Abstract
In this study, a total of 1125 actinobacteria were isolated from the selected mangrove species: Avicennia marina, Rhizopora mucronata and Ceriops tagal from three study stations viz., Minnie Bay, Carbyn’s Cove and Burmanallah. Among these three stations, the highest number of actinobacteria was recorded in Carbyn’s Cove (64.97%), followed by (25.51%) at Burmanallah and the minimum of (9.51%) was recorded in Minnie Bay. Maximum number of actinobacteria was recorded from Ceriops tagal (40.44%) than the other selected mangrove species Avicennia marina (34.13%) and Rhizopora mucronata (25.42%). Among the 1,125 mangrove-associated actinobacteria, 103 morphologically different isolates from the Minnie Bay station was selected for the further characterization studies. In antibacterial assay, 30.11% of the isolates revealed inhibitory activity against all tested clinical pathogens and 65% isolates displayed inhibitory activity against minimum of 04 tested clinical pathogens. Growth survival studies of the actinobacterial isolates also accomplished to withstand in varied NaCl and pH levels. Of 103 isolates, all were found to synthesize gelatinase enzyme, 73 isolates demonstrated amylolytic activity, 38 isolates exhibited proteolytic and 63 isolates displayed urease activity. Interestingly, 56 isolates exhibited excellent DNase activity and 71 isolates revealed positive for l-asparaginase production. To our recognition, 11 isolates exhibited constructive results in the production of 06 extracellular enzymes of industrial importance. Of 103 isolates, 48 isolates were confirmed by molecular level identification. Based on the phylogenetic analysis, the isolates were categorized under the genera: Streptomyces, Nocardiopsis, Salinispora and Actinomadura.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mangrove forests are among the world’s most productive eco-system that supplements coastal waters; yields commercial forest products, protect coastlines and support coastal fisheries. However, mangroves exist under condition of varying salinity, extreme tides, strong winds, high temperature and muddy anaerobic soils. Mangrove plants are somewhat woody that grow at the interface between land and sea in tropical and subtropical latitudes. These plants, and the associated microbes, plants and animals constitute the mangrove forest community and eco-system [1]. Mangroves provide a suitable habitat for commercial fish, crustaceans and wildlife species that contribute to sustaining the survival of local fish and shellfish populations [2].
Marine environments are largely untapped source for the isolation of new microorganisms with potentiality to produce bioactive secondary metabolites [3]. Among such microorganisms, actinobacteria are of special interest, since they are known to produce chemically diverse class of compounds with a wide range of biological activities [4]. The demand for new antibiotics continues to grow due to the rapid emerging of multiple antibiotic-resistant pathogens causing life-threatening infection [5].
Traditionally, actinobacteria have been isolated from the terrestrial sources only and the first report of mycelium-forming actinobacteria being recovered from marine sediments [6]. Recently, marine actinobacteria are recognized as a major source of novel antibiotic and anticancer agent with unusual structure and properties [7]. Actinobacteria represent a ubiquitous group of bacteria widely distributed in natural ecosystems.
Research in actinobacteria has gained prominence in recent years because of their potential in producing antibiotics [8]. Streptomycin, gentamicin, and rifamycin are some of the antibiotics which are being in use produced from actinobacteria. Actinobacteria play an important role in the field of agriculture also. Previous study showed that actinobacteria isolated from Malaysian soil have the potential to inhibit the growth of several plant pathogens [9]. Oskay et al. [10] also reported about the ability of actinomycetes isolated from Turkey’s farming soil to inhibit Erwinia amylovora, a bacterium that causes fire blight disease in apple and Agrobacterium tumefaciens, an agent for crown gall disease [9].
The Andaman & Nicobar (A & N) Islands marine ecosystems are mostly unexplored, and may provide a rich source of microorganisms producing novel and efficient antimicrobial compounds [3]. Only limited research on marine actinobacteria from A & N Islands has been reported. Rather, these Islands are an unexploited part of Indian seas and have rarely been explored for microbial diversity research and their metabolites. Hence, there is an immense possibility to prospect and functionally characterize novel marine actinobacteria to identify potential bioactive compounds [11]. No studies have been reported on the biodiversity of mangrove-associated actinobacteria from Port Blair, South Andaman, A & N Islands. Hence, there is a possibility to identify and characterize the mangrove-associated actinobacteria for novel bioactive compounds. There are 37 true species of mangroves distributed among 10 families and 14 genera throughout the A & N Islands [3]. The aim of the present study is to isolate and functionally characterize the mangrove-associated actinobacteria of industrial and pharmaceutical interest with the ultimate objective of discovering novel bioactive compounds.
Materials and methods
Sample collection
Mangrove root and associated sediment samples were collected from three stations of South Andaman region: Minnie Bay (Station-I), Carbyn’s Cove (Station-II) and Burmanallah (Station-III) (Fig. 1). Dominant mangrove species in all 3 stations, such as Avicennia marina, Rhizopora mucronata, and Ceriops tagal, were selected. The sediment and root samples were collected in January, 2019 during low tide and the sediment samples were collected from 5 to 10 cm. The samples were stored in ice boxes and transported immediately to the laboratory for further processing.
Isolation of mangrove-associated actinobacteria
One gram of sediment and root samples was processed for enumeration of associated actinobacteria. The samples were serially diluted (10–1-10–4) with sterile seawater. Isolation of actinobacteria was performed as described previously by [12] with starch casein agar (SCA) medium contained soluble starch 10 g, vitamin free casein 0.3 g, KNO3 2 g, NaCl 2 g, K2HPO4 2 g, MgSO4.7H2O 0.05 g, CaCO3 0.02 g, FeSO4.7H2O 0.01 g, agar 20 g, pH 7.0 ± 0.2 [13], Bennet’s agar (yeast extract 1.0 g, beef extract 4.0 g, casein enzyme hydrolysate 2.0 g, dextrose 10.0 g, agar 15.0 g, pH 7.3 ± 0.2) and Emerson agar (yeast extract 1.0 g, beef extract 1.0 g, peptic digest of animal tissue 2.0 g, dextrose 10.0 g, sodium chloride 2.5 g, agar 15.0 g pH 7.0 ± 0.2), with 50% aged sea water. The medium was supplemented with nalidixic acid 50 μg/mL (Hi-Media, Mumbai) and Nystatin 25 μg/mL (Hi-Media, Mumbai) to inhibit the fungal and fast-growing Gram-negative bacteria. The plates were incubated at room temperature for 21 days. The appearance and growth of marine actinobacteria were monitored regularly. The colonies were recognized by their characteristic chalky to leathery appearance on the culture plates. After incubation, morphologically diverse actinobacterial colonies were picked and further sub-cultured onto respective isolation medium. The colonies were purified using SCA and ISP-2 medium, once the pure colonies were obtained, each colony was further identified on the basis of its earthy like smell, colony morphology, color of hyphae and the presence or absence of aerial and substrate mycelium. The selected and identified colonies of actinobacteria were sub-cultured in SCA slants for further studies. The pure cultures were also preserved in 20% glycerol vials and stored at −80 °C for long-term preservation [14].
