptides including TSATNGTR_N2H6F0S0, N2H7F0S0 and N2H8F0S0 were down-regulated by fold changes of 0.12, 0.23 ± 0.03 and 0.25 ± 0.02, respectively. Lysophosphatidic acid (LPA) receptors are responsible for cancer cells malignant promotion. Compared with melanoma A375 cell line, LPAR5 showed lower expression in highly migratory A375-R11 cell line. A375-R11 cells showed higher survival rates than A375 cells when treated with anticancer drug cisplatin (CDDP) and dacarbazine (DTIC), suggesting LPAR5 was related to chemoresistance (Minami et al. 2019).
For KN motif and ankyrin repeat domain-containing protein 1 (Q14678, KANK1_HUMAN, N8), intact N-glycopeptide VNGSASGK_N2H8F0S0 was quantified to be up-regulated with a fold change of 3.32. KANK2 was positively correlated with microtubule (MT) poison sensitivity and decreased migration in the melanoma cell line MDA-MB-435S (Paradzik et al. 2020).
For titin (Q8WZ42, TITIN_HUMAN, N26369), the intact N-glycopeptide VNVSSSK_N2H8F0S0 was up-regulated (2.47) in MCF-7/ADR cells vs. MCF-7 cells. The molecular function of titin is related to ATP binding, calcium ion binding and calmodulin binding. TTN (titin) is the gene of titin, and its mutations are associated with chemotherapy prognosis. Nonsynonymous variants in TTN were observed to be occurred in pre- and post-platinum treatment of non-small cell lung cancer (NSCLC) patients with targeted and whole exome sequencing (WES) (Cobleigh et al. 1999). The variants of TTN were also identified in triple negative breast cancer (TNBC) patients with pre- and post-chemotherapy of doxorubicin and cyclophosphamide as studied by next generation sequencing (NGS) (Lips et al. 2015). TTN was associated with chemotherapy response, which suggested those mutations in TTN could be candidates for further chemotherapy response.
Conclusions
The site- and structure-specific differential N-glycosylation in MCF-7/ADR cells was identified and quantified by MS-based N-glycoproteomics. Specifically, with trypsin digestion, C18 desalting, ZIC-HILIC enrichment and SIDE labelling, the 1:1 mixture of intact N-glycopeptides from MCF-7/ADR and MCF-7 cells were analyzed by C18-RPLC-ESI–MS/MS (HCD with stepped collision energies). With target and decoy DB search using GPSeeker and the control of spectrum-level FDR ≤ 1%, 322 intact N-glycopeptides were identified together with 249 peptide backbone sequences, 248 putative N-glycosites and 234 intact N-glycoproteins; the N-glycan linkage structures of 165 IDs were confirmed with structure-diagnostic fragment ions. With the criteria of observation of Top 3 isotopic peaks of each precursor ion pair, fold change ≥ 1.5 and p value < 0.05, 20 differentially expressed intact N-glycopeptides were quantified in MCF-7/ADR relative to MCF-7 cells, where five of them were up-regulated and 15 of them were down-regulated. The corresponding intact N-glycoproteins are associated with drug resistance and may be considered as putative drug resistance biomarkers for further clinical research.
Availability of Data and Materials
The three RPLC-MS/MS (HCD) technical replicate datasets (.raw) are freely available at ProteomeXchange Consortium via the PRIDE partner repository (Vizcaino et al. 2013): PXD013836; supplemental information can be found with this article online at https://doi.org/10.1007/s43657-021-00029-8.
Code Availability
Not applicable.
