Abstract
Isotopic-labeling quantitative N-glycoproteomics characterization of cell-surface differentially expressed N-glycosylation in MCF-7/ADR cancer stem cells (CSCs) relative to MCF-7/ADR cells was carried out at the intact N-glycopeptide level with trypsin digestion, ZIC-HILIC enrichment, isotopic diethyl labeling, RPLC-MS/MS analysis of the 1:1 mixture, and GPSeeker DB search. With a spectrum-level false discovery rate of ≤ 1%, 1,336 intact N-glycopeptides from the combination of 301 unique peptide backbones and 169 putative N-glycan linkages (52 monosaccharide compositions) were identified; the corresponding intact N-glycoproteins and N-glycosites were 289 and 305, respectively, among which 176 N-glycosites were confirmed with GlcNAc-containing site-determining b/y fragment ion pairs. The N-glycan moieties in 546 intact N-glycopeptide IDs were identified with more than one structure-diagnostic fragment ions where multiple linkage structures exist for each of the monosaccharide compositions. With the criteria of ≥ 1.5-fold change and p value < 0.05, 72 cell-surface differentially expressed intact N-glycopeptides (DEGPs) were found in MCF-7/ADR CSCs relative to MCF-7/ADR cells, where 8 and 64 were downregulated and upregulated, respectively.

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the leading cancer in women, and multiple drug resistance leads to high mortality [1, 2]; formation of cancer stem cells (CSCs) is among one of the major drug resistance mechanisms. CSCs play important roles in tumor genesis, metastasis, therapeutic resistance, and thus recurrence [3, 4]. Cell-surface markers can be used for CSC profiling, diagnosis as well as therapeutic targets.
During the past decade, researchers have been searching glycoprotein markers of breast CSCs with various methods [5,6,7]. The expression level of c-KIT (a single-pass type I membrane protein) has been positively correlated with CSC enrichment in RH1-resistant breast cancer cells [8] and the phenotype of CD44high breast cancers [9]; c-KIT may contribute to cancer progression through the regulation of stemness and resistance to tyrosine kinase inhibitors [8]; blockage of BCL-2, MCL-1, BCL-xL, and BFL-1 by sabutoclax (a pan-active BCL-2 protein family antagonist) effectively reduced sphere formation of drug-resistant cells and eliminated the CSC subpopulation [10]. The same effect was also observed in the inhibition of PI3K by NVP-BKM120 [11]. With 3D sphere culture, Okita et al. found that cell-surface GPNMBhigh cells expressed high levels of CSC genes and EMT-TF genes; glycoprotein nmb (GPNMB) was proposed to be a cell-surface marker of dormant breast cancer cells with contribution to the acquisition of stem cell–like properties [12]. Inhibition of multidrug resistant protein-1 (MDR1, ABCB1) reduced camptothecin resistance among breast cancer CSCs where anticancer activity of the drug was detected by total internal reflection fluorescence [13].
Recently, we have successfully developed site- and structure-specific quantitative N-glycoproteomics pipeline and applied it to characterize putative cancer markers of HCC [14, 15] and CSC markers of MCF-7 [16]. Compared with MCF-7 cells, 91 and 53 of intact N-glycopeptides in MCF-7 CSCs were downregulated and upregulated, respectively; most of the well-known CSC markers (such as 13 cluster of differentiation proteins) were comprehensively characterized. Here we report our comparative N-glycoproteomics study of cell-surface N-glycoproteins of MCF-7/ADR CSCs vs. MCF-7/ADR cells at the intact N-glycopeptide level. With ZIC-HILIC enrichment, isotopic diethyl labeling, RPLC-MS/MS using HCD with stepped normalized collision energies (NCEs) analysis and GPSeeker DB search, differentially expressed cell-surface N-glycosylation in MCF-7/ADR CSCs was quantitatively characterized. With the criteria of > 1.5-fold change and p value < 0.05, 72 differentially expressed intact N-glycopeptides (DEGPs) corresponding to 12 intact N-glycoproteins were quantified.
Materials and methods
Materials
Dithiothreitol (DTT), iodoacetamide (IAA), 2,2,2-trifluoroethanol (TFE, ≥ 99%), sodium cyanoborohydride, acetaldehyde-13C2 (99 atom % 13C), ammonium hydroxide solution (28–30% NH3 basis), trifluoroacetic acid (TFA, 99%), formic acid (FA), and all HPLC solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA). Trypsin-EDTA (2.5 g/L, 0.25% EDTA) was obtained from Life Technologies (Grand Island, NY, USA). Acetaldehyde solution (40% in H2O) was obtained from General Reagent (Shanghai, China). Ultrapure water was produced on site by Millipore Simplicity System (Billerica, MA, USA).