Morphological and cultural characteristics
Morphological studies of the actinobacterial isolates were determined by following the International Streptomyces Project (ISP) procedures [15]. The yeast and malt extract agar (ISP-2), oat meal agar (ISP-3), inorganic salts-starch agar (ISP-4), glycerol-asparagine agar (ISP-5) media were used for morphological studies and color determinations. These observations include some basic tests, aerial mass color, reverse side pigment, melanoid pigments, spore chain morphology and spore morphology.
Aerial mass color
Aerial mycelium is one of the important characters for the identification of endophytic actinobacteria. The isolated strains were inoculated on ISP 2–7 medium and the plates were incubated at 28 °C for 6–7 days. After incubation, the nature of colonies were observed. Basically, color of the matured spore forming aerial mycelium is white, red, gray, blue and green. Sometimes the aerial mycelium is also present in combination of two colors. Accordingly, both the colors were also recorded. Sometimes aerial mass color of a strain may reveal intermediate tints; besides, both the color series should be noted.
Reverse side pigments
The actinobacterial strains were also classified based on their ability to produce pigments on the reverse side of the colony known as distinctive (positive) and not distinctive or none (negative). Pale yellow of chroma and yellowish brown color of the growth were also recorded as positive (P) and no color of the plates was recorded as negative (N).
Melanoid pigments
The isolated actinobacterial strains were inoculated on the ISP-5 medium and incubated at 28 °C for 4–5 days for identification of the melonoid pigmentation. After incubation, the positive strains of the cultures showed greenish brown, brown to black diffusible pigment or a distinct brown pigment was recorded as positive (P). The absence of the pigment was recorded as negative (N) [15]. The strains with greenish brown to black diffusible pigment or a distinct brown pigment were recorded as positive ( +). Absence of diffusible pigment was recorded as negative (−).
Spore surface morphology
Spore surface features can be observed under the scanning electron microscope (SEM). The coverslip culture technique prepared for observation under the light microscope may be used for this purpose. The electron grid is cleaned and adhesive tape is placed on the surface of the grid. The spore structures in actinobacteria can be reported as: smooth (sm), spiny (sp), warty (wa) and hairy (ha).
Physiological and biochemical characterization
Assimilation of carbon sources
The ability of different actinobacterial strains in utilizing various carbon compounds as source of energy was studied by following the method recommended in International Streptomyces Project. Carbon utilization medium was modified from [16]. The stock solution containing the 10 × concentration of different carbon sources i.e., d-glucose, l-arabinose, sucrose, d-fructose, d-xylose, raffinose, d-mannitol, cellulose, rhamnose, inositol was prepared on double distilled water and filtered using 0.22 mm pore size membrane filter and stored at 4 °C for further use. The strains were streaked in sterile basal medium with 1% of carbon sources and the plates were incubated at 28 °C for 7–15 days. The growth of actinobacteria was identified depending on the utilization of carbon sources. Positive results were recorded as positive (P), and negative results as negative (N).
Hydrogen-sulfide production
Tryptone–yeast extract agar slants with actinobacterial strains were incubated for 7 days at room temperature. Presence of the characteristic greenish brown, brown, bluish black or black color of the substrate, indicates H2S production after 7th, 10th and 15th day of incubation.
Sodium chloride tolerance assay
Different concentrations of sodium chloride (0, 5, 10, 15, 20, 25 and 35%) solution were added to the ISP-2 medium to check the sodium chloride tolerance capability of the isolates. This test was very much important to understand the native nature of mangrove-associated actinobacterial isolates. The isolates were streaked on the agar medium and incubated at 37 °C for 07–15 days. The presence or absence of growth was recorded on 7th day onwards.
pH tolerance assay
The pH of the ISP-2 medium was adjusted to (5, 6, 7, 8, 9, 10 and 11) with 0.1 N NaOH/0.1 N HCl. The isolates were inoculated and incubated at 28 °C for 7 days. After incubation, the positive and negative indications of the culture were determined.
Effect of temperature on growth
Ability of the actinobacterial isolates to grow at different temperatures was studied at 10 °C, 15 °C, 20 °C, 25 °C, 30 °C, 35 °C, 40 °C, 45 °C, 50 °C, 55 °C, 60 °C, 65 °C, 70 °C, 75 °C and 80 °C. The isolates were inoculated in to the ISP-1 broth and incubated at different temperatures [17]. The results were documented after 7–8 days of incubation.
Screening for antibacterial activity
Isolates collected from Minnie Bay were screened for antibacterial activity by cross-streak method [18]. Briefly, the isolates were streaked on SCA medium as a straight line in the left side corner of the Petri plate and were incubated at room temperature for 5 days. After observing a good ribbon-like growth of the actinobacterial isolates, overnight cultures of Proteus mirabilis MTCC1429, Bacillus halodurans MTCC443, Vibrio cholerae MTCC3904, Streptococcus pneumoniae MTCC1935, Enterococcus faecalis MTCC439, Pseudomonas aeruginosa MTCC424, Bacillus subtilis MTCC441, Staphylococcus aureus MTCC96, Shigella flexineri MTCC1457 and Micrococcus luteus MTCC1541 were streaked in separate plates at the right angle of actinobacterial cultures. The plates were again incubated at 28 °C for 48 h and the zone of inhibition was observed. SCA plates without actinobacteria, but with simultaneous streaking of test organisms were maintained as control.
