Abstract
γ-Aminobutyric acid (GABA) is a bioactive compound with diverse physiological functions. It has a wide range of applications in food and medicine and mainly biosynthesized through glutamate decarboxylase (GAD) catalysis. Bacillus subtilis is recognized for its robust secretion capabilities and high food safety standards, making it a prevalent choice for recombinant protein expression. In traditional industrial enzyme production in B. subtilis, antibiotics are required to sustain plasmid stability. However, the incorporation of antibiotics fails to align with the criteria applicable to enzymes for use in the food industry. To eliminate the need for antibiotics in the production of GAD preparations, we constructed a marker-free recombinant strain B. subtilis WS9D-GAD. This strain was developed based on antibiotic-free multi-copy gene expression vector pUBDAL-amyL and d-alanine racemase (dal)—deficient B. subtilis WS9D, ultimately enabling the food-grade expression of GAD. To enhance GAD expression levels, we integrated the gadA expression cassette into the genome of B. subtilis using the Cre/lox system method. Additionally, strain WS9C6D-GAD was generated through the co-expression of free plasmid and genome integration, which carried the free plasmid pUBDAL-gadA and featured six copies of the gadA expression cassette within its genome. The enzyme activity during shake flask fermentation reached 28.15 U mL−1, while the enzyme activity in high-density 3-L fermenter culture reached 199.49 U mL−1, marking the highest level of food-grade GAD expression with a multitude of potential applications. This study presents an effective strategy for the expression of food-grade industrial enzymes in B. subtilis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
γ-Aminobutyric acid (GABA) is a four-carbon non-protein amino acid that occurs widely in animals, plants, and microorganisms [1]. It serves diverse physiological roles, including diuretic effects, blood pressure regulation, intestinal microflora modulation, anxiety alleviation, and hormone secretion regulation [2,3,4,5]. Consequently, GABA is extensively applied in various aspects, including food, medicine, and other sectors [6]. GABA preparation methods encompass plant enrichment, chemical synthesis, enzymatic or whole-cell biocatalysis, and microbial fermentation. Notably, enzymatic or whole-cell biocatalysis holds significant promise due to its gentle reaction conditions, high catalytic efficiency, and environmental friendliness [7]. Glutamate decarboxylase (GAD), a crucial enzyme in GABA biosynthesis, is a type II decarboxylase with pyridoxal 5-phosphate (PLP) as a coenzyme. It catalyzes the irreversible decarboxylation of l-glutamic acid to generate GABA. Currently, lactic acid bacteria and yeast are predominantly employed for GABA production in the food industry, yet face challenges related to low yield [8, 9]. Furthermore, research indicates that GAD exhibits high expression levels in Escherichia coli [10]. Although recombinant enzymes and cells exhibit the capability for GABA synthesis, E. coli is a Gram-negative bacterium that synthesizes endotoxin during the advanced stages of fermentation, which constrains its application in the food industry [11].
Bacillus subtilis, recognized as a “Generally Recognized as Safe” (GRAS) microorganism, is extensively employed in industrial enzyme production due to its robust protein secretion capacity, well-defined genetic profile, ease of isolation and culture, and negligible codon bias [12, 13]. In the manufacturing of the majority of industrial enzymes, within the framework of recombinant bacterial culture utilizing free plasmid expression, antibiotics are introduced as a selective pressure to discern recombinant strains. This procedure is implemented to avert significant loss of recombinant plasmids during subculturing, thereby securing efficient production of the target product and maintaining stability in recombinant plasmids [14]. Nevertheless, the utilization of antibiotics not only elevates the production cost by necessitating the procurement of extra antibiotics in the preparation of industrial enzyme fermentation but also requires subsequent separation and removal efforts during product purification, thereby escalating challenges and costs in downstream processing. Significantly, the inclusion of antibiotics introduces potential food safety hazards, contravening the stipulated requirements for industrial enzymes in the food industry. [15]. Hence, there is a need to develop a recombinant strain that facilitates the efficient expression of exogenous proteins without the reliance on antibiotic screening markers.