Cell culture of MCF-7/ADR cells and MCF-7/ADR CSCs
Drug-resistant cell line MCF-7/ADR was kindly donated by Dr. Yongmei Yin (Nanjing Medical University, China) and cultured using DMEM (Thermo Scientific Hyclone, MA, USA) supplied with 10% fetal bovine serum (Thermo Scientific Gibco, MA, USA) and 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C and 5% CO2. To maintain a highly drug-resistant cell population, MCF-7/ADR cells were periodically reselected by growing them in the presence of 1000 ng/mL Adriamycin. Experiments were performed using the cells incubated without DOX for 48 h. CD24- and CD44-microbeads antibodies (Miltenyi Biotec, Germany) were used for cell sorting of breast cancer stem cells (BCSCs) [17]. Briefly, 107 total MCF-7/ADR cells were incubated with the above antibodies on ice for 40 min. After washing with cold PBS, CD44+CD24−/low BCSCs named MCF-7/ADR CSCs were purified from MCF-7/ADR cell lines. The characteristics of MCF-7/ADR CSCs were regularly detected by flow cytometry and maintained into ultra-low attachment six well plates (Corning, New York, USA) in MammoCult™ Human Medium Kit (Stem cell technologies, Vancouver, Canada) according to manufacturer’s guideline [18].
Sample preparation of the 1:1 mixture of the labeled cell-surface intact N-glycopeptides of MCF-7/ADR CSCs and MCF-7/ADR cells
Cells (MCF-7/ADR or MCF-7/ADR CSCs) were dispersed in 1 mL of trypsin-EDTA (2.5 g/L) and incubated at 37 °C for 30 min (stopping reagent 1% TFA). The pellets were centrifuged at 14,000 rpm and 4 °C for 15 min, and the supernatant tryptic digests were collected. The digests were washed through house-made C4-tip to remove excess trypsin and EDTA, and then desalted using house-made C18-tip and eluted with 400 μL of 50% ACN with 0.1% TFA and 400 μL of 80% ACN with 0.1% TFA. Desalted peptides were concentrated and stored at − 20 °C for further use.
Intact N-glycopeptides were enriched using house-made ZIC-HILIC (zwitterionic type of hydrophilic interaction chromatography) pipette tip containing 30 mg ZIC-HILIC particles (Merck Millipore, 5 μm, 200 Å). Briefly, desalted peptides were redissolved in 80% ACN with 5% TFA and loaded onto ZIC-HILIC tip which were pre-equilibrated with 0.1% TFA and 80% ACN with 5% TFA. The ZIC-HILIC tip was then washed using 800 μL 80% ACN with 5% TFA. Enriched cell-surface intact N-glycopeptides were eluted with 300 μL 0.1% TFA and 100 μL 50 mM NH4HCO3, and intact N-glycopeptide concentration was determined by BCA (SK3021, Sangon Biotech, Shanghai, China).
Stock solution of NaBH3CN (0.6 M), CH3CHO (20%, w/w), 13CH313CHO (20%, w/w), NH4OH (4%, v/v) and formic acid (2%, v/v) were freshly made. N-Terminal and lysine amino groups of intact N-glycopeptides were diethylated with CH3CHO and NaBH3CN. Briefly, two identical aliquots of cell-surface intact N-glycopeptides of MCF-7/ADR cells and MCF-7/ADR CSCs were re-suspended in 100 μL TFE and 8 μL 20% CH3CHO or 13CH313CHO was added and mixed. Subsequently, 8 μL freshly prepared 0.6 M NaBH3CN was added and incubated at 37 °C for 1 h, and the reaction was quenched with 8 μL 4% (v/v) NH4OH and incubated for 1 min followed by addition of 16 μL 2% (v/v) FA. After concentrated, the labeled cell-surface intact N-glycopeptides were desalted using house-made C18-tip and eluted with 250 μL of 50% ACN with 0.1% TFA and 250 μL of 80% ACN with 0.1% TFA. Desalted labeled intact N-glycopeptides were concentrated and re-suspended in ultrapure water for further analysis.
C18-RPLC-MS/MS (HCD) analysis of the 1:1 mixture
A total of 6 μg cell-surface intact N-glycopeptide mixtures enriched from MCF-7/ADR CSCs or MCF-7/ADR cells were used as starting material for a RPLC-MS/MS analysis. The intact N-glycopeptides were trapped on a 5-cm-long trap column (360 μm o.d. × 200 μm i.d.) and separated on a 70-cm-long analytical column (360 μm o.d. × 75 μm i.d.) both packed with C18 particles (Phenomenex, 5 μm, 300 Å) on a Dionex Ultimate 3000 RSLC nano-HPLC system (Thermo Fisher Scientific). Buffer A is mixture of 99.8% H2O and 0.2% FA; buffer B is mixture of 95.0% ACN, 4.8% H2O, and 0.2% FA. A constant flow of 300 nL/min was set for elution and conducted at the following gradient. The gradient was 4 h in total for 1:1 cell-surface intact N-glycopeptides: 2% buffer B for 10 min for sample-loading and 2–40% B in 190 min, followed by an increase to 95% B in 10 min, held for another 5 min and held for 2% B for the last 20 min for equilibration.
