Abstract
Metabolic biosensors are increasingly used in metabolic engineering and synthetic biology. In this study, using Saccharomyces cerevisiae as a model system, we developed a methodology to identify promoter elements that are responsive to glucaric acid. Through transcriptome analysis, it was found that multiple genes were upregulated when cells were exposed to high concentrations of glucaric acid. From the promoters of these candidate genes, the YCR012W promoter (PYCR012W) was observed to specifically respond to glucaric acid in a dose-dependent manner. To gain further insight into the binding site of glucaric acid-responsive activators, we truncated the promoter and revealed that the -564 to -464 bp regions of PYCR012W was essential for glucaric acid-responsive expression. To investigate the glucaric acid-responsive transcription factors, we predicted the transcription factor binding sites in the -564 to -464 bp region of PYCR012W and found that two transcription factors, Ash1p and Cbf1p, might be linked to glucaric acid responses. The strategies used in this study outline a method for the identification and development of metabolic biosensors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the past decade, advances in synthetic biology have greatly contributed to the establishment of novel metabolic engineering tools [1, 2]. Synthetic biology and metabolic engineering have led to a substantial increase in microbial production of value-added compounds. Bioinformatics, synthetic biology and systems biology have supported the engineering of microbial cell factories. To improve the performance of engineered strains, new biosensor devices have been built and tested. In vivo metabolite biosensors can sense and respond to important intermediates or target products in engineering pathways [3], and are powerful phenotypic tools for medicine and applied biology research. Biosensors are increasingly being applied in metabolic engineering, especially high-throughput screening of metabolic strains [4,5,6,7]. In addition, dynamic regulation pathways have been constructed based on biosensors to optimise engineering pathways and increase the production of target chemicals [8,9,10]. Furthermore, genetically encoded sensors can be used for live cell imaging studies to monitor the production, growth and viability of engineered strains in a time-resolved manner at single-cell resolution [11]. Biosensor-mediated strain metabolic engineering has received great attention in recent years [12, 13].
There are many types of biosensors based on molecular recognition elements, including enzyme sensors, microbial sensors, organelle sensors, tissue sensors and immunosensors. Currently, biosensors in microbial cells are usually used as high-throughput screening tools to detect intermediate metabolites, including proteins, amino acids, polymer precursors and organic acids. Fortunately, nature has evolved a variety of intracellular molecular sensors, such as transcription factors, allosteric proteins, enzymes, riboswitches and metabolite-responsive promoters, to sense intracellular and extracellular chemicals, and these can be used to detect biosynthesis intermediates. There have been many reports on the development of genetically encoded biosensors based on transcription factors. For instance, Skjoedt et al. [7] studied the prokaryotic transcription factor BenM, a biosensor for cis,cis-muconic acid (CCM) in Saccharomyces cerevisiae. In another example, Leonie Baumann et al. developed a whole-cell short- and medium-chain fatty acid biosensor based on responsive promotor pPDR12, which is regulated by the transcription factor War1p [14, 15]. At present, E. coli is one of the main hosts for the industrial production of value-added chemicals [16]. As a widely studied model strain, E. coli has built many successful biosensors as a host [17]. S. cerevisiae has advantages over E. coli in terms of genetic tractability, robustness, and resistance to phage [18]. However, the development of genetic biosensors in S. cerevisiae still lags far behind bacteria, likely because yeast cells lack engineered control elements for sensing and regulating metabolic pathways [19]. To date, there are only limited examples of biosensors developed in yeast for measuring and adjusting the concentrations of metabolites.
Glucaric acid (GA), a derivative of glucose, is a naturally occurring dibasic acid that is considered an important value-added chemical derived from biomass [20] due to its useful applications in food, chemical, and pharmaceutical industries [21]. GA is found in many fruits and vegetables, and it is mainly produced from glucose through chemical oxidation [22] using nitric acid as solvent and oxidant [23]. However, the production of GA by chemical oxidation has disadvantages of low yield, large quantities of by-products, and it is not environmentally friendly. With the development of metabolic engineering, biological production of GA is being studied. E. coli and S. cerevisiae have been engineered as GA producer strains in various ways through metabolic engineering [20, 24], and engineered S. cerevisiae strains possess potential for overproduction of GA [25]. In vivo metabolite biosensors can result in better yeast producer strains through dynamic control of cell metabolism. Therefore, researchers can use biosensors as high-throughput screening tools to select efficient GA-producing strains. In 2016, Rogers et al. [26] developed a series of biosensor systems in E. coli, including a GA biosensor based on co-expression of the transcription factor cdaR and a green fluorescent protein reporter gene (gfp) under the control of a 521 bp promoter. Since Saccharomyces cerevisiae has the advantage of low pH-induced acid toxicity [24, 27], yeast cells are an attractive alternative host due to inherently high acid resistance [28] as well as genetic tractability and robustness. Therefore, it is necessary to develop new biosensor systems in Saccharomyces cerevisiae.