Screening of actinobacteria for extracellular enzymes
Amylase activity/Starch hydrolysis
Mangrove-associated actinobacteria were inoculated in the ISP-2 medium supplemented with 1% starch and incubated at 28 °C for 5–7 days. After incubation, the plates were flooded with Lugol’s iodine solution, starch in the presence of iodine will impart a blue black color to the medium indicating the absence of starch splitting enzymes representing a negative result. However, the presence of clear colorless zone surrounding growth of the organism represents a positive result for starch hydrolysis [19].
Lipolytic activity
The lipolytic activity of isolates was performed by the method reported in Shirling and Gottileb [15]. The assay was performed in ISP-2 medium with 1% Tween 20 and the plates were incubated at room temperature for 7 days. After incubation, the presence of visible halo around colonies was recorded as positive reaction.
Cellulase activity
Cellulase activity of the isolates was carried out as described previously by [20]. Briefly, the ISP-2 medium was supplemented with 1% of carboxy methyl cellulose (CMC) and the isolates were cross-streaked in the plates and incubated for 7 days at room temperature. The positive results were recorded by the presence of clear zone around the isolates after flooded with Lugol’s iodine solution.
Gelatinase activity
Gelatin hydrolysis was conducted by sub-culturing the actinobacterial isolates on gelatin agar medium and inoculated at 28 °C for 7 days. After incubation, the plates were flooded with 10 ml of mercuric chloride solution and zone of hydrolysis was recorded [21].
Urease activity
Urease activity of the isolates was carried out as described by Leon et al. [20]. Briefly, the isolates were inoculated in the Christenson base broth supplemented with 40% (w/v) urea solution and incubated at room temperature for 7 days. The change in the medium color from yellow to pink indicates the positive reaction.
DNAse activity
DNase assay was performed with the actinobacterial isolates using DNase agar medium (Hi-Media, Mumbai) with 0.2% DNA and 0.1% toluidine blue [20]. The capability of isolates to hydrolyze DNA was confirmed by the blue precipitate formation around the colonies.
l-Asparaginase activity
Production of L-asparaginase by mangrove-associated actinobacteria was performed in the medium reported previously by Gulati et al. [22]. Production medium containing, asparagine dextrose salts agar (asparagine, 1.0%,dextrose, 0.2%; K2HPO4, 0.1%; MgSO4, 0.05%; agar, 1.5%) was supplemented with different concentrations of phenol red as the indicator and control plates were maintained without asparagine and phenol red. Diameter of clear pink zone around the colony was measured and documented after 7 days of incubation.
Casein hydrolysis
The proteolytic activity of isolates was studied using milk casein agar (g/L) peptone 1.0, sterile skimmed milk (5%), and agar 20.0. The isolates were streaked on milk casein agar and incubated at 28 °C for 5–7 days. Following incubation, isolates secreting protease enzyme will exhibit a zone of proteolysis, which is demonstrated by clear zone surrounding their growth. This loss of opacity is the result of a hydrolytic reaction yielding soluble, non-colloidal amino-acids and it represents a positive for casein hydrolysis [19].
Molecular identification of mangrove-associated actinobacteria
DNA isolation
Genomic DNA of mangrove-associated actinobacteria was isolated by following the modified procedure of Kutchma et al. [23]. Briefly, 2 ml of 72 h culture broth was centrifuged at 8,000 rpm for 5 min at room temperature and the pellets were washed with 1 ml TE buffer, suspended in TE buffer containing lysozyme with the final concentration of 4 mg/ml. The mixture was incubated at 37 °C water bath for 3 h. Subsequently, 75 μL of 10% SDS and 125 μL of 5 M NaCl were added to cell pellet and incubated at 37 °C for 30 min. Reaction tubes were later incubated at − 40 °C for 5 min and subsequently to 65 °C water bath for 3 min. This step was repeated 3 times and the supernatant was collected by centrifugation at 8000 rpm for 10 min at room temperature. To the supernatant, 50 μg/mL Proteinase K and 200 μg/ml RNase were added and incubated at 37 °C for 30 min. Equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) was added to the solution and mixed by inversion. After centrifugation at 8000 rpm for 5 min, upper aqueous phase containing DNA was recovered and precipitated with 2 volumes of 95% ethanol by centrifugation at 13,000 rpm for 15 min. DNA pellet was dissolved in 50 μl of TE buffer and stored at − 40 °C for further use.
PCR amplification of 16S rDNA
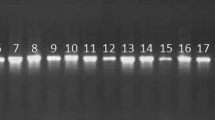
Amplification of 16S rDNA was performed using universal primers 16Sf (5′ AGAGTTTGATCCTGGCTCAG 3′) and 16Sr (5′ GGTTACCTTGTTACGACTT 3′). Final volume of reaction was 25 μl, which comprised Taq buffer (10 ×), dNTP‟s (200 μM) (MBI Fermentas, USA), forward and reverse primer (0.5 μM), MgCl2 (1.0 mM), Taq DNA polymerase (1.25 U; MBI Fermentas), template (1 μl) and remaining with autoclaved Milli Q water. PCR was performed with the initial denaturation at 98 °C for 3 min, followed by 30 cycles of reaction with denaturation at 94 °C for 1 min; annealing at 53 °C for 1 min; extension at 72 °C and final extension at 72 °C for 10 min. PCR amplicons were analyzed on 1.5% agarose gel along with DNA molecular weight marker (MBI Fermentas). Positive amplicons as judged by size were purified using QIAquick PCR purification kit (Qiagen, Germany) and sequenced on an ABI PRISM 377 genetic analyzer (Applied Biosystems, USA).
Phylogenetic analysis
16S rDNA sequences of the mangrove-associated actinobacteria were aligned manually in GenBank database with BLAST [24] and the sequences with 100–98% homology were considered for molecular taxonomy analysis. Multiple alignments of 16S rDNA sequences in this study and the sequences in GenBank database were performed with CLUSTAL × program [25]. Phylogenetic trees were constructed by neighbor-joining and maximum-parsimony tree making methods in Molecular Evolutionary Genetic Analysis (MEGA version 5.0) [26] and bootstrap values based on 1,000 replication [27].