Q Exactive MS (Thermo Fisher Scientific, San Jose, CA, USA) with nano-ESI source was adopted to detect the eluted intact N-glycopeptides online. MS spectra were acquired in the 700–2000 m/z mass range. A mass resolution of 70 k (m/z 200) was set for MS1 spectra while 17.5 k for MS2 spectra. Fragmentation was obtained in a data-dependent mode (Top20) with optimal stepped higher energy collisional dissociation (HCD) normalized collision energies (NCEs) of 20.0%, 30.0%, and 30.0%. The automatic gain control (AGC) target value and maximum injection time were placed at 2 × 105 and 50 ms for MS1 and at 5 × 105 and 250 ms for MS2 scans. Isolation window and dynamic exclusion were set at 3.0 m/z and 20.0 s. The temperature of the ion transfer capillary and the spray voltage were set to 280 °C and 2.8 kV. With the above setting, three technical replicates (TR1, TR2, and TR3) were acquired.
Database search, identification, and quantitation of intact N-glycopeptides
Database search engine GPSeeker was used for intact N-glycopeptide identification with FDR control. Four theoretical customized human intact N-glycopeptides databases of two directions (forward and reverse) and two labels (light and heavy diethylation) (i.e., LF, LR, HF, HR) were first created, and each dataset was searched against the four databases independently. The precursor ions in the MS spectra and the fragment ions in the MS/MS spectra are searched with the following parameters: isotopic abundance cutoff (IPACO), 40%; isotopic peak m/z deviation (IPMD), 20 ppm; and isotopic abundance deviation (IPAD), 50%, respectively. Initial GPSMs were obtained with the following refinement criteria: Y1 ions, Top4; minimal percentage of matched fragment ions of N-glycosite-containing peptides (PMPs), ≥ 10%; minimal matched product ions of N-glycan (g-MPs), ≥ 1; minimal matched product ions of peptide (p-MPs), ≥ 5; TopN hits, N = 2 (Top1 hits have the lowest P score).
For each dataset, the target and decoy GPSMs were combined and ranked with increasing P score, and a cutoff P score was then chosen to achieve spectrum-level FDR ≤ 1%. Target GPSMs from three technical replicates with P scores lower than the cutoff value were grouped and repetitive IDs were removed with the criteria of “peptide sequence, N-glycosite, and N-glycan linkage”and the final list of intact N-glycopeptide IDs was generated.
Relative quantitation of the identified intact N-glycopeptides was carried out using GPSeekerQuan. A mass tolerance of 20 ppm (the same with IPMD) and mass difference of 4.01344 Da were adopted for the search of the paired isotopic envelopes of the precursor ions in the MS spectra. For each intact N-glycopeptide ID, Top3 isotopic peaks in each isotopic envelope were adopted and all the six isotopic peaks from the light and heavy diethylation were required to be observed; the relative abundance ratio of each intact N-glycopeptide ID between MCF-7/ADR CSCs and MCF-7/ADR cells was computed with the summed abundance to the Top3 isotopic peaks, and observation of the ratios in no less than two of the three technical replicates was required. The p value was calculated using the t test [19] for the intact N-glycopeptide IDs quantitated at least twice; differentially expressed intact N-glycopeptides (DEGPs) were defined as intact N-glycopeptide IDs with a fold change of no less than 1.5 and p value no bigger than 0.05.
Multiplexing was allowed for the search and identification, and multiple intact N-glycopeptides with different peptide backbones of high sequence similarity may be simultaneously identified from the same MS and MS/MS spectra. Because this is a cell-surface study, intact N-glycopeptides with the corresponding N-glycoproteins solely annotated as “nucleus” proteins are excluded from identification and quantitation.