Biosensors have important value for various aspects of strain engineering, dynamic control, and single cell analysis. In all cases, a metabolite-sensing protein (e.g., transcription factors, enzymes or periplasmic-binding proteins) is activated upon binding to an effector molecule, and this controls the expression of an actuator component (e.g., fluorescent reporters, regulatory switches or selection markers) [29]. In this context, transcription factors control microbial physiology at the transcription level, and play a major role in the design of biosensors. In bacteria, transcriptional activation can take place through a transcriptional activator binding to an operator site in a promoter, thereby improving its ability to guide RNA polymerase to initiate transcription [30,31,32]. A lack of transcription factors also limits the successful construction of genetic sensors in S. cerevisiae.
To solve the problem of the lack of GA biosensors in eukaryotic hosts, key metabolite levels could be converted into fluorescent signals to monitor the concentrations of intracellular compounds in living cells, and thereby provide effective tools for high-throughput genetic screening. High-throughput transcriptome sequencing can comprehensively and quickly identify genes in different expression systems under specific conditions. When intermediate metabolites accumulate to toxic levels, the resulting stress response typically alters promoter activity [9], and this can be used to develop promoter-based biosensors [5, 33].
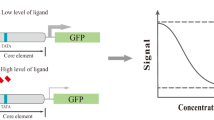
In the present study, transcriptome analysis was performed, and based on the cellular stress response, the GA response element for the biosensor device was determined (Fig. 1). Specifically, S. cerevisiae cells were first exposed to high concentrations of GA and then were subjected to RNA sequencing (RNA-Seq) analysis. Genes upregulated in the presence of GA were identified, and the responses of the promoters of candidate genes to GA were investigated by GFP reporter. From these candidates, two promoters with specific responses to GA were selected. By truncating the promoter region, the GA-active region was determined, and transcription factors that could serve as GA biosensors were identified. The findings have implications for further improving GA biosensors in future work.
Materials and methods
Strains and growth media
The yeast strains used in this study are listed in Table S1. E. coli JM109 was used as the host for plasmid cloning. E. coli cells were cultured at 37 °C with shaking at 200 rpm in LB medium (1% w/v tryptone, 0.5% w/v yeast extract, 1% w/v NaCl) containing 100 mg/L ampicillin, when needed. Yeast strains were maintained on YPD (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose) or SD-URA (1.7 g/L yeast nitrogen base, 20 g/L glucose, 5 g/L (NH4)2SO4, with amino acid mixture without uracil) at 30 °C. Solid medium was prepared by adding 2% (w/v) agar.
To test the response to GA, S. cerevisiae strains were cultured in synthetic dextrose (SD-URA) medium supplemented with GA. In the late logarithmic growth period, cells were collected for fluorescence intensity measurement.
Transcriptome analysis and quantitative real-time PCR analysis
To fully investigate the GA-responsive genes in S. cerevisiae, an RNA-Seq transcriptome comparison experiment was carried out. Yeast cells were first cultivated in YPD medium at 30 °C for ~ 16 h, and cells were then transferred into 100 mL of fresh YPD, cultured to OD600 0.6 − 0.8, and divided into equal parts. Cells were then inoculated into fresh medium with or without GA, cultured for another 24 h, collected, and total RNA was extracted using the hot phenol method [34]. Total RNA was then isolated, RNA quality was checked using a BioTek instrument (Winooski), and transcriptome sequencing was performed by Genewiz (Suzhou, China). Each sample was carried out in three replicates. Gene expression levels were analysed as described in our previous study [35]. GO [36] and KEGG pathway [37] enrichment analyses were performed by Genewiz.
To confirm the RNA-Seq results, the RNA used for the above RNA-Seq analysis was used to synthesize the first-strand cDNA using SuperRT cDNA Synthesis Kit (Cwbiotech, China) following the manufacturer’s instructions. The synthesised cDNA performed to detect the relative expression levels of four genes using SYBR Premix Ex Taq (Cwbiotech, China) in a Thermo Scientific CFX96 instrument and the results were analyzed by the − ΔΔCt method [38]. The ACT1 gene was used as an internal control. Each reaction was carried out in triplicate.