Change history
17 January 2022
A Correction to this paper has been published: https://doi.org/10.1007/s43657-021-00035-w
References
Ahn Y, Kang U-B, Kim J, Lee C (2010) Mining of serum glycoproteins by an indirect approach using cell line secretome. Mol Cells 29(2):123–130. https://doi.org/10.1007/s10059-010-0008-0
Aksorn N, Chanvorachote P (2019) Integrin as a molecular target for anti-cancer approaches in lung cancer. Anticancer Res 39(2):541–548. https://doi.org/10.21873/anticanres.13146
Alagesan K, Khilji SK, Kolarich D (2017) It is all about the solvent: on the importance of the mobile phase for ZIC-HILIC glycopeptide enrichment. Anal Bioanal Chem 409(2):529–538. https://doi.org/10.1007/s00216-016-0051-6
Ali A, Creevey L, Hao Y, McCartan D, O’Gaora P, Hill A, Young L, McIlroy M (2015) Prosaposin activates the androgen receptor and potentiates resistance to endocrine treatment in breast cancer. Breast Cancer Res. https://doi.org/10.1186/s13058-015-0636-6
Allen JE, Prabhu VV, Talekar M, van den Heuvel APJ, Lim B, Dicker DT, Fritz JL, Beck A, El-Deiry WS (2015) Genetic and pharmacological screens converge in identifying FLIP, BCL2, and IAP proteins as key regulators of sensitivity to the TRAIL-inducing anticancer agent ONC201/TIC10. Can Res 75(8):1668–1674. https://doi.org/10.1158/0008-5472.can-14-2356
Arumugam A, Subramani R, Nandy SB, Terreros D, Dwivedi AK, Saltzstein E, Lakshmanaswamy R (2019) Silencing growth hormone receptor inhibits estrogen receptor negative breast cancer through ATP-binding cassette sub-family G member 2. Exp Mol Med. https://doi.org/10.1038/s12276-018-0197-8
Bogush TA, Kalyuzhny SA, Chetyrkina MR, Yastrebova MA, Scherbakov AM, Ryabinina OM, Mamichev IA, Bogush EA, Kamensky AA (2018) Molecular mechanisms underlying drug resistance of the MCF7/ADR breast cancer cell line. Mosc Univ Chem Bull 73(5):248–250. https://doi.org/10.3103/S0027131418050036
Buelow DR, Anderson JT, Pounds SB, Shi L, Lamba JK, Hu S, Gibson AA, Goodwin EA, Sparreboom A, Baker SD (2021) DNA methylation-based epigenetic repression ofSLC22A4Promotes resistance to cytarabine in acute myeloid leukemia. CTS Clin Transl Sci 14(1):137–142. https://doi.org/10.1111/cts.12861
Chano T, Mori K, Scotlandi K, Benini S, Lapucci C, Manara MC, Serra M, Picci P, Okabe H, Baldini N (2004) Differentially expressed genes in multidrug resistant variants of U-2OS human osteosarcoma cells. Oncol Rep 11(6):1257–1263. https://doi.org/10.3892/or.11.6.1257
Choi K-M, Hwang SD, Joo M-S, Hwang JY, Kwon M-G, Jeong J-M, Seo JS, Lee JH, Lee H-C, Park C-I (2020) Two short antimicrobial peptides derived from prosaposin-like proteins in the starry flounder (Platichthys stellatus). Fish Shellf Immunol 105:95–103. https://doi.org/10.1016/j.fsi.2020.05.075
Clarke MF (2019) Clinical and therapeutic implications of cancer stem cells. N Engl J Med 380(23):2237–2245. https://doi.org/10.1056/NEJMra1804280
Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ (1999) Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 17(9):2639–2648. https://doi.org/10.1200/jco.1999.17.9.2639
Dominguez-Bautista JA, Klinkenberg M, Brehm N, Subramaniam M, Kern B, Roeper J, Auburger G, Jendrach M (2015) Loss of lysosome-associated membrane protein 3 (LAMP3) enhances cellular vulnerability against proteasomal inhibition. Eur J Cell Biol 94(3–4):148–161. https://doi.org/10.1016/j.ejcb.2015.01.003