Results and discussion
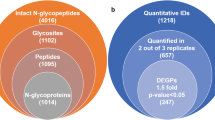
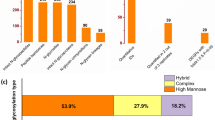
With ZIC-HILIC enrichment, isotopically diethyl labeling, cell-surface intact N-glycopeptides from MCF-7/ADR cells and MCF-7/ADR CSCs were mixed in 1:1 ratio and then online analyzed using C18-RPLC-nanoESI-MS/MS (HCD with stepped NCEs) to obtain three technical replicates (TR1, TR2, and TR3). The MS-only base-peak chromatograms from the three technical replicates are shown in Electronic Supplementary Material (ESM) Fig. S1. With target and decoy database searches using intact N-glycopeptide search engine GPSeeker, spectrum-level FDR control (≤ 1%) and duplicate removal, identified in total from the three technical replicates were 1,336 intact N-glycopeptides corresponding to 169 putative N-glycan linkages (see ESM Table S1) and 52 monosaccharide compositions (see ESM Table S2) on 305 N-glycosites of 301 peptide backbones (289 intact N-glycoproteins) (Fig. 1, left). For each of these 1,336 intact N-glycopeptides, the detailed tabular information of dataset number, spectrum index, retention time, precursor ion (experimental and theoretical m/z, z, IPMD), accession number, peptide sequence, N-glycosite, monosaccharide composition, N-glycan primary structure in the format of one-line text, − log(P score), G-bracket, and GF score are listed in ESM Table S3. IPMD means isotopic peak m/z deviation; G-bracket is defined as GlcNAc-containing site-determining peptide backbone b/y fragment ion pairs each of which can independently localize the N-glycosite; GF score for a specific N-glycan sequence/linkage structure is defined as number of structure-diagnostic fragment ions (from the N-glycan moiety) each of which can independently distinguish the structure from other isomeric structures with the same monosaccharide compositions. Among the 169 putative N-glycan linkages, the percentage of high-mannose, hybrid, and complex N-glycosylation are 27.8%, 6.7%, and 65.5%, respectively.
Intact N-glycopeptides identified (FDR ≤ 1%; left) and differentially expressed intact N-glycopeptides (DEGPs) quantified (two out of the three technical replicates, ≥ 1.5-fold change, p value < 0.05; right) from C18-RPLC-MS/MS (HCD with stepped NCEs) analysis of 1:1 mixture of isotopically diethylated cell-surface intact N-glycopeptides of MCF-7/ADR CSCs and MCF-7/ADR cells
Among the 305 putative N-glycosites, 297 are the only putative N-glycosites on the peptide backbones each with a single N-X-S/T (X ≠ P) sequon where 175 are confirmed with G-bracket score no less than one; one is confirmed with G-bracket score no less than one where two putative N-glycosites exist. For the 176 confirmed N-glycosites, 142 have not been annotated in UniProt as of July 25, 2019 (see ESM Table S4). For example, N-glycosite N20 of isoform 2 of transcription elongation factor A protein 1 is newly annotated N-glycosite in this study, where intact N-glycopeptide KNASTR_N2H8F0S0 was identified with G-brackets of singly charged b2*, b3*, b4*, and b5* (* = GlcNAc).
N-glycosylation micro-heterogeneity was observed for most of the identified intact N-glycoproteins. For N-glycosite N84 of killer cell immunoglobulin–like receptor 2DL1, intact N-glycopeptides ANFSISR with the linkages of -01Y41Y41M(31M21M)61M and -01Y41Y41M(31M)61M(31M)61M were identified with 21 and 26 structure-diagnostic fragment ions, respectively.
Among the 1,336 intact N-glycopeptide IDs, 546 were identified with GF scores ≥ 1. Intact N-glycopeptide VENGSGPK_N3H3F1S0 was identified with two fucose sequence/position isomers of 01Y(61F)41Y41M(31 M)61M61Y (core) and 01Y41Y41M(31M)61M61Y31F (branch); these two structures were confirmed with 6 (BI1, BI2, iI3II3Y, iI4II3Y, YI3, YI4) and 10 (BI2, BI3, YI1, YI2, iI3II3Y, iI4II3Y, YI3, YI4, iI5II3Y, YI5) structure-diagnostic fragment ions, respectively. These two isomeric intact N-glycopeptides were identified from N-glycosite N2121 of basement membrane–specific heparan sulfate proteoglycan core protein (PGBM_HUMAN, P98160).
The abundance of the Top3 isotopic peaks of the paired precursor ions from the MCF-7/ADR cells and MCF-7/ADR CSCs in the MS spectra for the 1,336 intact N-glycopeptide IDs was searched with GPSeekerQuan for relative quantitation. With the criteria of observation of all the six isotopic peaks, 312 IDs were quantified at least once and 78 at least twice out of the three technical replicates. Further with the criteria of ≥ 1.5-fold change and p < 0.05, 72 intact N-glycopeptides were found differentially expressed (Fig. 1, right), where 8 were downregulated and 64 upregulated (see ESM Table S5). For example, intact N-glycopeptide TNLSGR_N2H8F0S0 from N-glycosite N735 of N-glycoprotein serine/threonine- protein phosphatase 4 regulatory subunit 3A (P4R3A_HUMAN, Q6IN85) was found to be downregulated (0.54 ± 0.09) in MCF-7/ADR CSCs relative to MCF-7/ADR cells (Fig. 2); intact N-glycopeptide ELNATSR_N3H3F1S0 from N-glycosite N2236 of N-glycoprotein centrosome-associated protein 350 (CE350_HUMAN, Q5VT06) was found to be upregulated (6.91 ± 0.62) in MCF-7/ADR CSCs relative to MCF-7/ADR cells (Fig. 3).