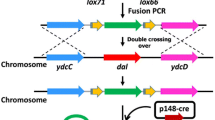
The current food-grade expression systems include toxin-antitoxin pairs [16], bacteriocin resistance [17], two-component cloning screening [18] and auxotroph complementation [19] as food-grade screening markers, along with genome-integrated expression to facilitate food-grade industrial enzyme expression. Among these, the auxotrophic complementation marker stands out due to its non-reliance on added screening media and its non-toxic nature, thereby avoiding adverse effects on organisms, Stable expression of foreign genes can be achieved using the method of genomic integrated expression. Notably, neither of these approaches requires the inclusion of antibiotics. These two methods are therefore examined in this study. The literature has documented continuous and repeatable genome editing systems for B. subtilis, comprising a gene editing system reliant on reverse screening markers [20, 21], a Cre/lox system based on site-specific recombination [22], and a gene editing system rooted in CRISPR technology [23]. The Cre/lox site-specific recombination system has been widely used in genetic engineering for gene knockout and exogenous gene integration due to its flexibility in genome editing, high specificity and no restriction to promoters [24, 25].
The Cre/lox site-specific recombination system is of tyrosine family origin and is derived from a 1029 bp gene sequence derived from the genome of E. coli bacteriophage P1 [26]. It operates with remarkable specificity on the 34 bp lox site independently, facilitating the deletion of genomic fragments and genome repositioning. By taking advantage of this feature, two homologous recombination sites lox71 and lox66, which can be recognized by Cre recombinase, were added at both ends of the resistance gene [22]. Subsequently, the resistance gene employed for screening homologous recombination events can be excised, resulting in the creation of a mutant devoid of the resistance marker gene and successfully edited on the genome can be obtained.
The addition of antibiotics during the GAD fermentation process limits the application of GABA in the food industry. This study aims to attain the high-efficiency expression of glutamate decarboxylase at a food-grade standard, establishing a robust groundwork for the industrial production of food-grade GABA. In this study, food-grade expression of GAD was accomplished using an alanine-deficient strain that employs the alanine racemase gene dal as a screening marker, obviating the need for additional screening media. To enhance the stability and efficiency of GAD expression, we employed the Cre/lox system for genome integration and then transformed the free expression plasmid into the integrated expression strain.
Materials and methods
Strains, plasmids, and media
The strains and plasmids used in this study are listed in Table 1. Recombinant expression plasmids were crafted using the frameworks of pHY300PLK-gadA-pdxH and pUBDAL-amyL. Specifically, the gadA (Gene ID:948027) originated from Escherichia coli str. K-12 substrain. MG1655. For cloning and plasmid preparation, B. subtilis SCK6D served as the host organism. For gene expression, the hosts employed were B. subtilis WS9D, B. subtilis WS9C, and B. subtilis WS9C6D.
The media compositions were as described: The Luria–Bertani (LB) medium consisted of 5 g L−1 of yeast extract, 10 g L−1 of tryptone, and 10 g L−1 of NaCl. The Terrific Broth (TB) medium contained 12 g L−1 tryptone, 24 g L−1 yeast extract, 5 mL·L−1 glycerol, 16.4 g L−1 K2HPO4·3H2O, and 2.3 g L−1 KH2PO4. GAD 3-L basic medium for high-density fermentation contained 5 g L−1 glycerin, 24 g L−1 industrial yeast powder, 12 g L−1 soytone, 2 g L−1 Na2SO3, 4 g L−1 NaH2PO4·H2O, 14.6 g L−1 K2HPO4, 1 g L−1 MgSO4·7H2O, 1 g L−1 (NH4)2-H-citrate, 2.68 g L−1 (NH4)2SO4, and 3 mL·L−1 trace element solution. The Composition of feeding medium is 40 mL·L−1 trace element solution, 300 g·L−1 glycerin, 63.36 g·L−1 (NH4)2HPO4, and 7.89 g·L−1 MgSO4·7H2O [27].
Chemicals and reagents
The Tianprep Mini Plasmid Kit and TIANgel Midi Purication Kit were purchased from Tiangen (Beijing, China); The 2× Phanta Flash Master Mix was purchased from Vazyme (Nanjing, China). The Q5® High-Fidelity DNA Polymerase was purchased from New England Biolabs (Ipswich, MA, USA); The One-Step PAGE Gel Fast Preparation Kit was purchased from Vazyme (Nanjing, China); Protein Marker and DNA molecular weight marker were purchased from Yeasen (Shanghai, China); γ-aminobutyric acid standard was purchased from Sigma-Aldrich (St. Louis, MO, USA); The OPA was purchased from Agilent (California, USA); The pyridoxine was purchased from VITA (Shanghai, China); All chemicals were reagent grade and obtained from China National Pharmaceutical Group Corp (Beijing, China).