Results
Diversity of mangrove-associated actinobacteria in Port Blair Bays
A total of 1,125 actinobacteria were isolated from the selected mangrove species, Avicennia marina, Rhizopora mucronata and Ceriops tagal from the three study stations, namely Minnie Bay (Station-I), Carbyn’s Cove (Station-II) and Burmanallah (Station-III). Enumeration and isolation of actinobacteria were performed using three different isolation media, starch casein agar, Bennet’s agar and Emerson agar with 50% aged sea water. Among these three stations, the highest number of actinobacteria was recorded in Carbyn’s Cove 731 nos. (64.97%), followed by 287 (25.51%) at Burmanallah and the lowest number of 107 (9.51%) was recorded in Minnie Bay (Fig. 2). Maximum number of actinobacteria was recorded from Ceriops tagal 455 (40.44%) than the other selected mangrove species (Avicennia marina 384 (34.13%), Rhizopora mucronata 286 (25.42%)) (Fig. 3). Among the 1,125 actinobacteria, maximum isolates 635 (56.44%), were isolated with Emerson’s agar than the starch casein agar and Bennet’s agar (Fig. 4). Of 103 actinobacteria isolated from Minnie Bay station, maximum number was isolated with Bennet’s agar (46 (44.66%) than the other two isolation media. A total number of 91 (88.34%) isolates were recovered from Ceriops tagal, in which Bennet’s agar scored maximum count 42 (46.15%) followed by 31 (34.06%) with Emerson’s agar and the lowest count was recorded in starch casein agar 18 (19.78%). Among 10 isolates from Avicennia marina, Bennet’s agar and Emerson’s agar scored maximum count 4 (40.00%) and the lowest count was recorded in starch casein agar 2 (20.00%). A total of 06 actinobacteria were isolated from the Rhizopora mucronata, in which, starch casein agar scored maximum count 4 (66.66%) followed by 2 (33.33%) isolates with Bennet’s agar and no isolates were recorded in the Emerson’s agar. Among the 1,125 mangrove-associated isolates, 103 morphologically different isolates from the Minnie Bay station were selected for the further characterization studies.
Phenotypic characterization and cultural characteristics
Cultural characteristics of the mangrove-associated actinobacteria were studied based on the guidelines of International Streptomyces Project. Among the 103 mangrove-associated actinobacteria from Minnie Bay, 97.08% (n = 100) of the isolates exhibited good growth on tryptone–yeast extract agar (ISP-1), 86.40% (n = 89) exhibited excellent growth on yeast extract-malt extract-dextrose agar (ISP-2), 73.78% (n = 76) of the isolates were exhibited luxuriant growth on glycerol–asparagine agar (ISP-5), 1.94% (n = 2) and 0.00% (n = 0) of the isolates exhibited moderate growth on starch inorganic salts agar (ISP-4) and tyrosine agar (ISP-7) and poor mycelial growth rate was recorded in peptone yeast extract iron agar (ISP-6). The substrate mycelium ranges from greenish white (ISP-1 and ISP-2) to bluish white (ISP-3, ISP-4, ISP-5 and ISP-7) or whitish to brown in starch casein agar.
Aerial mass color
The color of substrate mycelium was determined by observing the culture plates after 7 to 10 days. Of 103 isolates, eight color series were observed, white, whitish ash, bluish white, greenish white, ash, bluish green, creamy and brown. Whitish ash color series was dominated in 27 (26.21%) followed by ash-colored actinobacteria 27 (26.21%), white 20 (19.41%), bluish white 8 (7.76%), greenish white 7 (6.79%), bluish green 7 (6.79%), creamy 6 (5.82%) and only one brown-colored actinobacteria was isolated and maintained. Among 27 of each whitish ash and ash-colored actinobacteria from the Minnie Bay station, maximum 21 (20.38%) number of isolates were recorded from the Ceriops tagal, followed by Avicennia marina 3 (2.91%) and 3 (2.91%) from Rhizopora mucronata. Of 10 ash-colored actinobacteria, 26 (25.24%) were from Ceriops tagal, 1 (0.97%) from Avicennia marina, and none of the ash-colored actinobacteria were recorded from the Rhizopora mucronata. Of 08 bluish white actinobacteria, 07 (6.79%) from Ceriops tagal and 01 (0.97%) from Avicennia marina and no ash-colored actinobacteria were recorded from the Rhizopora mucronata. One brown-colored actinobacteria was also isolated from Ceriops tagal.
Reverse side pigment, melanoid pigments and soluble pigments
The isolates were divided into two groups according to their ability to produce pigments on the reverse side of the colony, namely distinctive ( +) and not distinctive or none (−). Reverse side pigments and melanoid pigmentation were observed by the formation of greenish brown, brownish black or distinct brown pigment. Colors observed for non-distinctive were pale yellow, olive or yellowish brown color considered as negative. Most of the isolates revealed pigment formation. A total of 39 (37.86%) isolates revealed the distinctive ( +) pigment production, and the melanoid pigment production was positive for 37 (35.92%) of isolates. Maximum numbers of pigment-producing-associated actinobacteria were isolated from the Ceriops tagal (33.98%), followed by Avicennia marina (0.97%) and Rhizopora mucronata (0.97%). Soluble pigment production was also observed among the isolates and the results were recorded as present ( +) and absent (−). Among them, 20 isolates were shown positive result for soluble pigment production. Maximum number of soluble pigment-producing actinobacteria was isolated from Ceriops taga1 18 (17.47%), followed by 2 (1.94%) from Rhizopora mucronata and none of the pigment-producing actinobacteria were isolated from Avicennia marina.
Physiological and biochemical characteristics
Assimilation of carbon sources
Ability of actinobacterial isolates in utilizing various carbon compounds as source of energy was experimented by following the method recommended in International Streptomyces Project. After comparing growth with negative and positive control, it was observed that raffinose was the most assimilated carbon source by all actinobacterial isolates and the arabinose and xylose were least assimilated carbon source. Of all actinobacterial isolates, 30.09% of the isolates revealed good growth in all carbon sources and 08.73% of the isolates recorded least utilization of the carbon sources. However, MBCT 17A, MBCTS 35B, MBCT 17B and MBCTS38 have shown very good assimilation of arabinose and xylose as a carbon source in comparison to other strains and other carbon sources. Of 90 associated actinobacteria from Ceriops tagal, 77.77% isolates revealed positive for raffinose, 74.44% were positive for inositol, 73.33% of isolates were positive for glucose and cellulose, 72.22% of the isolates for sucrose and mannitol 70% and 60% of the isolates were positive for xylose and fructose. Minimum of (45.55%) isolates recorded positive for arabinose utilization. Of all actinobacteria from Avicennia marina (n = 9), 55.55% of isolates were positive for arabinose, 44.44% of isolates were positive for sucrose, fructose, xylose inositol, raffinose, mannitol and cellulose, minimum of (33.33%) isolates were positive for glucose utilization. Isolates from Rhizopora mucronata (n = 4), 75% of isolates were positive for sucrose fructose, inositol and raffinose, 50% were positive for glucose, xylose, mannitol and cellulose. All isolates revealed negative for arabinose utilization.