Downregulation (0.54 ± 0.07) of cell-surface intact N-glycopeptide TNLSGR_N2H8F0S0 in MCF-7/ADR CSCs relative to MCF-7/ADR; the N-glycosite is N735 on N-glycoprotein serine/threonine-protein phosphatase 4 regulatory subunit 3A (P4R3A_HUMAN, Q6IN85). a–c The paired precursor ions in the three technical replicates. d The graphical fragmentation map of the N-glycan moiety with the matched fragment ions. e The annotated MS/MS spectrum with the matched fragment ions
Upregulation (6.91 ± 0.62) of cell-surface intact N-glycopeptide ELNATSR_N3H3F1S0 in MCF-7/ADR CSCs relative to MCF-7/ADR; the N-glycosite is N2236 on N-glycoprotein centrosome-associated protein 350 (CE350_HUMAN, Q5VT06). a–c The paired precursor ions in the three technical replicates. d The graphical fragmentation map of the N-glycan moiety with the matched fragment ions where YI1 and ZI1 are structure-diagnostic ones. e The MS/MS spectrum with the matched fragment ions
The 64 upregulated intact N-glycopeptides come from 8 N-glycosites of 8 intact N-glycoproteins. Centrosome-associated protein 350 (CE350) displayed the highest upregulation (20.22 ± 3.44) followed by OGFD3, FA5, T132D, CBPC1, B4GN3, GVIN1, and MUC16.
On N-glycosite of 2,236 of intact N-glycoprotein centrosome-associated protein 350 (CE350), intact N-glycopeptides with the same peptide backbone of ELNATSR and 6 different N-glycan monosaccharide compositions (N3H3F0S0, N3H3F1S0, N4H3F0S0, N4H3F1S0, N4H4F0S0, N4H5F2S0) were quantified to be uniformly upregulated with a fold change range of 4.73–20.22. CE350 plays an essential role in centriole growth. Overexpression of CAP350 is reported to cooperate with FGFR1 oncogene partner in microtubule anchoring as analyzed by Western blotting [20].
On N-glycosite 215 of intact N-glycoprotein 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 3 (OGFD3), intact N-glycopeptides with the same peptide backbone of INSTEAR and 5 different N-glycan monosaccharide compositions were quantified to be uniformly upregulated with a fold change range of 4.73–20.22. OGFD3 is a single-pass type II membrane protein.
On N-glycosite 1,559 of intact N-glycoprotein coagulation factor V heavy chain (FA5), intact N-glycopeptides with the same peptide backbone of TNINSSR and 10 different N-glycan monosaccharide compositions were quantified to be uniformly upregulated with a fold change range of 2.68–10.84. Coagulation factor V was previously reported overexpressed in human breast tumors as detected by immunohistochemistry [21].
On N-glycosite 72 of intact N-glycoprotein transmembrane protein 132D (T132D), intact N-glycopeptides with the same peptide backbone of NSSLQSR and 7 different N-glycan monosaccharide compositions were quantified to be uniformly upregulated with a fold change range of 3.54–10.84. T132D is a single-pass type I membrane protein which has been reported as a cell-surface marker for oligodendrocyte differentiation as analyzed by Western blot and immunohistochemistry [22].
On N-glycosite 14 of intact N-glycoprotein ATP/GTP-binding protein 1 (AGTPBP1), intact N-glycopeptides with the same peptide backbone of SLTNNSR and 3 different N-glycan monosaccharide compositions (N2H5F0S0, N3H3F0S0, and N4H3F0S0) were quantified to be upregulated at the fold changes of 8.23 ± 2.48, 9.17 ± 0.04, and 4.88 ± 0.60, respectively.
On N-glycosite 494 of intact N-glycoprotein beta-1,4-N-acetylgalactosaminyltransferase 3 (B4GN3), intact N-glycopeptides NSTASFPGR_N4H3F0S0 was quantified to be upregulated with a fold change of 6.89 ± 0.90. B4GN3 is a single-pass type II membrane protein, and increase of B4GN3 mRNA in Adriamycin-resistant cells was detected by Northern blot analysis [23].
On N-glycosite 1,848 of intact N-glycoprotein interferon-induced very large GTPase 1 (GVIN1), intact N-glycopeptide NQSQER_N4H5F0S1 was quantified to be upregulated at the fold changes of 3.89 ± 0.12.
On N-glycosite 8,055 of intact N-glycoprotein mucin-16 (AGTPBP1, CA125), intact N-glycopeptides with the peptide backbone THPSSNR and N-glycan monosaccharide compositions of N2H6F0S0 and N2H7F0S0 were quantified to be upregulated at the fold changes of 1.96 ± 0.34 and 1.52 ± 0.04, respectively. Serum/tissue CA125 was measured as overexpressed by solid-phase enzyme-linked immunosorbent assay in women with ovarian/breast tumors of different stages [24].