Construction of expression plasmid and recombinant strain
The sequences of all primers are provided in Supporting Information Table S1. The gadA gene expression cassette was ligated with the pUBDAL-amyL vector, resulting in the construction of pUBDAL-gadA. Using pHY300PLK-gadA-pdxH as the template and P1/P2 as the primers, the gadA expression cassette including the pyridoxine phosphate oxidase (PNPO) gene pdxH was amplified by PCR. PCR amplification was conducted using pUBDAL-amyL as the template and P3/P4 primers to amplify the pUBDAL vector backbone fragment. Following the acquisition of the two fragments, a PCR polymer was generated via POE-PCR [28], and this PCR product was subsequently introduced into B. subtilis SCK6 competent cells. The recombinant expression plasmid pUBDAL-gadA was generated through recombination utilizing the endogenous enzyme system of B. subtilis. Subsequent to sequencing, electrotransformation into B. subtilis WS9D was performed to yield the recombinant strain WS9D-GAD.
Construction of B. subtilis WS9C6 (ΔnprE::gadA, ΔaprE::gadA, ΔnprB::gadA, ΔsrfA::gadA, Δmpr::gadA, Δbpr::gadA) strain
In pursuit of enhancing GAD expression stability in B. subtilis, the gadA expression cassette was integrated into the B. subtilis genome. PCR amplification of the left and right arms of the nprE gene was performed using B. subtilis WS9 as the template and P5/P6 as the primers. The tetracycline (Tet) resistance gene was PCR-amplified using pHY300PLK-gadA-pdxH as the template and P7/P8 as the primers, resulting in the lox72-Tet-lox66 expression cassette. The gadA expression cassette was amplified by PCR using pUBDAL-gadA as a template and P9/P10 as primers. Subsequently, the three fragments were recovered and subjected to overlapping PCR to create the gadA integration cassette, connecting it with the resistance gene and the homologous arm. The resulting cassette was introduced into WS9C-competent cells and then plated onto tetracycline (20 μg mL−1) resistant plates for selection. The transformed strains were verified and identified through PCR analysis with P10/P11 primers and subsequently sent to Suzhou Jinweizhi Biotechnology Co., Ltd for sequencing. The resulting strain, WS9C1-Tet, was successfully engineered with the integration of the nprE gene locus, gadA expression cassette, and the lox71-Tet-lox66 expression framework.
To further enhance the integrated copy number, it is necessary to eliminate the Tet resistance gene from the genome of the recombinant strain WS9C1-Tet. The Tet resistance gene can be deleted by the Cre/lox system and recombined between lox71 and lox66 using Cre recombinase [20]. The recombinant strain WS9C1 was used as the host into which the plasmid pE194-Cre was transferred into it. The expression of the Cre enzyme was induced by IPTG at a final concentration of 1 mM, and the plasmid pE194-Cre was removed at 51 °C for 12 h. Ultimately, the recombinant strain WS9C1, devoid of the resistance gene and pE194-Cre plasmid, was subjected to screening using the bacterial photocopy method. The newly constructed strain served as the host for the subsequent integration rounds, applying the same method. Sequential integration of the nprE, aprE, nprB, srfA, mpr, and bpr sites led to the formation of the recombinant strain WS9C6.
Knockout of alanine racemase (dal) gene
Employing WS9C6 as the host and the dal gene as the designated integration site, the gadA expression cassette was successfully integrated into the dal gene through the Cre/lox method. At the same time, the insertion of the gadA expression cassette resulted in the loss of activity of the dal gene, effectively achieving the knockout objective. Ultimately, the alanine-deficient recombinant strain WS9C6D was successfully generated.
Expression of GAD recombinant bacteria
The culture conditions at the shake flask level are as follows: The recombinant bacteria stored at −80 °C were inoculated into LB liquid medium with a volume fraction of 0.2%, and the constant temperature of the shaker was 37 °C, 200 r min−1 for 10–12 h. Subsequently, the culture was transferred to a TB fermentation medium at a 5% volume fraction and maintained on a thermostatic shaker at 37 °C and 200 r min−1 for 2 h. Pyridoxine (PN) was added to reach a final concentration of 0.2 mM. The introduction of the cost-effective precursor PN to the culture medium facilitates its conversion into PLP via the PLP remedial pathway catalyzed by PNPO oxidase. This enzymatic conversion results in the generation of coenzyme PLP, which plays a crucial role in preserving the stability of GAD and facilitating its proper folding. [29]. Subsequently, fermentation was conducted at 33 °C with agitation at 200 r min−1 for 36 h. The fermentation solution was used to determine the total OD600 of the bacteria. The bacterial suspension, with a cell density (OD600) of 5, underwent centrifugation at 12,000 rpm for 5 min to harvest the bacteria. Subsequently, the cells were resuspended in a citric acid-disodium hydrogen phosphate buffer (pH 5.0, 50 mM) containing 0.15 mM PLP. The cells were ultrasonically broken in ice water (power 110 W, 10 min), centrifuged at 12,000 rpm for 5 min, and the GAD enzyme activity was determined in the broken supernatant.