Plasmid construction
The plasmid pGFP33 with a URA expression cassette was adopted to construct the GFP reporter plasmid. First, the candidate promoters were amplified using the S. cerevisiae BY4741 genome as the template with the relevant primer pairs listed in Table S2. Plasmid pGFP33 was then linearised using restriction enzymes SphI and EcoRI and used for cloning the selected candidate promoters. Each amplified candidate promoter was cloned into the linearised pGFP33 plasmid using an ABClonal kit (Nanjing Well-Offer Biotechnology Co., Ltd) following the manufacturer’s instructions.
The GFP reporter plasmids constructed above were transformed into yeast cells using the standard lithium acetate method [39]. All primers were synthesised by Genewiz (USA). All DNA polymerases and DNA restriction enzymes were purchased from Takara (Japan). SanPrep Column Plasmid Mini-prep Kits were from Sangon Biotech (Shanghai, China) for preparing plasmid DNA from E. coli.
Optical density and fluorescence measurements
Yeast cells containing the GFP reporter plasmids with different promoters were first cultured in the corresponding media as described above. The OD600 value and the fluorescence (λexcitation 480 nm, λemission 530 nm) of the biosensor output were measured using 200 μL yeast cultures at an appropriate dilution using a BioTek microtiter plate reader (Winooski, VT, USA) with diluted medium as the blank. Specific fluorescence was measured and normalised by dividing the measured fluorescence value by the measured OD600 value.
Statistical analysis
All experiments included biological triplicates unless otherwise stated. Student’s t tests were performed to estimate significance, and p < 0.05 was considered statistically significant.
Results
Transcriptome analysis of the responses of S. cerevisiae to glucaric acid
To fully investigate the influence of GA on S. cerevisiae at the molecular level, RNA-Seq was used to study the genome-wide responses to GA. Before RNA-Seq analysis, log phase yeast cells were first treated with 10 g/L GA and three replicates of each simple was used for the RNA-seq analysis. After RNA-Seq analysis, two upregulated genes of YFR032C and YHR175W, and two down-regulated genes of YMR011W and YNL192W, were selected to confirm the RNA-seq results. It was indicated that the expression levels of YFR032C and YHR175W were significantly increased by the supplement of GA, while the expression levels of YMR011W and YNL196W were both significantly decreased (Supplementary Fig. 1) in response to GA. All these results were consistent with the results of RNA-Seq analysis. According to the RNA-seq results, when genes were upregulated twofold (i.e., log2 fold change [Log2FC] ≥ 1) or downregulated twofold (Log2FC ≤ −1) and the false discovery rate (FDR) was < 0.05, they were considered as differentially expresses genes (DEGs). There were totally 591 DEGs were identified, of which 239 were upregulated and 352 were downregulated (Fig. 2a, Table S3), compared with controls without GA.
Comprehensive overview of DEGs in strain BY4741 with or without GA. a Volcano plot of DEGs. Each point represents a gene, red dots represent significantly upregulated genes (Log2FC ≥ 1), blue dots represent significantly downregulated genes (Log2FC ≤ −1). b KEGG pathway enrichment of DEGs. The y-axis indicates the pathway name, the x-axis indicates the rich factor corresponding to the pathway, − log10(p value) is represented by dot colour, and the number of DEGs is represented by dot size
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed to investigate the main pathways in which these DEGs were involved. It was showed that the most significant DEGs were related to carbon metabolism, glycolysis/gluconeogenesis, ribosome biogenesis in eukaryotes, oxidative phosphorylation, and biosynthesis of amino acids (Fig. 2b). In summary, genes involved in various pathways play a significant role in regulating carbon metabolism and responding to GA in S. cerevisiae.
Identification and verification of glucaric acid-responsive promoters
To construct the biosensor for GA in S. cerevisiae, among the 239 upregulated genes, the promoters of the 36 genes that were most upregulated in response to GA based on the RNA-Seq data were selected as candidate genes (Fig. 3). Subsequently, reporter assays were performed to evaluate whether expression of these 36 genes was indeed upregulated by GA. Each promoter (-800 to 1 bp) was cloned upstream of the gene encoding GFP in plasmid pGFP33. The constitutive TDH3 promoter (PTDH3) was fused to GFP (PTDH3-GFP) and considered as a positive control. The resultant plasmids were introduced into S. cerevisiae (BY4741opi1Δ) [40] to evaluate the promoter strength in response to GA.