Hydrogen sulfide production
Production of hydrogen sulfide by the mangrove-associated actinobacteria was observed after 7th, 10th and 15th day of incubation. By comparing the presence of bluish black and black color slants to the control slants, observations were taken. Among 103 isolates, 53.39% shown positive results and 46.60% isolates revealed negative results for hydrogen sulfide production. Of 90 isolates from Ceriops tagal, 53 isolates were positive for H2S production and minimum number (01) was recorded from the Rhizophora mucronata.
Sodium chloride tolerance test
To study the sodium chloride tolerance range among the isolated actinobacteria, 06 concentrations of sodium chloride were supplemented in the test medium along with the control medium with only distilled water and neither seawater nor sodium chloride was added. Of 103 isolates, all revealed negative result (no growth) at 0% NaCl concentration. With 0.5% sodium chloride, of 103 isolates, 91 (88.34%) exhibited good growth, 04 (3.88%) shown moderate growth, and 08 (7.76%) showed negative result. With 1.0% NaCl concentration, 87 (84.46%) showed good growth, 10 (9.70%) isolates exhibited moderate growth and 06 (5.82%) isolates revealed negative result. It was also observed that, 84 (81.55%) isolates revealed good growth at 2.0% sodium chloride, whereas 15 (14.56%) showed moderate growth and 04 (3.88%) disclosed negative result in the medium. In 5.0% NaCl concentration, 61 (59.22%) showed good growth, 34 (33.00%) exhibited moderate growth and 08 (7.76%) gave negative result. With 10% NaCl concentration, 46 isolates (44.66%) exhibited good growth, 15 (14.56%) isolates shown moderate growth and 57 (55.33%) revealed negative result. In 15% NaCl concentration, only 17 (16.50%) isolates were able to exhibit good growth and 6 (5.82%) showed moderate growth and 86 (83.49%) were not able to tolerate the higher concentration of sodium chloride (Fig. 5).
Ability to grow in different pH
All isolates revealed acceptable pH tolerance growth at different ranges of pH 5.0 to pH 11.0. Of the total isolates, 21.35% revealed positive growth on all pH ranges. This depicts their tolerance to grow in acidic and alkaliphilic conditions. At pH 5.0, 7.76% isolates revealed negative result and all the isolates were revealed positive growth in the pH range of 8.0–11.0. Isolates from the Ceriops tagal revealed optimum growth at the pH range of 8.0–9.0. Isolates from Avicennia marina exhibited excellent growth at the pH range of 7.0–9.0 and the isolates from the Rhizopora mucronata revealed optimum growth at the pH range of 7.0–11.0 (Fig. 6).
Effect of temperature on growth
Different types of growth were recorded in 103 mangrove-associated actinobacteria, depending on the temperature level. All isolates exhibited good growth in the temperature range of 25–30 °C and 66.01% of the isolates were found grown at 50 °C. Of the total isolates from Minnie Bay, 33.00% were observed with moderate to fair growth at 50–60 °C. All isolates revealed negative growth in the temperature range of 55–80 °C. Excellent pigment production was observed by the isolates; MBAMS 2, MBCTS 39B, MBCT 17A, MBCT 17B, MBCTS 35A, MBCT 102 and MBRM 92 in different temperature ranges 20 °C, 30 °C, 35 °C, 15 °C, 30 °C, 25 °C and 25 °C (Fig. 7). Of the total actinobacteria from Ceriops tagal, 83.43% isolates revealed excellent growth at 25–35 °C, 78.64% with moderate growth at 15 °C, 20 °C, 40 °C and 50 °C. Deprived growth rate was recorded in 10 °C and 15 °C. Among the Avicennia marina-associated actinobacteria, all isolates revealed excellent growth at 25–30 °C, 77.77% isolates showed moderate growth at 15–20 °C, poor growth rate was recorded in the temperature range of 35 °–50 °C. Of the total actinobacteria from Rhizopora mucronata, 100% of isolates revealed excellent growth at 20–35 °C, 21% isolates with moderate growth at 15 °C and 35 °–40 °C, poor growth rate was recorded at 10 °C and 45 °–80 °C temperature range.
Screening for antibacterial activity
Antibacterial assay was conducted by cross-streak method against 09 bacterial pathogens viz., Proteus mirabilis MTCC1429, Bacillus halodurans MTCC443, Vibrio cholerae MTCC3904, Enterococcus faecalis MTCC439, Pseudomonas aeruginosa MTCC424, Bacillus subtilis MTCC441, Staphylococcus aureus MTCC96, Shigella flexineri MTCC1457 and Micrococcus luteus MTCC1541. The extent of antibacterial activity against 09 clinical pathogens was varied among the actinobacterial isolates (Fig. 8). Of 103 isolates, 19.41% exhibited appreciable inhibitory activity against Gram-negative bacteria and 50.48% against Gram-positive bacteria. Remaining 30.11% revealed excellent antibacterial activity against both Gram-positive and Gram-negative bacteria. Total proportion of inhibition by actinobacteria against pathogenic bacteria was noted as Proteus mirabilis—75.72%, Bacillus halodurans—76.69%, Enterococcus faecalis—91.26%, P. aeruginosa—95.14%, Bacillus subtilis—88.34%, S. flexneri—71.84%, V. cholerae—88.34%, Micrococcus luteus—90.29%, S. aureus—86.40%. A total of 93 (90.29%) actinobacteria disclosed antibacterial activity toward any one of the tested bacteria and 10 (9.70%) actinobacteria presented no antagonistic activity. Among the actinobacteria, isolate from Ceriops tagal (93.33%) has shown good antibacterial activity against the tested bacterial pathogens. Growth of P. aeruginosa was inhibited by most of the strains, S. flexneri was the least-inhibited pathogen and moderate inhibition rate was observed against Bacillus halodurans. Actinobacteria from Avicennia marina (88.88%) have shown good antimicrobial activity against the all tested bacterial pathogens. Growth of Bacillus subtilis was inhibited by most of the strains and Proteus mirabilis was the least-inhibited pathogenic strain and the moderate inhibition rate was observed against Bacillus halodurans. Actinobacteria from Rhizopora mucronata (75%) have shown good antibacterial activity against the all tested bacterial pathogens. Growth of P. aeruginosa was inhibited by most of the strains and Shigella flexineri was the least-inhibited pathogenic strain and the moderate inhibition rate was observed against Bacillus subtilis.