The culture conditions of high-density fermentation in a 3-L fermenter are as follows: The recombinant GAD recombinant strain was activated on the LB plate, and the monoclonal strain was inoculated into the LB liquid medium at 37 °C for 10 h and transferred to the 50 mL LB liquid medium with 0.2% inoculation amount. After an expanded culture at 37 °C for 10 h, it was inoculated into a 3-L fermenter containing 0.9 L of basal medium for high-density culture. The control parameters of the fermentation process are as follows: the fermentation temperature is 37 °C, the pH is 7.0, the dissolved oxygen is coupled to the rotation speed of the stirrer shaft, and the dissolved oxygen is set at 30%, and the dissolved oxygen rebounds (i.e., the rotation speed suddenly decreases and the dissolved oxygen suddenly increases). At this point, the feeding medium was started. Cell density (OD600) and GAD production were determined during fermentation.
Determination of GAD activity
The GAD enzyme activity assay was performed as follows 360 µL of 0.1 M monosodium glutamate monohydrate and 0.15 M PLP were dissolved in 50 mM pH 4.5 citric acid-disodium hydrogen phosphate buffer to prepare a substrate solution. The solution was preheated in a 37 °C water bath for 10 min. Subsequently, 40 µL of the disrupted supernatant was added and allowed to react for an additional 10 min. Following this, 600 µL of pH 10 boric acid buffer was introduced into the boiling water bath for an additional 10 min to terminate the reaction. The quantification of reaction products in the sample was performed using the HPLC-OPA pre-column derivatization method.
The HPLC detection conditions were as follows: GL Inertsil ODS-3 liquid chromatography column with a UV detector was used at a column temperature of 35 °C, with a flow rate of 0.8 mL min−1 and an injection volume of 10 µL. The mobile phase consisted of two components, mobile phase A and mobile phase B, and gradient elution was employed for detection. Mobile phase A: anhydrous sodium acetate 4.52 g L−1, 200 μL L−1 triethylamine, 5 mL L−1 tetrahydrofuran, pH 7.2 adjusted by acetic acid. Mobile phase B: anhydrous sodium acetate 22.6 g L−1/acetonitrile/methanol = 1:2:2 (v/v/v) The gradient elution program was as follows: 0–12.0 min, B phase was 8–60% (v/v), 12–14.0 min, B phase was 60–100% (v/v), 14–16.5 min, B phase maintained 100% (v/v), 16.5–18.5 min, B phase was 100–8% (v/v), 18.5–22.0 min, B phase maintained 8% (v/v); an enzyme activity unit was defined as the amount of enzyme required to catalyze the conversion of the substrate to 1 µmol GABA in 1 min.
Results and discussion
Food-grade expression of GAD based on dal screening marker
In the growth and metabolism of B. subtilis, d-alanine plays a crucial role in cell wall synthesis. In common fermentation media, alanine is mostly l-type, and d-alanine racemase is required to convert l-alanine to d-alanine [30]. Deletion of the d-alanine racemase gene (dal) in the B. subtilis genome has been shown directly or indirectly impact d-alanine supply, leading to incomplete cell wall growth or even the failure to synthesize, resulting in the strain of inability to grow [31]. At the same time, the insertion of the essential gene dal into the plasmid can complement the d-alanine-deficient, resulting in a screening pressure, which enables the screening of recombinant bacteria and the maintenance of the plasmid without the application of antibiotics. While a limited number of studies have demonstrated the feasibility and effectiveness of GAD expression at a food-grade level [32]. Nonetheless, a significant disparity remains between the production of food-grade GAD and the demands of the industrial sector.
In this study, to achieve the food-grade expression of GAD, we employed the alanine auxotrophic strain WS9D preserved in the laboratory was used as the host. We fused the gadA expression cassette with the pUBDAL vector that has complemented the d-alanine racemase production pathway (Fig. 1a). Consequently, we developed the expression strain WS9D-GAD, thereby establishing a comprehensive food-grade GAD expression system. The results of SDS-PAGE in shake flask fermentation are presented in Fig. 1b. revealing a distinct protein band within the 45–60 kDa range, in alignment with the anticipated 52.7 kDa molecular weight, and the enzyme activity of fermentation was 21.68 U mL−1.