The 8 downregulated intact N-glycopeptides come from 4 N-glycosites of 4 intact N-glycoproteins; unconventional myosin-VIIa (MYO7A) displayed the lowest downregulation (0.05 ± 0.01) followed by MAGI3, ZN829, P4R3A, MYB, and KI2L1.
On N-glycosite 1,237 of intact N-glycoprotein unconventional myosin-VIIa (MYO7A), intact N-glycopeptide TFVNGTR_N4H3F1S0 was quantified to be downregulated at the fold change of 0.05 ± 0.01.
On N-glycosite 84 of intact N-glycoprotein killer cell immunoglobulin–like receptor 2DL1 (KI2L1), intact N-glycopeptide ANFSISR_N4H3F1S0 was quantified to be downregulated at the fold change of 0.07 ± 0.03. KI2L1 is a single-pass type I membrane protein and annotated to be inhibiting the activity of natural killer cells thus preventing cell lysis [25]. Intact N-glycopeptide ANFSISR_N4H3F1S0 from N-glycosite 84 of intact N-glycoprotein transcriptional activator Myb (MYB) was simultaneously quantified because the two peptide backbones are equally scored, and multiplexing is allowed. MYB was annotated as playing an important role in the control of proliferation and differentiation of hematopoietic progenitor cells. Deregulation of c-Myb expression was previously reported to be involved in the development of leukemia, and breast and colon cancer [26]; however, MYB is annotated as a nucleus protein, so it is less likely to be present in this cell-surface study.
On N-glycosite 735 of intact N-glycoprotein serine/threonine-protein phosphatase 4 regulatory subunit 3A (P4R3A), intact N-glycopeptide TNLSGR_N2H8F0S0 was quantified to be downregulated at the fold change of 0.54 ± 0.09. P4R3A is a regulatory subunit of serine/threonine-protein phosphatase 4.
On N-glycosite 249 of intact N-glycoprotein membrane-associated guanylate kinase inverted 3 (MAGI3), intact N-glycopeptide EAINGSGNAENRER_N4H3F0S0 was quantified to be downregulated at the fold change of 0.65 ± 0.01. MAGI3 acts as a scaffolding protein at cell-cell junctions. The expression of MAGI-3 was reported downregulated in the transient transfection of the hepatocyte growth factor activator inhibitor-1 vector into SiHa and HeLa cells [27].
Intact N-glycopeptides EINGSK_N2H8F0S0 (atlastin-3, ATLA3) and TNLSGR_N2H8F0S0 (serine/threonine-protein phosphatase 4 regulatory subunit 3A, PPP4R3A) were previously quantified to be upregulated (3.21 ± 0.61) and downregulated (4.11 ± 0.19) in MCF-7 CSCs (vs. MCF-7 cells) (Fig. 4) [16]. However, in this study of MCF-7/ADR CSCs (vs. MCF-7/ADR cells), no significant differential expression was observed with the fold changes of 1.18 ± 0.12 and 1.87 ± 0.23. Atlastin-3 is annotated as GTPase tethering membranes through formation of trans-homo oligomers and mediating homotypic fusion of endoplasmic reticulum membranes, whereas PPP4R3A may regulate the activity of PPP4C at centrosomal microtubule organizing centers.
Gene ontology analysis using PANTHER (protein annotation through evolutionary relationship) classification system (http://pantherdb.org/) of the intact N-glycoproteins corresponding to the 72 differentially expressed cell-surface intact N-glycopeptides in MCF-7/ADR CSCs showed that most of them are localized on the membrane or cytoplasm (based on UniProt database) (Fig. 5), where they mostly participate in the cellular and metabolic processes, while their molecular functions are mainly binding and catalytic activity.
Formation and proliferation of CSCs is the major hurdle of cancer therapy, and differentially expressed cell-surface N-glycoproteins on CSCs can not only serve as sorting and detection markers but also as drug targets. The intact N-glycoproteins corresponding to the differentially expressed intact N-glycopeptides of MCF-7/ADR CSCs (relative to MCF-7/ADR cells) identified in this study are valuable putative biomarkers for further verification and validation in real clinical samples (such as tumor tissues and patient sera) with this N-glycoproteomics or alternative orthogonal method. The cell-surface isotopic-labeling comparative N-glycoproteomics workflow reported in this study can be extended to any other type of CSCs.