Expression of 36 promoter reporters in response to GA. The GFP fluorescence reporter was under the control of the promoters of selected genes. The promoter of each candidate gene was included up to ~ 800 bp upstream of its coding sequence. S. cerevisiae strains were cultured in SD-URA medium supplemented with GA, and cells were collected after 15 h. The three selected promoters are indicated with arrows. Results are the means ± standard deviation (SD) of three biological replicates. The asterisk of * and ** shows statistically significant differences of p < 0.05 and p < 0.01, respectively
The engineered strains were cultivated in medium supplemented with 10 g/L GA to test their response to GA (Fig. 3). As expected, in response to 10 g/L GA, the positive control of PTDH3-GFP showed no response to GA. Analysis of GFP expression levels under the control of the 36 promoters that were upregulated by GA in the RNA-Seq analysis showed that expression of two different reporter cassettes, PYCR012W-GFP and PYGL009C-GFP, was significantly increased following exposure to 10 g/L GA. YCR012W, known as phosphoglycerate kinase 1 (PGK1), is the key enzyme in glycolysis and gluconeogenesis that catalyses the high-energy phosphoryl transfer of the acyl phosphate of 1,3-bisphosphoglycerate to ADP to produce ATP [41]. YGL009C, encoding isopropyl malate isomerase Leu1, catalyses the second step in the leucine biosynthesis pathway [42]. Therefore, PYCR012W and PYGL009C were identified as indicators of GA concentration.
Next, to determine their dose response and specificity, the PYCR012W and PYGL009C promoters were tested for their response to GA at various concentrations (0 g/L, 10 g/L, 20 g/L, 30 g/L, 40 g/L and 50 g/L) using PYCR012W-GFP and PYGL009C-GFP reporters (Fig. 4). Expression of GFP controlled by either PYCR012W or PYGL009C exhibited a specific response to GA in a dose-dependent manner in cultures at 12 h, 24 h and 36 h. Therefore, we concluded that PYCR012W and PYGL009C could be used to construct GA-responsive biosensors. Because the linear relationship of PYCR012W-GFP in response to GA was much better than that of PYGL009C-GFP (Supplementary Fig. 2), we chose PYCR012W for our further study.
Dose–response tests of a PYCR012W-GFP and b PYGL009C-GFP to GA. S. cerevisiae strains were cultured in SD-URA medium supplemented with different concentrations of glucaric acid (0 g/L, 10 g/L, 20 g/L, 30 g/L, 40 g/L and 50 g/L) and collected after 12 h, 24 h or 36 h. Results are the means ± standard deviation (SD) of three biological replicates. The asterisk of * and ** shows statistically significant differences of p < 0.05 and p < 0.01, respectively
Identification of the glucaric acid-responsive element
To determine the smallest region of the PYCR012W promoter controlling the response to GA, PYCR012W was truncated at various places upstream of the 5’ site. The results showed that when the promoter of PYCR012W was truncated 200 bp upstream of the 5’ site (-764 to -664 bp and -764 to -564 bp), the expression of PYCR012W-T1 and PYCR012W-T2 could still be induced by GA. However, when the promoter region of PYCR012W was truncated 300 bp upstream of the 5’ site, the induction of the GFP reporter was inhibited in response to GA (Fig. 5a). Accordingly, deletion of sequences more than 300 bp upstream of the 5’ site resulted in loss of the response to GA, and the reporter gene expression was decreased in all cases (-764 to -464 bp, -764 to -364 bp and -764 to 264 bp for PYCR012W-T3, PYCR012W-T4 and PYCR012W-T5, respectively). Hence, it appears that the -564 to -464 bp region in PYCR012W may contain the binding site of the GA-responsive activator.
Identification of the smallest region of promoter PYCR012W required for the response to GA. a Responses of PYCR012W and its truncated mutants to GA. S. cerevisiae strains were cultured in SD-URA medium supplemented with 10 g/L GA and cells were collected after 15 h. b GA dose–response results for truncated versions of PYCR012W (YCR012W-T1, YCR012W-T2 and YCR012W-T3) was measured using a plate reader. S. cerevisiae strains were cultured in SD-URA medium supplemented with different concentrations of GA (0 g/L, 10 g/L, 20 g/L,30 g/L, 40 g/L and 50 g/L) and cells were collected after 15 h. Results are the means ± standard deviation (SD) of three biological replicates. The asterisk of * and ** shows statistically significant differences of p < 0.05 and p < 0.01, respectively
To determine the dose response and specificity of the truncated versions of the PYCR012W promoter, the three promoters PYCR012W-T1, PYCR012W-T2 and PYCR012W-T3 were used to test the response to GA at various concentrations (0 g/L, 10 g/L, 20 g/L, 30 g/L, 40 g/L and 50 g/L). The results showed that the PYCR012W-T1 promoter responded to GA in a much more significant dose-dependent manner than that of PYCR012W-T2 and PYCR012W-T3 (Fig. 5b).