Elsnerova K, Bartakova A, Tihlarik J, Bouda J, Rob L, Skapa P, Hruda M, Gut I, Mohelnikova-Duchonova B, Soucek P, Vaclavikova R (2017) Gene expression profiling reveals novel candidate markers of ovarian carcinoma intraperitoneal metastasis. J Cancer 8(17):3598–3606. https://doi.org/10.7150/jca.20766
Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J, Johnatty SE, Chen X, Harnett P, George J, Williams RT, Flemming C, Lambrechts D, Despierre E, Lambrechts S, Vergote I, Karlan B, Lester J, Orsulic S, Walsh C, Fasching P, Beckmann MW, Ekici AB, Hein A, Matsuo K, Hosono S, Nakanishi T, Yatabe Y, Pejovic T, Bean Y, Heitz F, Harter P, du Bois A, Schwaab I, Hogdall E, Kjaer SK, Jensen A, Hogdall C, Lundvall L, Engelholm SA, Brown B, Flanagan J, Metcalf MD, Siddiqui N, Sellers T, Fridley B, Cunningham J, Schildkraut J, Iversen E, Weber RP, Berchuck A, Goode E, Bowtell DD, Chenevix-Trench G, deFazio A, Norris MD, MacGregor S, Haber M, Henderson MJ, Australian Ovarian Canc Study G (2014) ABCA Transporter Gene Expression and Poor Outcome in Epithelial Ovarian Cancer. JNCI J Natl Cancer Inst. https://doi.org/10.1093/jnci/dju149
Hill JJ, Tremblay T-L, Pen A, Li J, Robotham AC, Lenferink AEG, Wang E, O’Connor-McCourt M, Kelly JF (2011) Identification of vascular breast tumor markers by laser capture microdissection and label-free LC-MS. J Proteome Res 10(5):2479–2493. https://doi.org/10.1021/pr101267k
Hoshiba T, Tanaka M (2013) Breast cancer cell behaviors on staged tumorigenesis-mimicking matrices derived from tumor cells at various malignant stages. Biochem Biophys Res Commun 439(2):291–296. https://doi.org/10.1016/j.bbrc.2013.08.038
Hu Y, Pan J, Shah P, Ao M, Thomas SN, Liu Y, Chen L, Schnaubelt M, Clark DJ, Rodriguez H, Boja ES, Hiltke T, Kinsinger CR, Rodland KD, Li QK, Qian J, Zhang Z, Chan DW, Zhang H, Clinical Proteomic Tumor Anal C (2020) Integrated proteomic and glycoproteomic characterization of human high-grade serous ovarian carcinoma. Cell Rep. https://doi.org/10.1016/j.celrep.2020.108276
Imperio GE, Javam M, Lye P, Constantinof A, Dunk CE, Reis FM, Lye SJ, Gibb W, Matthews SG, Ortiga-Carvalho TM, Bloise E (2019) Gestational age-dependent gene expression profiling of ATP-binding cassette transporters in the healthy human placenta. J Cell Mol Med 23(1):610–618. https://doi.org/10.1111/jcmm.13966
Ito S, Nagata K (2017) Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Semin Cell Dev Biol 62:142–151. https://doi.org/10.1016/j.semcdb.2016.11.005
Jiang T, Liu B, Wu D, Zhang F (2020) BCLAF1 induces cisplatin resistance in lung cancer cells. Oncol Lett. https://doi.org/10.3892/ol.2020.12090
Li B, Jiang J, Assaraf YG, Xiao H, Chen Z-S, Huang C (2020) Surmounting cancer drug resistance: new insights from the perspective of N-6-methyladenosine RNA modification. Drug Resist Updates. https://doi.org/10.1016/j.drup.2020.100720
Lips EH, Michaut M, Hoogstraat M, Mulder L, Besselink NJM, Koudijs MJ, Cuppen E, Voest EE, Bernards R, Nederlof PM, Wesseling J, Rodenhuis S, Wessels LFA, Ctr Personalized Canc T (2015) Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. https://doi.org/10.1186/s13058-015-0642-8