Conclusions and perspectives
Following trypsin digestion, ZIC-HILIC enrichment, and isotopic diethyl labeling, the 1:1 mixture of intact N-glycopeptides from MCF-7/ADR CSCs and MCF-7/ADR cells was analyzed with C18-RPLC-MS/MS (HCD with stepped NCEs) on a Q Exactive Orbitrap MS. With forward-target DB search using GPSeeker and control of spectrum-level FDR ≤ 1%, 1,336 intact N-glycopeptides were identified; 176 out of the 301 N-glycosites were confirmed with GlcNAc-containing site-determining b/y fragment ion pairs, and the N-glycan moieties in 546 intact N-glycopeptide IDs were confirmed with structure-diagnostic fragment ions. The abundance of the Top3 isotopic peaks of the precursor ions was used for relative quantitation; with the criteria of ≥ 1.5-fold change and p value < 0.05, 72 cell-surface differentially expressed intact N-glycopeptides (DEGPs) were found in MCF-7/ADR CSCs relative to MCF-7/ADR cells, where 8 and 64 were downregulated and upregulated, respectively. Both upregulated EINGSK_N2H8F0S0 (atlastin-3, ATLA3) and downregulated TNLSGR_N2H8F0S0 (serine/threonine-protein phosphatase 4 regulatory subunit 3A, PPP4R3A) in MCF-7 CSCs (relative to MCF-7 cells) were found to be leveled off in MCF-7/ADR CSCs (relative to MCF-7/ADR cells).
References
Yang T, Xu FF, Sheng Y, Zhang W, Chen Y. A targeted proteomics approach to the quantitative analysis of ERK/Bcl-2-mediated anti-apoptosis and multi-drug resistance in breast cancer. Anal Bioanal Chem. 2016;408(26):7491–503. https://doi.org/10.1007/s00216-016-9847-7.
Zhang RX, Zhuang XY, Zong L, Liu S, Liu ZQ, Song FR. Investigations on the cell metabolomics basis of multidrug resistance from tumor cells by ultra-performance liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2016;408(21):5843–54. https://doi.org/10.1007/s00216-016-9696-4.
Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234(6):8381–95. https://doi.org/10.1002/jcp.27740.
Rajan P, Srinivasan R. Targeting cancer stem cells in cancer prevention and therapy. Stem Cell Rev. 2008;4(3):211–6. https://doi.org/10.1007/s12015-008-9037-x.
Kim BJ, Han C, Moon H, Kwon J, Jang IS, Lim SK, et al. Monitoring of post-mortem changes of saliva N-glycosylation by nano LC/MS. Anal Bioanal Chem. 2018;410(1):45–56. https://doi.org/10.1007/s00216-017-0702-2.
Kailemia MJ, Park D, Lebrilla CB. Glycans and glycoproteins as specific biomarkers for cancer. Anal Bioanal Chem. 2017;409(2):395–410. https://doi.org/10.1007/s00216-016-9880-6.
Hu WT, Su XM, Zhu ZK, Go EP, Desaire H. GlycoPep MassList: software to generate massive inclusion lists for glycopeptide analyses. Anal Bioanal Chem. 2017;409(2):561–70. https://doi.org/10.1007/s00216-016-9896-y.
Chen SY, Dong Q, Hu SS, Cai JX, Zhang WP, Sun JY, et al. Proteomic analysis of the proteins that are associated with the resistance to paclitaxel in human breast cancer cells. Mol BioSyst. 2014;10(2):294–303. https://doi.org/10.1039/c3mb70428a.
Tsang JYS, Huang YH, Luo MH, Ni YB, Chan SK, Lui PCW, et al. Cancer stem cell markers are associated with adverse biomarker profiles and molecular subtypes of breast cancer. Breast Cancer Res Tr. 2012;136(2):407–17. https://doi.org/10.1007/s10549-012-2271-6.
Hu YH, Yague E, Zhao J, Wang LY, Bai JC, Yang QX, et al. Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer. Cancer Lett. 2018;423:47–59. https://doi.org/10.1016/j.canlet.2018.02.036.
Hu Y, Guo R, Wei J, Zhou Y, Ji W, Liu J, et al. Effects of PI3K inhibitor NVP-BKM120 on overcoming drug resistance and eliminating cancer stem cells in human breast cancer cells. Cell Death Dis. 2015;6:ARTN e2020. https://doi.org/10.1038/cddis.2015.363.
Chen C, Okita Y, Watanabe Y, Abe F, Fikry MA, Ichikawa Y, et al. Glycoprotein nmb is exposed on the surface of dormant breast cancer cells and induces stem cell-like properties. Cancer Res. 2018;78(22):6424–35. https://doi.org/10.1158/0008-5472.Can-18-0599.
Arumugam P, Song JM. Quantitative evaluation of ABC transporter-mediated drug resistance based on the determination of the anticancer activity of camptothecin against breast cancer stem cells using TIRF. Integr Biol-Uk. 2016;8(6):704–11. https://doi.org/10.1039/c6ib00021e.
Xiao KJ, Tian ZX. GPSeeker enables quantitative structural N-glycoproteomics for site- and structure-specific characterization of differentially expressed N-glycosylation in hepatocellular carcinoma. J Proteome Res. 2019;18(7):2885–95. https://doi.org/10.1021/acs.jproteome.9b00191.