Construction and expression of recombinant plasmids. a Construction of pUBDAL-gadA. b SDS-PAGE of GAD. M, GoldBand Plus 3-color Regular Range Protein Marker (8–180 kDa). Lane 1 and Lane 2 represent the intracellular supernatant and cell wall-disrupted precipitate of the B. subtilis WS9D-GAD strain, respectively
In this study, we conducted high-density fermentation of the recombinant strain WS9D-GAD within a 3-L fermenter to assess the expression of glutamate decarboxylase in relation to food quality. We controlled the culture medium and adjusted several parameters, introducing PN into the fermenter at 24 h intervals from the seed liquid until reaching a final concentration of 3 mM. As shown in Fig. 2, the intracellular GAD activity of the recombinant strain WS9D-GAD reached 109.71 U mL−1 after 42 h, which was 5.06 times higher than the shake flask level. It can be seen from the enzyme production curve that the GAD enzyme activity reached a peak at 42 h, but the expression of GAD was unstable during the later fermentation process. The utilization of the plasmid-based gene expression system was widespread in the fermentation of recombinant bacteria. Nonetheless, the instability of plasmids may result in diminished expression of the recombinant product [33]. The stability of the pUBDAL-gadA plasmid was 48% after 50 h, and the plasmid loss rate was higher. This may be attributed to two factors. First, during the replication process, plasmid undergoes internal structural rearrangements, leading to the generation of unstable single strands that result in inconsistent plasmid expression in the host. Second, as the fermentation cycle extends, additional d-alanine production pathways develop within the strain, reducing the selective pressure for d-alanine. Consequently, this compromises plasmid stability, resulting in lower expression levels in the later stages than in the early stages.
Food-grade expression of GAD based on genomic integration
To address the issue of unstable expression associated with free plasmids, genomic integration is a prevalent method for expressing foreign proteins [34]. Genomic integration involves employing homologous recombination to insert the target fragment into the genome for expression purposes. Subsequently, gene expression is replicated alongside chromosome replication, resulting in the acquisition of a recombinant strain exhibiting stable expression. Within a certain range, gene expression gradually increases with the increase of copy number, eliminating the need for additional antibiotics. To improve the expression stability of the GAD, the GAD was integrated into the genome of B. subtilis using the Cre/lox method (Fig. 3). Research has demonstrated that, in comparison to integrated vectors, linearized fragments facilitate easier integration into the genome. The process of homologous recombination is straightforward, and it eliminates the need for the repeated construction of vectors [35].
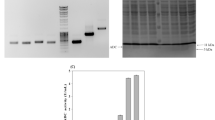
In this study, WS9C was used as the host, in which the srfC, spoIIAC, amyE, nprE, aprE, nprB, bpr, mpr and epr genes were knocked out, and the comk gene was integrated at the amyE site. This host has high transformation efficiency and strong recombinant protein expression ability, and the knockout and integration of the above gene sites have no significant effect on the growth of the strain. Initially, we employed Cre/lox system to integrate the GAD into the nprE site of the WS9C strain, yielding the recombinant strains WS9C1. Subsequently, we further integrated the GAD enzyme into the aprE, nprB, srfA, mpr, and bpr sites, yielding the recombinant strains WS9C2, WS9C3, WS9C4, WS9C5, and WS9C6, respectively. The results of PCR verification during the construction of the recombinant strains are presented in Fig. 4a. The size of the left and right homologous arm gene fragments is approximately 1000 bp, the gadA expression cassette measures 3385 bp, and the lox72 fragment is 34 bp in size. The gel electrophoresis pattern of the successfully integrated strain with the gadA integration cassette exhibited a size of approximately 5419 bp, while the original strain measured 2100 bp. Notably, lanes 2, 4, 6, 8, 10, and 12 correspond to the target fragments resulting from homologous recombination, and their sizes align with the theoretical values, confirming the successful integration of the gadA integration cassette at different sites. In shake flask experiments, the recombinant strains WS9C1 through WS9C6 displayed enzyme activities of 2.57, 4.28, 5.80, 7.85, 10.47 and 12.89 U mL−1, respectively (Fig. 4b). Notably, there was a gradual increase in GAD enzyme activity as the number of integrated copy sites increased. The results of SDS-PAGE analysis of enzyme solutions from shake flask fermentation are presented in Supporting Information Fig. S2. GAD enzyme electrophoresis band became thicker with the increase of enzyme activity, and no inclusion body was formed.