Screening of actinobacteria for extracellular enzymes
No studies on characterization of extracellular enzyme from mangrove-associated actinobacteria of A & N Islands have been reported. Of 103 isolates, all the isolates were found to synthesize gelatinase enzyme, 73 isolates demonstrated amylolytic activity, 38 isolates exhibited proteolytic and 63 isolates displayed urease activity (Table 1). Interestingly, 56 isolates exhibited excellent DNase activity and 71 isolates revealed positive for L-asparaginase. Of 103 isolates, all revealed negative results for lipolytic activity and cellulose enzyme production (Fig. 9). Of 90 isolates from Ceriops tagal, all were found to synthesize gelatinase, 65 isolates demonstrated amylolytic activity, 34 isolates exhibited proteolytic and 55 isolates displayed urease activity. Interestingly, 51 isolates exhibited excellent DNase activity and 64 isolates revealed positive for l-asparaginase. Of 09 isolates from Avicennia marina, all isolates were found to synthesize gelatinase, 04 isolates demonstrated amylolytic activity, 01 isolate exhibited proteolytic and 06 isolates displayed urease activity. Interestingly, 04 isolates exhibited excellent DNase activity and 05 isolates revealed positive for L-asparaginase. Of 04 isolates from Rhizopora mucronata, all 04 isolates were found to synthesize gelatinase, 04 isolates demonstrated amylolytic activity, 03 isolates exhibited proteolytic and 02 isolates displayed urease activity. Interestingly, 01 isolate exhibited excellent DNase activity and 02 isolates revealed positive for L-asparaginase. Isolates MBCT 13, MBCT 14, MBCT 15, MBCTS 29, MBCTS 35B, MBCTS 40, MBCTS 46, MBCTS 47 and MBCTS 56 exhibited elevated enzymatic activity for all 06 industrial enzymes. Consequently, these potent isolates were subjected for the detailed characterization on industrially potent enzymes like amylase, gelatinase and protease. Of 103 isolates, all were found positive for gelatinase enzyme production and maximum number of amylase producing isolates was isolated from Rhizopora mucronata and maximum casinase, DNase and l-asparaginase producers were isolated from Ceriops tagal. These results on enzymatic production confirm the ability of mangrove-associated actinobacteria to synthesize the valuable enzymes of industrial importance.
Phylogenetic analysis of mangrove-associated actinobacteria
Phylogenetic relationships of the mangrove-associated actinobacteria were ascertained based on the 16S rDNA sequence similarity with reported strains using BLAST sequence similarity search. Upon analysis, it was established that the deduced 16S rDNA sequences of Streptomyces sp. NIOT_MBAMS1 were highly homologous (96.34%) with reported sequences of Streptomyces lannensis (GenBank: LC12345). Sequence analysis also specified that 16S rDNA sequences of Streptomyces sp. NIOT_MBAMS1 were closely related to the phylogenetic neighbors: Streptomyces sp., Streptomyces chromofuscus and Streptomyces bulli with sequence similarity of 93% and 92%, respectively. Phylogenetic analysis based on neighbor-joining tree (Fig. 10) further revealed that strain NIOT_MBAMS1 formed a distinct branch with Streptomyces lannensis. 16S rDNA sequences of Streptomyces sp. NIOT_MBCT12 were highly homologous (96%) with reported sequences of Streptomyces erythrogriseus (GenBank: EU301830). Sequence analysis also indicated that 16S rDNA sequence of Streptomyces sp. NIOT_MBCT12 was highly homologous to the phylogenetic neighbors: Streptomyces variabilis, Streptomyces sp., Streptomyces griseocarnancaus and Streptomyces griseorubans with sequence similarity of 92 and 89%. Neighbor-joining tree also disclosed that strain NIOT_MBCT12 forms a single cluster with Streptomyces erythrogriseus. The sequences of Nocardiopsis sp. NIOT_MBCT19 also established 93% homology with the previous report of Nocardiopsis lucentensis (GenBank: KC493980.1). BLAST analysis also indicated that 16S rDNA sequences of Nocardiopsis sp. NIOT_MBCT19 were found extremely related to the phylogenetic neighbors: Nocardiopsis sp., Nocardiopsis flavescens and Nocardiopsis dassonvillei with the similarity between 91 and 86%. Neighbor-joining tree also disclosed a distinct cluster between NIOT_MBCT19 and Nocardiopsis lucentensis. Upon analysis, it was established that the deduced 16S rDNA sequences of Streptomyces sp. NIOT_ MBCTS29 were highly homologous (91.24%) with reported sequences of Streptomyces violascens (GenBank: KU973983.1). Sequence analysis also specified that 16S rDNA sequences of Streptomyces sp. NIOT_ MBCTS29 were closely related to the phylogenetic neighbors: Streptomyces hydrogenans, Streptomyces koyangensis and Streptomyces argenteolus with sequence similarity of 89 and 83%, respectively. Phylogenetic analysis based on neighbor-joining tree further revealed that strain NIOT_ MBCTS29 formed a distinct branch with Streptomyces violascens.
The sequences of Nocardiopsis sp. NIOT_ MBCTS36 also established 98% homology with the previous report of Nocardiopsis aegyptia (GenBank: KR476435.1). BLAST analysis also indicated that 16S rDNA sequence of Nocardiopsis sp. NIOT_ MBCTS36 was found extremely related to the phylogenetic neighbors: Nocardiopsis sp., Nocardiopsis dassonvillei and Nocardiopsis synnemataformans with the similarity between 91 and 86%. Neighbor-joining tree also disclosed a distinct cluster between NIOT_ MBCTS36 and Nocardiopsis aegyptia.