Xiao K, Tian Z. Site- and structure-specific quantitative N-glycoproteomics using RPLC-pentaHILIC separation and the intact N-glycopeptide search engine GPSeeker. Current protocols in protein science. 2019;97(1):e94. https://doi.org/10.1002/cpps.94.
Wang Y, Xu FF, Xiao KJ, Chen Y, Tian ZX. Site- and structure-specific characterization of N-glycoprotein markers of MCF-7 cancer stem cells using isotopic-labelling quantitative N-glycoproteomics. Chem Commun. 2019;55(55):7934–7. https://doi.org/10.1039/c9cc04114a.
Guler G, Balci S, Costinean S, Ussakli CH, Irkkan C, Suren D, et al. Stem cell-related markers in primary breast cancers and associated metastatic lesions. Mod Pathol. 2012;25(7):949–55. https://doi.org/10.1038/modpathol.2012.37.
Zhong H, Davis A, Ouzounova M, Carrasco RA, Chen C, Breen S, et al. A novel IL6 antibody sensitizes multiple tumor types to chemotherapy including trastuzumab-resistant tumors. Cancer Res. 2016;76(2):480–90. https://doi.org/10.1158/0008-5472.CAN-15-0883.
Chen Z, Yu Q, Hao L, Liu F, Johnson J, Tian Z, et al. Site-specific characterization and quantitation of N-glycopeptides in PKM2 knockout breast cancer cells using DiLeu isobaric tags enabled by electron-transfer/higher-energy collision dissociation (EThcD). Analyst. 2018;143(11):2508–19. https://doi.org/10.1039/c8an00216a.
Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J Proteome Res. 2005;4(2):628–32. https://doi.org/10.1021/pr049770q.
Tinholt M, Garred O, Borgen E, Beraki E, Schlichting E, Kristensen V, et al. Subtype-specific clinical and prognostic relevance of tumor-expressed F5 and regulatory F5 variants in breast cancer: the CoCaV study. J Thromb Haemost. 2018;16(7):1347–56. https://doi.org/10.1111/jth.14151.
Nomoto H, Yonezawa T, Itoh K, Ono K, Yamamoto K, Oohashi T, et al. Molecular cloning of a novel transmembrane protein MOLT expressed by mature oligodendrocytes. J Biochem. 2003;134(2):231–8. https://doi.org/10.1093/jb/mvg135.
Fukumoto H, Nishio K, Ohta S, Hanai N, Saijo N. Reversal of adriamycin resistance with chimeric anti-ganglioside G(M2) antibody. Int. J. Cancer. 1996;67(5):676–80. https://doi.org/10.1002/(Sici)1097-0215(19960904)67:5<676::Aid-Ijc14>3.0.Co;2-3.
Nazmeen A, Maiti S, Mandal K, Roy SK, Ghosh TK, Sinha NK, et al. Better predictive value of cancer antigen125 (CA125) as biomarker in ovary and breast tumors and its correlation with the histopathological type/grade of the disease. Med Chem. 2017;13(8):796–804. https://doi.org/10.2174/1573406413666170424155452.
Yu MC, Su LL, Zou L, Liu Y, Wu N, Kong L, et al. An essential function for beta-arrestin 2 in the inhibitory signaling of natural killer cells. Nat Immunol. 2008;9(8):898–907. https://doi.org/10.1038/ni.1635.
Bujnicki T, Wilczek C, Schomburg C, Feldmann F, Schlenke P, Muller-Tidow C, et al. Inhibition of Myb-dependent gene expression by the sesquiterpene lactone mexicanin-I. Leukemia. 2012;26(4):615–22. https://doi.org/10.1038/leu.2011.275.
Nakamura K, Abarzua F, Hongo A, Kodama J, Nasu Y, Kumon H, et al. The role of hepatocyte growth factor activator inhibitor-1 (HAI-1) as a prognostic indicator in cervical cancer. Int J Oncol. 2009;35(2):239–48. https://doi.org/10.3892/ijo_00000333.
Funding
This research was financially supported by the National Natural Science Foundation of China (21775110, 21575104) and Shanghai Science and Technology Commission (14DZ2261100).
Author information
Authors and Affiliations
Contributions
Conceptualization, Z. Tian, Y. Chen; cell culture, F. Xu, Y. Chen; LC-MS, Y. Wang; data analysis, Y. Wang, Z. Tian; draft, Z. Tian, Y. Wang, F. Xu.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 3.70 mb)
Rights and permissions
About this article
Cite this article
Wang, Y., Xu, F., Chen, Y. et al. A quantitative N-glycoproteomics study of cell-surface N-glycoprotein markers of MCF-7/ADR cancer stem cells. Anal Bioanal Chem 412, 2423–2432 (2020). https://doi.org/10.1007/s00216-020-02453-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02453-7