Minami K, Ueda N, Maeda H, Ishimoto K, Otagaki S, Tsujiuchi T (2019) Modulation of chemoresistance by lysophosphatidic acid (LPA) signaling through LPA(5) in melanoma cells treated with anticancer drugs. Biochem Biophys Res Commun 517(2):359–363. https://doi.org/10.1016/j.bbrc.2019.07.092
Moon JH, Rho YS, Lee SH, Koo BS, Lee HJ, Do SI, Cho JH, Eun YG, Park MW, Shin HA, Lim YC (2019) Role of integrin beta 1 as a biomarker of sternness in head and neck squamous cell carcinoma. Oral Oncol 96:34–41. https://doi.org/10.1016/j.oraloncology.2019.07.001
Mysling S, Palmisano G, Hojrup P, Thaysen-Andersen M (2010) Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal Chem 82(13):5598–5609. https://doi.org/10.1021/ac100530w
Naboulsi W, Megger DA, Bracht T, Kohl M, Turewicz M, Eisenacher M, Voss DM, Schlaak JF, Hoffmann A-C, Weber F, Baba HA, Meyer HE, Sitek B (2016) Quantitative tissue proteomics analysis reveals versican as potential biomarker for early-stage hepatocellular carcinoma. J Proteome Res 15(1):38–47. https://doi.org/10.1021/acs.jproteome.5b00420
Neue K, Mormann M, Peter-Katalinic J, Pohlentz G (2011) Elucidation of glycoprotein structures by unspecific proteolysis and direct nanoESI mass spectrometric analysis of ZIC-HILIC-enriched glycopeptides. J Proteome Res 10(5):2248–2260. https://doi.org/10.1021/pr101082c
Okabe M, Szakacs G, Reimers MA, Suzuki T, Hall MD, Abe T, Weinstein JN, Gottesman MM (2008) Profiling SLCO and SLC22 genes in the NCI-60 cancer cell lines to identify drug uptake transporters. Mol Cancer Ther 7(9):3081–3091. https://doi.org/10.1158/1535-7163.mct-08-0539
Padden J, Megger DA, Bracht T, Reis H, Ahrens M, Kohl M, Eisenacher M, Schlaak JF, Canbay AE, Weber F, Hoffmann A-C, Kuhlmann K, Meyer HE, Baba HA, Sitek B (2014) Identification of novel biomarker candidates for the immunohistochemical diagnosis of cholangiocellular carcinoma. Mol Cell Proteom 13(10):2661–2672. https://doi.org/10.1074/mcp.M113.034942
Paradzik M, Humphries JD, Stojanovic N, Nestic D, Majhen D, Dekanic A, Samarzija I, Sedda D, Weber I, Humphries MJ, Ambriovic-Ristov A (2020) KANK2 Links alpha V beta 5 focal adhesions to microtubules and regulates sensitivity to microtubule poisons and cell migration. Front Cell Dev Biol. https://doi.org/10.3389/fcell.2020.00125
Pellon-Maison M, Montanaro MA, Lacunza E, Garcia-Fabiani MB, Soler-Gerino MC, Cattaneo ER, Quiroga IY, Abba MC, Coleman RA, Gonzalez-Baro MR (2014) Glycerol-3-phosphate acyltranferase-2 behaves as a cancer testis gene and promotes growth and tumorigenicity of the breast cancer MDA-MB-231 cell line. Plos One. https://doi.org/10.1371/journal.pone.0100896
Qi W, Zhang Q (2020) Gene’s co-expression network and experimental validation of molecular markers associated with the drug resistance of gastric cancer. Biomark Med 14(9):761–773. https://doi.org/10.2217/bmm-2019-0504
Reinhard J, Wagner N, Kraemer MM, Jarocki M, Joachim SC, Dick HB, Faissner A, Kakkassery V (2020) Expression changes and impact of the extracellular matrix on etoposide resistant human retinoblastoma cell lines. Int J Mol Sci. https://doi.org/10.3390/ijms21124322
Saraswathy M, Gong S (2013) Different strategies to overcome multidrug resistance in cancer. Biotechnol Adv 31(8):1397–1407. https://doi.org/10.1016/j.biotechadv.2013.06.004