Identification of glucaric acid-responsive transcription factors
Currently, most biosensors have been constructed using transcription factors, and this method is limited by the availability of previously well-characterized transcription factors [43]. Therefore, it is particularly important to determine the transcription factors of GA biosensors. Herein, to identify the GA-responsive transcription factors, we first predicted the transcription factor binding sites in the -564 to -464 bp region of PYCR012W using yeastract [44] (http://yeastract.com/findregulators.php). As a result, we chose 12 non-essential genes encoding potential GA-responsive transcription factors (Table 1). To verify the target transcription factors, the PYCR012W-GFP reporter plasmid was introduced into these gene-deleted mutant strains to test whether deletion of these genes encoding the indicated transcription factors influenced the PYCR012W-GFP expression in response to GA (Fig. 6a). The results showed that the deletion of the 12 genes did not affect their cell growth (Supplementary Fig. 3). The GFP expression levels were increased without supplementation of GA for all mutants for seven genes (GLN3, PHO4, GAT1, GZF3, NRG1, XBP1 and FKH1), indicating these transcription factors might inhibit the expression of YCR012W. In gcn4Δ, skn7Δ, pho4Δ, gat1Δ, xbp1v, fkh1Δ and fkh2Δ mutants, expression of PYCR012W-GFP could be still induced by addition of GA, indicating that these transcription factors were not responsive to GA. Interestingly, deletion of the two transcription factors of Ash1 and Cbf1 tended to decrease the fluorescence signal of PYCR012W-GFP in response to GA. These results indicate that Ash1p and Cbf1p might be transcription factors mediating GA biosensors.
Identification of GA-responsive transcription factors. a The expression of PYCR012W-GFP in the 12 mutants for the predicted transcription factors. The biosensor plasmid PYCR012W-GFP was transformed into the gene-deleted strain of the corresponding candidate transcription factor. S. cerevisiae strains were cultured in SD-URA medium supplemented with GA and cells were collected after 15 h. Results are the means ± standard deviation (SD) of three biological replicates. The asterisk of ** shows statistically significant differences of p < 0.01. b The predicted mode of how Ash1p and Cbf1p regulate the expression of PYCR012W-GFP
Discussion
System engineering has gradually advanced our understanding of interactions between gene sequences [45]. This makes it possible to rationally design gene circuits for biotechnological applications. Since most valuable fine chemicals (such as GA) lack suitable biosensors, it is important to develop methods to discover potential biosensors. Most biosensor devices originate from prokaryotes [17, 26], and there are few reports on constructing biosensors that respond to cellular metabolites in eukaryotes [5, 46], even though eukaryotes possess advantages in the field of metabolic engineering [25, 40] and synthetic biology [18]. It is particularly important to develop a strategy for studying biosensors in eukaryotes. In the present study, we used high-throughput genomics to analyse the DEGs in S. cerevisiae under GA treatment. From the 36 genes displaying the most significant upregulation by GA, we found that the PYCR012W promoter showed a significant response in the presence of GA using GFP as the reporter gene. Next, we truncated the promoter region to determine the smallest region required for the GA response, and we found that the -564 to -464 bp region in PYCR012W might be the binding site of GA-responsive activators. By predicting the transcription factors that might bind to this region, we identified two transcription factors, Ash1p and Cbf1p, that respond to GA.
In general, transcriptome sequencing technology is used to measure differences in gene expression levels under specific conditions. Recently, RNA-Seq technology has also been used to develop transcription factor-based biosensors. For instance, Liangpo Li et al. developed the transcriptome-assisted metabolite-sensing (TAMES) strategy to identify the sensing module cusR that responds positively to the metabolite 3-dehydroshikimate (DHS) [47]. In another example, Shi et al. [5] performed transcriptome analysis to discover and engineer a 1-butanol biosensor. In the present study, we also obtained promoters that respond to GA via transcriptome sequencing. As a tool for high-throughput screening, another key indicator of biosensors is their linear range. Within the concentration range of 0 − 50 g/L GA, PYCR012W-GFP and its truncated version PYCR012W-T1-GFP exhibited dose-dependent responses to GA. The PYCR012W promoter was the best GA responsive element identified in this work.