16S rDNA sequence of Streptomyces sp. NIOT_MBCT43 was highly homologous (97%) with reported sequences of Streptomyces teitici (GenBank: KY909252). Sequence analysis also indicated that 16S rDNA sequence of Streptomyces sp. NIOT_MBCT43 was highly homologous to the phylogenetic neighbors: Streptomyces sp., and Streptomyces sanyensis with sequence similarity of 93 and 90%. Neighbor-joining tree also disclosed that strain NIOT_MBCT43 forms a single cluster with Streptomyces teitici. Of 103 isolates, 48 isolates were confirmed by molecular level identification. Based on the phylogenetic analysis, the isolates were categorized under the genera: Streptomyces, Nocardiopsis, Salinispora and Actinomadura.
Discussion
Mangrove root and associated sediment samples were collected from three stations of South Andaman region. Dominant mangrove species in three sampling stations, such as Avicennia marina, Rhizopora mucronata, and Ceriops tagal, were selected in this study. Among the three different media used, Bennet’s agar and Emerson’s agar plates displayed morphologically different colonies than in starch casein agar plates. A total of 103 different actinobacterial isolates were selected based on their morphological differences. Of 103 actinobacteria, 96 isolates revealed good growth with varying aerial mass color on ISP-2 agar medium. All the isolates displayed the presence of aerial and substrate mycelium. Previous studies reported Streptomyces as major actinobacteria from mangrove eco-system.
The aerial mass color of maximum isolates was whitish gray, blueish white, gray and one strain has displayed brown color. Vanajakumar et al. [28] have also reported the white color actinobacteria in dominant forms. Colonial growth on agar plate, morphological observation of colonial characteristics, such as color of vegetative growth, presence of color in aerial mycelium and spores and presence of diffusible pigments, was also recorded. Of 103 isolates, 07 isolates were recorded as pigment-producing actinobacteria and 5 were able to produce melanoid. The spore chain morphology was studied by scanning electron microscope (SEM) in three strains among them; one strain has shown spiny structure with smooth surface. SEM analysis is specific for examining spore surface morphology of the actinobacteria [29]. In antibacterial assay, 30.11% of the isolates revealed inhibitory activity against all tested clinical pathogens, and 65% of the isolates displayed inhibitory activity against minimum of 04 tested clinical pathogens. It is observed that the new drugs or antibiotics are urgently needed to halt and reverse the relentless spread of antibiotic-resistant pathogens, which may cause life-threatening infections. Filamentous actinobacteria belong to the order Actinomycetales, especially Micromomospora and Streptomyces strains have the unique and proven capacity to produce novel antibiotics [30]. It is also noted that under-explored habitats, such as desert biomass and marine ecosystems, are a very rich sources of novel actinobacteria with the potential for biosynthesis of novel bioactive compounds, including antibiotics [31].
Studies on utilization of carbon sources by actinobacteria viz., arabinose, xylose, inositol, mannitol, fructose, rhamnose, sucrose and raffinose were analyzed for classification. These carbon sources were supplemented separately in each ISP medium. Another major milestone in the identification of actinobacteria was the assimilation of carbon sources by the isolates. Maximum isolates revealed luxuriant growth (MBCT 17A, MBCTS 35B, MBCT 17B and MBCTS 38) and some strains displayed minimal growth. Pandey et al. [32] reported that for optimum production of antibiotics, certain carbon sources are required. They were also suggested that, pH is an important factor for the production of antibiotics by actinobacteria. pH and sodium chloride tolerance test also carried out for all 103 isolates, growth survival studies of the actinobacterial isolates also accomplished to withstand in varied NaCl and pH levels. Hence, the actinobacteria were isolated from the mangroves-associated environment, the strains were expected to tolerate at diversified salinity levels. Previous reports also disclosed that, majority of the actinobacterial species isolated from marine sediments were moderate alkaliphilic and moderate halophilic in nature [33]. To cope with the external stress, these organisms have developed an adaptive metabolic feature to survive under extreme conditions [34]. Nesterenkonia alba sp. nov., an alkaliphilic actinobacteria was reported to grow optimally at pH 9–10 [35]. Singh et al. [34] also reported a halophilic marine actinobacteria, Nocardiopsis litoralis sp. nov., isolated from sea anemone. Vasavada et al. [36] revealed that, culture medium, pH, salinity, carbon and nitrogen may affect the growth and antibiotic production by actinobacteria. Carbohydrate utilization was determined by growth on carbon utilization medium [16] supplemented with 1% carbon source at 28 °C. Temperature range for growth was determined in inorganic salt starch agar medium (ISP-1) by cultivating the actinobacteria at different temperatures (10 °C, 15 °C, 25 °C, 35 °C, 40 °C, 45 °C, 50 °C, 55 °C, 60 °C, 65 °C, 70 °C, 75 °C and 80 °C).
Conclusion
Actinobacteria are physiologically diverse group in synthesizing various enzymes and metabolic products of industrial interest and are well recognized to produce most valuable pharmaceuticals and agrochemicals [37]. Marine actinobacteria isolated from East and West Coast of India were reported in the production of various industrial enzymes [34]. Upon characterization for industrially potential enzymes, results from the potential isolates in our study revealed highly competent enzyme activity with that of previous reports. Bernfield [38] isolated several actinobacteria from marine sediments of the Central and West Coast of Peru with multi-enzyme activity. These results on enzymatic production authenticated the capability of our isolate to over synthesize the valuable enzymes of industrial importance.
Availability of data and materials
Not Applicable.
References
Kathiresan NK, Bingham BL. Biology of mangroves and mangrove ecosystems. Adv Mar Biol. 2001;40:81–251.
Brown BE. Integrated coastal management: South Asia. Newcastle, Newcastle upon Tyne: Department of Marine Sciences and Coastal Management, Univ; 1997.
Baskaran R, Vijayakumar R, Mohan PM. Enrichment method for the isolation of bioactive actinomycetes from mangrove sediments of Andaman islands, India. Malays J Microbiol. 2011;7:1–7.
Bredholt H, Fjaervik E, Jhonsen G, Zotechev SB. Actinomycetes from sediments in the Trondhein Fjrod, Norway: diversity and biological activity. Mar Drugs. 2008;6:12–24.