Shi R, Peng H, Yuan X, Zhang X, Zhang Y, Fan D, Liu X, Xiong D (2013) Down-regulation of c-fos by shRNA sensitizes adriamycin-resistant MCF-7/ADR cells to chemotherapeutic agents via P-glycoprotein inhibition and apoptosis augmentation. J Cell Biochem 114(8):1890–1900. https://doi.org/10.1002/jcb.24533
Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344(11):783–792. https://doi.org/10.1056/nejm200103153441101
Song X, Liao Z, Zhou C, Lin R, Lu J, Cai L, Tan X, Zeng W, Lu X, Zheng W, Chen J, Su Z (2017) HSP47 is associated with the prognosis of laryngeal squamous cell carcinoma by inhibiting cell viability and invasion and promoting apoptosis. Oncol Rep 38(4):2444–2452. https://doi.org/10.3892/or.2017.5893
Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Perez-Riverol Y, Reisinger F, Rios D, Wang R, Hermjakob H (2013) The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41(D1):D1063–D1069. https://doi.org/10.1093/nar/gks1262
Vogel C, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, Shak S, Stewart SJ, Genentech HSI (2001) First-line, single-agent Herceptin (R) (trastuzumab) in metastatic breast cancer: a preliminary report. Eur J Cancer 37:S25–S29. https://doi.org/10.1016/s0959-8049(00)00405-6
Wang Y, Tian Z (2020) New energy setup strategy for intact N-glycopeptides characterization using higher-energy collisional dissociation. J Am Soc Mass Spectrom 31(3):651–657. https://doi.org/10.1021/jasms.9b00089
Wang Q, Yao J, Jin Q, Wang X, Zhu H, Huang F, Wang W, Qiang J, Ni Q (2017) LAMP1 expression is associated with poor prognosis in breast cancer. Oncol Lett 14(4):4729–4735. https://doi.org/10.3892/ol.2017.6757
Wang Y, Xu F, Xiao K, Chen Y, Tian Z (2019) Site- and structure-specific characterization of N-glycoprotein markers of MCF-7 cancer stem cells using isotopic-labelling quantitative N-glycoproteomics. Chem Commun 55(55):7934–7937. https://doi.org/10.1039/c9cc04114a
Wang Y, Xiao K, Tian Z (2020a) Quantitative N-glycoproteomics using stable isotopic diethyl labeling. Talanta. https://doi.org/10.1016/j.talanta.2020.121359
Wang Y, Xu F, Chen Y, Tian Z (2020b) A quantitative N-glycoproteomics study of cell-surface N-glycoprotein markers of MCF-7/ADR cancer stem cells. Anal Bioanal Chem 412(11):2423–2432. https://doi.org/10.1007/s00216-020-02453-7
Wei J, Han R, Su X, Chen Y, Shi J, Cui X, Zhang H, Gong Y, Chu X, Chen J (2019) Identification of biomarkers and their functions in dasatinib-resistant pancreatic cancer using bioinformatics analysis. Oncol Lett 18(1):197–206. https://doi.org/10.3892/ol.2019.10281
Wojtowicz K, Szaflarski W, Januchowski R, Zawierucha P, Nowicki M, Zabel M (2012) Inhibitors of N-glycosylation as a potential tool for analysis of the mechanism of action and cellular localisation of glycoprotein P. Acta Biochim Pol 59(4):445–450. https://doi.org/10.1016/B978-0-12-394291-3.00009-5
Wu Y, Chen X, Wang S, Wang S (2019) Advances in the relationship between glycosyltransferases and multidrug resistance in cancer. Clin Chim Acta 495:417–421. https://doi.org/10.1016/j.cca.2019.05.015
Wu A, Zhang S, Liu J, Huang Y, Deng W, Shu G, Yin G (2020) Integrated analysis of prognostic and immune associated integrin family in ovarian cancer. Front Genet. https://doi.org/10.3389/fgene.2020.00705