Transcription factors enhance the ability of promoters to transcribe by binding to operator sites in E. coli [30,31,32]. For example, a CCM biosensor was constructed based on the transcription factors BenM in S. cerevisiae [7]. In this way, transcription factors play a major role in the design of biosensors. Herein, we truncated the promoter to determine the smallest region required for the GA response of PYCR012W. Two proteins, Ash1p and Cbf1p, are suggested to be transcription factors for GA. Among them, yeast centromere binding protein Cbf1p, of the helix–loop–helix protein family, is required for chromosome stability and methionine prototrophy [48]. Cbf1p is required for the complete derepression of inositol-1-phosphate synthase (Ino1) transcription [49]. By co-expressing INO1 from S. cerevisiae with inositol oxygenase (MIOX) and urate dehydrogenase (UDH) genes, GA can be produced from glucose [40]. Cbf1p is a GA transcription factor-related enzyme in the GA biosynthetic pathway. However, this work reports a response to the addition of GA, which has not yet been applied to high-throughput screening. Therefore, it is difficult to construct a more sensitive biosensor that can be applied to high-throughput screening.
When the promoter of PYCR012W was truncated 200 bp upstream of the 5’ site (-764 to -564), its expression could still be induced by GA (Fig. 5a). However, when the promoter region of PYCR012W was less than 464 bp, the induction of the GFP reporter was inhibited in response to GA. Therefore, we got the conclusion that the 100 bp of -564 to -464 bp region in PYCR012W might contain the binding site(s) of the GA-responsive activator. According to the predicted results, the binding sites of Ash1p and Cbf1p in PYCR012W is -564 to -560 bp and -544 to -539 bp, respectively. In the following study, we found that mutant for Ash1 or Cbf1 could both totally inhibit the induction of PYCR012W-GFP by GA. These results indicate that Ash1p and Cbf1p might be transcription factors mediating GA biosensors. In addition, it has been reported that the expression of Cbf1 is negatively regulated by Ash1 in a previous study [50]. From this point of view, the expression of Cbf1p will be increased in ash1Δ mutant. Considering the induction of PYCR012W-GFP is still be inhibited by GA in ash1Δ mutant, we speculate that Ash1p and Cbf1p may work together in regulating the expression of PYCR012W in response to GA (Fig. 6b).
In summary, we developed a general method for identifying chemical-responsive promoter elements and thus to help develop and engineer their related biosensors. The method is suitable for discovering molecules that can exert pressure on cells, because a change in promoter activity is a common stress response. Researchers could use these stress response promoters to develop specific biosensors. Our findings will lay a foundation for the subsequent construction of a more sensitive GA biosensors.
References
Ng CY, Khodayari A, Chowdhury A, Maranas CD. Advances in de novo strain design using integrated systems and synthetic biology tools. Curr Opin Chem Biol. 2015;28:105–14. https://doi.org/10.1016/j.cbpa.2015.06.026.
Wendisch VF. Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development. Curr Opin Biotechnol. 2014;30:51–8. https://doi.org/10.1016/j.copbio.2014.05.004.
Ellis JM, Wolfgang MJ. A genetically encoded metabolite sensor for malonyl-CoA. Chem Biol. 2012;19(10):1333–9. https://doi.org/10.1016/j.chembiol.2012.08.018.
Dietrich JA, Shis DL, Alikhani A, Keasling JD. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth Biol. 2013;2(1):47–58. https://doi.org/10.1021/sb300091d.
Shi S, Choi YW, Zhao H, Tan MH, Ang EL. Discovery and engineering of a 1-butanol biosensor in Saccharomyces cerevisiae. Bioresour Technol. 2017;245(Pt B):1343–51. https://doi.org/10.1016/j.biortech.2017.06.114.
Pfleger BF, Pitera DJ, Newman JD, Martin VJ, Keasling JD. Microbial sensors for small molecules: development of a mevalonate biosensor. Metab Eng. 2007;9(1):30–8. https://doi.org/10.1016/j.ymben.2006.08.002.
Skjoedt ML, Snoek T, Kildegaard KR, Arsovska D, Eichenberger M, Goedecke TJ, et al. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat Chem Biol. 2016;12(11):951–8. https://doi.org/10.1038/nchembio.2177.
Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30(4):354–9. https://doi.org/10.1038/nbt.2149.
Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, et al. Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol. 2013;31(11):1039–46. https://doi.org/10.1038/nbt.2689.
Xu P, Wang W, Li L, Bhan N, Zhang F, Koffas MA. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli. ACS Chem Biol. 2014;9(2):451–8. https://doi.org/10.1021/cb400623m.