Integration verification and expression of the gadA integration cassette. a PCR results of colonies screened using the integrated gadA expression cassette: M, 10,000 bp standard DNA molecular weight marker; the colony PCR results of the original fragments of the reference strains WS9 nprE, aprE, nprB, srfA, mpr, and bpr gene loci were labeled as 1, 3, 5, 7, 9, 11, respectively. The results of colony PCR for the nprE, aprE, nprB, srfA, mpr, and bpr gene site in the integrated strain WS9C6-GAD were labeled as 2, 4, 6, 8, 10, 12, respectively. b Total enzyme activity (purple bars) and growth assessment (yellow bars) of each recombinant strain. Each strain was assessed after 36 h of incubation at 33 °C
Genome integration and recombinant plasmid co-expression improved the food-grade expression level of GAD
To enhance GAD expression, we employed a strategy involving genome integration and co-expression of recombinant plasmids. Genome integration serves as a stable and enduring method for gene expression, complementing the transient yet efficient expression facilitated by exogenous plasmids. The combined expression levels resulting from genomic integration and plasmid co-expression have the potential to surpass those achieved through single-genome integration or free plasmid expression alone, ensuring a stable and elevated expression of GAD. In this study, we utilized the WS9C6 strain with six genome integrations as the host. Through homologous recombination, we inserted the gadA expression cassette into the dal gene, resulting in the inactivation of the dal gene and the creation of a d-alanine deficient strain known as WS9C6D (Fig. S2). Additionally, we introduced the plasmid pUBDAL-gadA into this strain, yielding the recombinant strain WS9C6D-GAD, which effectively achieved genome integration and co-expression of the recombinant plasmid within B. subtilis.
At 36 h of shake flask fermentation, the intracellular enzyme activity of the recombinant strain WS9C6D-GAD was 28.15 U mL−1. And the OD600 reached 87.6 after 68 h of 3-L fermenter the GAD activity reached the highest value of 199.49 U·mL−1 (Fig. 5), representing an impressive 81.83% increase compared to the original food-grade expression strain WS9D-GAD, which was the highest expression level of food-grade expression of glutamate decarboxylase. In comparison with the 3-L fermenter fermentation of the recombinant strain WS9D-GAD, the highest enzyme activity exhibited a substantial 80% increase. The stability of the recombinant plasmid pUBDAL-gadA in WS9C6D-GAD was 54% at 59 h. However, during the middle and late stages of high-density fermentation, WS9C6D-GAD demonstrated relative stability, with a gradual upward trend. This phenomenon can be attributed to continuous and stable genome integration expression throughout the fermentation process, compensating for reduced expression resulting from plasmid instability.
Conclusion
In this study, we successfully generated a strain, WS9D-GAD, for expressing glutamate decarboxylase, utilizing d-alanine as the select marker, within the auxotrophic strain of B. subtilis. However, this strain encounters plasmid instability during the middle and late stages of 3-L fermenter fermentation, leading to reduced glutamate decarboxylase expression levels. To enhance expression stability, we employed the Cre/lox system to achieve genome integration and resistance-free expression of GAD. Building upon this foundation, we successfully accomplished genome integration and plasmid-free co-expression to enhance GAD expression levels. As a result, in the high-density fermentation culture of a 3-L fermenter, the GAD activity in the recombinant strain increased from 109.71 to 199.49 U mL−1, achieving the highest known level of food-grade GAD expression within a food-grade expression host. This successful attainment of efficient food-grade GAD expression holds significant promise for the industrial-scale production of food-grade γ-aminobutyric acid.
Data availability
The authors declare the availability of data supporting the findings in this study within the article. Raw or derived data associated with these findings can be obtained upon request from the corresponding author.
References
Hong J, Kim K. Crystal structure of γ-aminobutyrate aminotransferase in complex with a PLP-GABA adduct from Corynebacterium glutamicum. Biochem Biophys Res Commun. 2019;514:601–6. https://doi.org/10.1016/j.bbrc.2019.04.194.
Hong AR, Kim YA, Bae JH, et al. A possible link between parathyroid hormone secretion and local regulation of GABA in human parathyroid adenomas. J Clin Endocrinol Metab. 2016;101:2594–601. https://doi.org/10.1210/jc.2015-4329.
Xu N, Wei L, Liu J. Biotechnological advances and perspectives of gamma-aminobutyric acid production. World J Microbiol Biotechnol. 2017;33:64. https://doi.org/10.1007/s11274-017-2234-5.