Xiao K, Tian Z (2019a) GPSeeker enables quantitative structural N-glycoproteomics for site- and structure-specific characterization of differentially expressed N-glycosylation in hepatocellular carcinoma. J Proteome Res 18(7):2885–2895. https://doi.org/10.1021/acs.jproteome.9b00191
Xiao K, Tian Z (2019b) Site- and structure-specific quantitative N-glycoproteomics using RPLC-pentaHILIC separation and the intact N-glycopeptide search engine GPSeeker. Curr Protoc Protein Sci 97(1):e94–e94. https://doi.org/10.1002/cpps.94
Xu S-F, Zheng Y, Zhang L, Wang P, Niu C-M, Wu T, Tian Q, Yin X-B, Shi S-S, Zheng L, Gao L-M (2019) Long non-coding RNA LINC00628 interacts epigenetically with the LAMA3 promoter and contributes to lung adenocarcinoma. Mol Therapy Nucl Acids 18:166–182. https://doi.org/10.1016/j.omtn.2019.08.005
Xu F, Wang Y, Xiao K, Hu Y, Tian Z, Chen Y (2020) Quantitative site- and structure-specific N-glycoproteomics characterization of differential N-glycosylation in MCF-7/ADR cancer stem cells. Clin Proteom. https://doi.org/10.1186/s12014-020-9268-7
Xue Y, Xie J, Fang P, Yao J, Yan G, Shen H, Yang P (2018) Study on behaviors and performances of universal N-glycopeptide enrichment methods. Analyst 143(8):1870–1880. https://doi.org/10.1039/c7an02062g
Xue B, Xiao K, Wang Y, Tian Z (2020) Site- and structure-specific quantitative N-glycoproteomics study of differential N-glycosylation in MCF-7 cancer cells. J Proteom. https://doi.org/10.1016/j.jprot.2019.103594
Yang H, Zhao J, Wu C, Ye C, Zou D, Wang S (2018) Facile synthesis of colloidal stable MoS2 nanoparticles for combined tumor therapy. Chem Eng J 351:548–558. https://doi.org/10.1016/j.cej.2018.06.100
Yang J, Cui R, Liu Y (2020) MicroRNA-212–3p inhibits paclitaxel resistance through regulating epithelial-mesenchymal transition, migration and invasion by targeting ZEB2 in human hepatocellular carcinoma. Oncol Lett. https://doi.org/10.3892/ol.2020.11884
Yuan C, Xiong Z, Shi J, Peng J, Meng X, Wang C, Hu W, Ru Z, Xie K, Yang H, Chen K, Zhang X (2020) Overexpression of PPT2 represses the clear cell renal cell carcinoma progression by reducing epithelial-to-mesenchymal transition. J Cancer 11(5):1151–1161. https://doi.org/10.7150/jca.36477
Funding
This research was financially supported by National Natural Science Foundation of China (21775110, 22074105) and Shanghai Science and Technology Commission (14DZ2261100).
Author information
Authors and Affiliations
Contributions
Conceptualization: ZT and YC; cell culture: FX and YC; LC–MS: HY; software: KX; data analysis: HY, ZT, and KX; draft: HY; manuscript revision: ZT.
Corresponding authors
Ethics declarations
Conflicts of Interest
The authors declare there is no conficts of interest regarding the publication of this paper.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
The original online version of this article was revised due to wrong figures 3 and 5.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, H., Xu, F., Xiao, K. et al. N-Glycoproteomics Study of Putative N-Glycoprotein Biomarkers of Drug Resistance in MCF-7/ADR Cells. Phenomics 1, 269–284 (2021). https://doi.org/10.1007/s43657-021-00029-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43657-021-00029-8