Mustafi N, Grunberger A, Mahr R, Helfrich S, Noh K, Blombach B, et al. Application of a genetically encoded biosensor for live cell imaging of L-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS ONE. 2014;9(1): e85731. https://doi.org/10.1371/journal.pone.0085731.
Rogers JK, Taylor ND, Church GM. Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol. 2016;42:84–91. https://doi.org/10.1016/j.copbio.2016.03.005.
Zhang J, Jensen MK, Keasling JD. Development of biosensors and their application in metabolic engineering. Curr Opin Chem Biol. 2015;28:1–8. https://doi.org/10.1016/j.cbpa.2015.05.013.
Baumann L, Rajkumar AS, Morrissey JP, Boles E, Oreb M. A yeast-based biosensor for screening of short- and medium-chain fatty acid production. ACS Synth Biol. 2018;7(11):2640–6. https://doi.org/10.1021/acssynbio.8b00309.
Kren A, Mamnun YM, Bauer BE, Schuller C, Wolfger H, Hatzixanthis K, et al. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol Cell Biol. 2003;23(5):1775–85. https://doi.org/10.1128/MCB.23.5.1775-1785.2003.
Pontrelli S, Chiu TY, Lan EI, Chen FY, Chang P, Liao JC. Escherichia coli as a host for metabolic engineering. Metab Eng. 2018;50:16–46. https://doi.org/10.1016/j.ymben.2018.04.008.
Wan X, Ho TYH, Wang B. Engineering prokaryote synthetic biology biosensors. In: Thouand G, editor. Handbook of cell biosensors. Cham: Springer International Publishing; 2019. p. 1–37.
Walmsley RM, Keenan P. The eukaryote alternative: advantages of using yeasts in place of bacteria in microbial biosensor development. Biotechnol Bioprocess Eng. 2000;5(6):387–94. https://doi.org/10.1007/bf02931936.
Yang X, Liu J, Zhang J, Shen Y, Qi Q, Bao X, et al. Quorum sensing-mediated protein degradation for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab Eng. 2021;64:85–94. https://doi.org/10.1016/j.ymben.2021.01.010.
Moon TS, Yoon SH, Lanza AM, Roy-Mayhew JD, Prather KL. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl Environ Microbiol. 2009;75(3):589–95. https://doi.org/10.1128/AEM.00973-08.
Walaszek ZW, Szemraj JS, Hanausek MH, Adams AA, Sherman US, Research N. d-Glucaric acid content of various fruits and vegetables and cholesterol-lowering effects of dietary d-glucarate in the rat. Nutr Res. 1996;16:4.
Sr. DKKH. Method of oxidation using nitric acid. 2007.
Smith TN, Hash K, Davey CL, Mills H, Williams H, Kiely DE. Modifications in the nitric acid oxidation of D-glucose. Carbohydr Res. 2012;350:6–13. https://doi.org/10.1016/j.carres.2011.12.024.
Gupta A, Hicks MA, Manchester SP, Prather KL. Porting the synthetic D-glucaric acid pathway from Escherichia coli to Saccharomyces cerevisiae. Biotechnol J. 2016;11(9):1201–8. https://doi.org/10.1002/biot.201500563.
Zhao Y, Li J, Su R, Liu Y, Wang J, Deng Y. Effect of magnesium ions on glucaric acid production in the engineered Saccharomyces cerevisiae. J Biotechnol. 2021;332:61–71. https://doi.org/10.1016/j.jbiotec.2021.03.020.
Rogers JK, Church GM. Genetically encoded sensors enable real-time observation of metabolite production. Proc Natl Acad Sci USA. 2016;113(9):2388–93. https://doi.org/10.1073/pnas.1600375113.
Shiue E, Prather KL. Improving D-glucaric acid production from myo-inositol in E. coli by increasing MIOX stability and myo-inositol transport. Metab Eng. 2014;22:22–31. https://doi.org/10.1016/j.ymben.2013.12.002.
Weber C, Farwick A, Benisch F, Brat D, Dietz H, Subtil T, et al. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biotechnol. 2010;87(4):1303–15. https://doi.org/10.1007/s00253-010-2707-z.
Mahr R, Frunzke J. Transcription factor-based biosensors in biotechnology: current state and future prospects. Appl Microbiol Biotechnol. 2016;100(1):79–90. https://doi.org/10.1007/s00253-015-7090-3.
Blazeck J, Alper HS. Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol J. 2013;8(1):46–58. https://doi.org/10.1002/biot.201200120.
Raman S, Rogers JK, Taylor ND, Church GM. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci U S A. 2014;111(50):17803–8. https://doi.org/10.1073/pnas.1409523111.