Pelaez F. The historical delivery of antibiotic from microbial natural products—can history repeat? Biochem Pharmacol. 2006;71:981–90.
Veiga M, Esparis A, Fabregas J. Isolation of cellulolytic actinomycetes from marine sediments. Appl Environ Microbiol. 1983;46:286–7.
Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycetes diversity from tropical Pacific ocean sediments. Appl Environ Microbiol. 2005;7:1039–48.
Kumar KS, Sahu MK, Kathiresan K. Isolation and characterization of streptomycetes producing antibiotic, from a mangrove environment. Asian J Microbiol Biotechnol Environ Exp Sci. 2005;7:457–64.
Jeffrey LSH, SahilahSon AMR, Tosiah S. Isolation and screening of actinomycetes from Malaysian soil for their enzymatic and antimicrobial activities. J Tropical Agric Food Sci. 2007;35:159–64.
Oskay AM, Usame T, Cem A. Antibacterial activity of some actinomycetes isolated from farming soils of Turkey. African J Biotechol. 2004;3:441–6.
Meena B, Anburajan L, Vinithkumar NV, Kirubagaran R. Novel marine actinobacteria from emerald Andaman & Nicobar islands: a prospective source for industrial and pharmaceutical byproducts. BMC Microbiol. 2013;13:145.
Ellaiah P, Kalyan D, Rao VS, Rao BV. Isolation and characterization of bioactive actinomycetes from marine sediments. Hindustan Antibiot Bull. 1996;38:48–52.
Kuster E, Williams S. Selection of media for the isolation of Streptomyces. Nature. 1964;202:928–9.
Weyland H. Actinomycetes in North sea and Atlantic ocean sediments. Nature. 1969;223:858.
Shirling EB, Gottileb D. Methods for characterization of Streptomyces species. Int J Syst Bactriol. 1966;16:312–40.
Pridham TG, Gottlieb D. The utilization of carbon compounds by some actinomycetes as an aid for species determination. J Bacteriol. 1948;56:107–14.
Williams ST, Cross T. Actinomycetes, methods in microbiology, vol. 4. New York: Academic Press; 1971.
Lemos ML, Toranzo AE, Barja JL. Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microb Ecol. 1985;11:149–63.
Salle AJ. Laboratory manual on fundamental principles of bacteriology. UK: McGraw Hill; 1948.
Leon J, Liza L, Soto I, Cuadra D, Patino L, Zerpa R. Bioactive actinomycetes of marine sediment from the central coast of Peru. Rev Peru Biol. 2007;14:259–70.
Gordon RE, Mihm JM. A comparative study of some strains received as Nocardiae. J Bacteriol. 1957;73:15–27.
Gulati R, Saxena RK, Gupta R. A rapid plate assay for screening L-asparaginase producing microorganisms. Lett Appl Microbiol. 1997;24:23–6.
Kutchama AJ, Roberts MA, Knaebel DB, Crawford DL. Small-scale isolation of genomic DNA from Streptomyces mycelia or spores. Biotechniques. 1998;24:452–6.
Altschul SF, Thomas LM, Alejandro AS, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Talbot GH, Bradley J, Edwards JE Jr, Gilbert D, Scheld M, Bartlett JG. Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America. Clin Infect Dis. 2006;42:657–68.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–9.
Vanajakumar G, Selvakumar N, Natarajan R. Antagonistic properties of actinomycetes isolated from mollusks of the porto Novo region. South India. 1995: 267–274.
Saurav K, Kannabiran K. Diversity and optimization of process parameters for the growth of Streptomyces VITSVK9 sp. isolated from Bay of Bengal, India. J Nat Environ Sci. 2010;1:56–65.
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thompson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002;417:141–7.
Hong K, Gao AH, Xie QY, Gao H, Zhuang L, Lin HP, Yu HP, Li J, Yao XS, Goodfellow M, Ruan JS. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar Drugs. 2009;7:24–44.
Pandey A, Shukla A, Majumdar SK. Utilization of carbon and nitrogen sources by Streptomyces kanamyceticus M27 for the production of an anti-bacterial antibiotic. Afr J Biotech. 2005;4:909–10.
Ramesh S, Rajesh M, Mathivanan N. Characterization of a thermostable alkaline protease produced by marine Streptomyces fungicidicus MML1614. Bioprocess Biosyst Eng. 2009;32:791–800.
Singh SP, Thumar JT, Gohel SD and Purohit MK. Molecular diversity and enzymatic potential of salt-tolerent alkaliphilic actinomycetes. In Mendez A, editors. Current research, technology and education topics in applied microbiology and microbial biotechnology; 2010.
Luo HY, Wang YR, Miao LH, Yang PL, Shi PJ, Fang CX, Yao B, Fan YL. Nesterenkonia alba sp. nov., an alkaliphilic actinobacterium isolated from the black liquor treatment system of a cotton pulp mill. Int J Syst Evol Microbiol. 2009;59:863–8.
Vasavada SH, Thumar JT, Singh SP. Secretion of a potent antibiotic by salt- tolerent and alkaliphilic actinomycete Streptomyces sannanensis strain RJT-1. Curr Sci. 2006;91:1393–7.
Okami Y. Marine microorganisms as a source of bioactive agents. Microb Ecol. 1986;12:65–78.
Bernfield P. Amylases α and β. In: Methods in enzymology. 1st edition. New York, USA: Academic Press; 1955. p. 149–58.
Acknowledgements
The author’s great fully acknowledge the financial support given by the Ministry of Earth Sciences, Government of India, New Delhi, to conduct the survey and research. The authors are thankful to Dr. M. A. Atmanand, Director, National Institute of Ocean Technology, Chennai for providing necessary facilities to perform this research.
Funding
Not Applicable.
Author information
Authors and Affiliations
Contributions
The research concept and the experiments were executed by BM, LA and MAJ. NVV and GD analyzed the data and reviewed the manuscript. All of the authors approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meena, B., Anburajan, L., Johnthini, M.A. et al. Exploration of mangrove-associated actinobacteria from South Andaman Islands, India. Syst Microbiol and Biomanuf 3, 702–718 (2023). https://doi.org/10.1007/s43393-022-00134-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00134-3