Piao X, Jiang S, Wang J, et al. Pingchuan formula attenuates airway mucus hypersecretion via regulation of the PNEC-GABA-IL13-Muc5ac axis in asthmatic mice. Biomed Pharmacother. 2021;140: 111746. https://doi.org/10.1016/j.biopha.2021.111746.
Strandwitz P, Kim KH, Terekhova D, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4:396–403. https://doi.org/10.1038/s41564-018-0307-3.
Sarasa SB, Mahendran R, Muthusamy G, Thankappan B, Selta DRF, Angayarkanni J. A brief review on the non-protein amino acid, gamma-amino butyric acid (GABA): its production and role in microbes. Curr Microbiol. 2020;77:534–44. https://doi.org/10.1007/s00284-019-01839-w.
Luo H, Liu Z, Xie F, et al. Microbial production of gamma-aminobutyric acid: applications, state-of-the-art achievements, and future perspectives. Crit Rev Biotechnol. 2021;41:491–512. https://doi.org/10.1080/07388551.2020.1869688.
Yuan H, Zhang W, Xiao G, Zhan J. Efficient production of gamma-aminobutyric acid by engineered Saccharomyces cerevisiae with glutamate decarboxylases from Streptomyces. Biotechnol Appl Biochem. 2020;67(2):240–8. https://doi.org/10.1002/bab.1840.
Jia M, Zhu Y, Wang L, Sun T, Pan H, Li H. pH auto-sustain-based fermentation supports efficient gamma-aminobutyric acid production by Lactobacillus brevis CD0817. Fermentation. 2022;8(5):208. https://doi.org/10.3390/fermentation8050208.
Huang Y, Su L, Wu J. Pyridoxine supplementation improves the activity of recombinant glutamate decarboxylase and the enzymatic production of gama-aminobutyric acid. PLoS ONE. 2016;11(7): e0157466. https://doi.org/10.1371/journal.pone.0157466.
He W, Mu W, Jiang B, Yan X, Zhang T. Food-grade expression of d-psicose 3-epimerase with tandem repeat genes in Bacillus subtilis. J Agric Food Chem. 2016;64:5701–7. https://doi.org/10.1021/acs.jafc.6b02209.
Song Y, Nikoloff JM, Zhang D. Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis. J Microbiol Biotechnol. 2015;25:963–77. https://doi.org/10.4014/jmb.1501.01028.
Meissner L, Kauffmann K, Wengeler T, Mitsunaga H, Fukusaki E, Büchs J. Influence of nitrogen source and pH value on undesired poly (γ-glutamic acid) formation of a protease producing Bacillus licheniformis strain. J Ind Microbiol Biotechnol. 2015;42:1203–15. https://doi.org/10.1007/s10295-015-1640-7.
Popov M, Petrov S, Kirilov K, Ivanov GNI. Segregational instability in E. coli of expression plasmids carrying human interferon gamma gene and its 3′-end truncated variants. Biotechnol Biotechnol Equip. 2009;23:840–3. https://doi.org/10.1080/13102818.2009.10818553.
Peubez I, Chaudet N, Mignon C, et al. Antibiotic-free selection in E. coli: new considerations for optimal design and improved production. Microb Cell Fact. 2010;9:65. https://doi.org/10.1186/1475-2859-9-65.
Yang S, Kang Z, Cao W, Du G, Chen J. Construction of a novel, stable, food-grade expression system by engineering the endogenous toxin-antitoxin system in Bacillus subtilis. J Biotechnol. 2016;219:40–7. https://doi.org/10.1016/j.jbiotec.2015.12.029.
Li R, Takala TM, Qiao M, Xu H, Saris PEJ. Nisin-selectable food-grade secretion vector for Lactococcus lactis. Biotechnol Lett. 2011;33:797–803. https://doi.org/10.1007/s10529-010-0503-6.
Emond E, Lavallée R, Drolet G, Moineau S, LaPointe G. Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl Environ Microbiol. 2001;67:1700–9. https://doi.org/10.1128/AEM.67.4.1700-1709.2001.
Xia Y, Chen W, Zhao J, Tian F, Zhang H, Ding X. Construction of a new food-grade expression system for Bacillus subtilis based on theta replication plasmids and auxotrophic complementation. Appl Microbiol Biotechnol. 2007;76:643–50. https://doi.org/10.1007/s00253-007-1035-4.
Wang Y, Weng J, Waseem R, Yin X, Zhang R, Shen Q. Bacillus subtilis genome editing using ssDNA with short homology regions. Nucleic Acids Res. 2012;40: e91. https://doi.org/10.1093/nar/gks248.