Siedler S, Stahlhut SG, Malla S, Maury J, Neves AR. Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab Eng. 2014;21:2–8. https://doi.org/10.1016/j.ymben.2013.10.011.
Liu Y, Li Q, Zheng P, Zhang Z, Liu Y, Sun C, et al. Developing a high-throughput screening method for threonine overproduction based on an artificial promoter. Microb Cell Fact. 2015;14:121. https://doi.org/10.1186/s12934-015-0311-8.
Domdey KKH. Preparation of high molecular weight RNA. Biochem Gene Express. 1991;194:8.
Zhu K, Li G, Wei R, Mao Y, Zhao Y, He A, et al. Systematic analysis of the effects of different nitrogen source and ICDH knockout on glycolate synthesis in Escherichia coli. J Biol Eng. 2019;13:30. https://doi.org/10.1186/s13036-019-0159-2.
Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):258–61. https://doi.org/10.1093/nar/gkh036.
Kanehisa HOSGKSWFM. KEGG kyoto encyclopedia of genes and genomes. 1999.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. https://doi.org/10.1006/meth.2001.1262.
Gietz BRDW, Robin A, Peg DNA. Transformation of yeast by lithium acetate/single-stranded carrier dna/polyethylene glycol method. Method Enzymol. 2002;350:87–96.
Chen N, Wang J, Zhao Y, Deng Y. Metabolic engineering of Saccharomyces cerevisiae for efficient production of glucaric acid at high titer. Microb Cell Fact. 2018;17(1):67. https://doi.org/10.1186/s12934-018-0914-y.
Packham EA, Graham IR, Chambers A. The multifunctional transcription factors Abf1p, Rap1p and Reb1p are required for full transcriptional activation of the chromosomal PGK gene in Saccharomyces cerevisiae. Mol Gen Genet. 1996;250(3):348–56. https://doi.org/10.1007/BF02174393.
Kohlhaw GB. Leucine biosynthesis in fungi: Entering metabolism through the back door. Microbiol Mol Biol Rev. 2003;67(1):1–15. https://doi.org/10.1128/Mmbr.67.1.1-15.2003.
Qian S, Cirino PC. Using metabolite-responsive gene regulators to improve microbial biosynthesis. Curr Opin Chem Eng. 2016;14:93–102. https://doi.org/10.1016/j.coche.2016.08.020.
Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, et al. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34(1):D446–51. https://doi.org/10.1093/nar/gkj013.
Zuo F, Marcotte H. Advancing mechanistic understanding and bioengineering of probiotic lactobacilli and bifidobacteria by genome editing. Curr Opin Biotechnol. 2021;70:75–82. https://doi.org/10.1016/j.copbio.2020.12.015.
Bian Y, Li L, Dong M, Liu X, Kaneko T, Cheng K, et al. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat Chem Biol. 2016;12(11):959–66. https://doi.org/10.1038/nchembio.2178.
Li L, Tu R, Song G, Cheng J, Chen W, Li L, et al. Development of a synthetic 3-dehydroshikimate biosensor in Escherichia coli for metabolite monitoring and genetic screening. ACS Synth Biol. 2019;8(2):297–306. https://doi.org/10.1021/acssynbio.8b00317.
Cai M, Davis R. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61(3):437–46.
Shetty A, Lopes JM. Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryot Cell. 2010;9(12):1845–55. https://doi.org/10.1128/EC.00144-10.
Batt G, Page M, Cantone I, Goessler G, Monteiro P, de Jong H. Efficient parameter search for qualitative models of regulatory networks using symbolic model checking. Bioinformatics. 2010;26(18):i603–10. https://doi.org/10.1093/bioinformatics/btq387.
Acknowledgements
This work was supported by the National Key R&D Program of China (2019YFA0905502), the National Natural Science Foundation of China (21877053), the Natural Science Foundation of Jiangsu Province (BK20181345), and the Open Foundation of Jiangsu Key Laboratory of Industrial Biotechnology (KLIB-KF201807).
Author information
Authors and Affiliations
Contributions
YD and YZ designed and conceived the idea for all experiments. RS and SL performed the experiments. RS analyzed the results and wrote the first draft of the manuscript. YD and YZ supervised the work and revised the manuscript. All authors have read, edited, and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Su, R., Li, S., Zhao, Y. et al. Identification and development of a glucaric acid biosensor in Saccharomyces cerevisiae. Syst Microbiol and Biomanuf 2, 623–633 (2022). https://doi.org/10.1007/s43393-022-00086-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00086-8