Wenzel M, Altenbuchner J. Development of a markerless gene deletion system for Bacillus subtilis based on the mannose phosphoenolpyruvate-dependent phosphotransferase system. Microbiology (NY). 2015;161:1942–9. https://doi.org/10.1099/mic.0.000150.
Yan X, Yu H, Hong Q, Li S. Cre/lox system and PCR-based genome engineering in Bacillus subtilis. Appl Environ Microbiol. 2008;74:5556–62. https://doi.org/10.1128/AEM.01156-08.
Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18:67–83. https://doi.org/10.1038/s41579-019-0299-x.
Togawa Y, Nunoshiba T, Hiratsu K. Cre/lox-based multiple markerless gene disruption in the genome of the extreme thermophile Thermus thermophilus. Mol Genet Genomics. 2018;293:277–91. https://doi.org/10.1007/s00438-017-1361-x.
Polizzi KM, Bommarius AS, Broering JM, Chaparro-Riggers JF. Stability of biocatalysts. Curr Opin Chem Biol. 2007;11:220–5. https://doi.org/10.1016/j.cbpa.2007.01.685.
Chen H, Luo J, Zheng P, et al. Application of Cre-lox gene switch to limit the cry expression in rice green tissues. Sci Rep. 2017;7:14505. https://doi.org/10.1038/s41598-017-14679-0.
Zhang K, Su L, Duan X, Liu L, Wu J. High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb Cell Fact. 2017;16:32. https://doi.org/10.1186/s12934-017-0649-1.
You C, Zhang X, Zhang YHP. Simple cloning via direct transformation of PCR product (DNA multimer) to Escherichia coli and Bacillus subtilis. Appl Environ Microbiol. 2012;78:1593–5. https://doi.org/10.1128/AEM.07105-11.
Ling Q. Heterologous expression and fermentation optimization of E. coli glutamate decarboxylase in B. subtilis, and its application in preparation of GABA. Jiangnan University; 2021. https://doi.org/10.27169/d.cnki.gwqgu.2020.000254.
Chen J, Jin Z, Gai Y, Sun J, Zhang D. A food-grade expression system for d-psicose 3-epimerase production in Bacillus subtilis using an alanine racemase-encoding selection marker. Bioresour Bioprocess. 2017;4:9. https://doi.org/10.1186/s40643-017-0139-7.
Liu D, Zhang L, Xue W, Wang Y, Ju J, Zhao B. Knockout of the alanine racemase gene in Aeromonas hydrophila HBNUAh01 results in cell wall damage and enhanced membrane permeability. Fems Microbiol Lett. 2015;362:fnv89. https://doi.org/10.1093/femsle/fnv089.
Wangyang D, Bo J, Wanmeng M, Tao Z. Construction of a recombinant Bacillus subtilis and food-grade expression of glutamate decarboxylase. J Food Sci Biotechnol. 2020;39:24–31. https://doi.org/10.3969/j.issn.1673-1689.2020.02.004.
Zhang R, Yang Y, Wang J, Lin Y, Yan Y. Synthetic symbiosis combining plasmid displacement enables rapid construction of phenotype-stable strains. Metab Eng. 2019;55:85–91. https://doi.org/10.1016/j.ymben.2019.06.011.
Liu Y, Liu L, Li J, Du G, Chen J. Synthetic biology toolbox and chassis development in Bacillus subtilis. Trends Biotechnol. 2019;37:548–62. https://doi.org/10.1016/j.tibtech.2018.10.005.
Lambert JM, Bongers RS, Kleerebezem M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol. 2007;73:1126–35. https://doi.org/10.1128/AEM.01473-06.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32272264, 31730067, 31901633), the Natural Science Foundation of Jiangsu Province (BE2023686), the Fundamental Research Funds for the Central Universities (JUSRP122012), Shenzhen Institute of Synthetic Biology Scientific Research Program (DWKF20210004).
Author information
Authors and Affiliations
Contributions
HL: Conceptualization, methodology, investigation, formal analysis, and writing original draft. XY: methodology, formal analysis, and writing review & editing. TL: methodology, formal analysis, and writing review & editing. JW: methodology, writing review, and funding acquisition. SC: Methodology, supervision, project administration. KZ: Conceptualization, methodology, formal analysis, supervision, and writing review & editing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lv, H., Yu, X., Liu, T. et al. The high level food-grade expression of glutamate decarboxylase in Bacillus subtilis through combination of genomic integration and free plasmid. Syst Microbiol and Biomanuf 4, 1086–1095 (2024). https://doi.org/10.1007/s43393-023-00232-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-023-00232-w