Abstract
Apparently healthy birds in protected areas in northeastern Brazil were investigated, whether shedding bacterial pathogens to the environment. We determined whether pathogens varied according to the level of the shared habitat human of each protected area, the type of vegetation, hosts’ group and different history traits as migration and foraging behavior, body mass, and sensitivity to human impacts. In addition, we also investigated whether the protected areas were preserving the wildlife from antibiotic-resistant bacteria. For that, oropharyngeal and cloacal swabs were collected from 507 individuals of 91 species. In the culture-dependent method, most of the bacterial isolates belonged to Enterobacterales, with the highest frequency of Klebsiella aerogenes (20.5%) and Escherichia coli (19.3%). There was no relationship between Enterobacterales occurrence according to the type of vegetation, hosts’ group and history traits as foraging behavior (foraging stratum and main trophic category), and body mass, and there was a low association between the protected area and Enterobacterales (φ = 0.17). For Mycoplasma, 10.8% of PCR-tested individuals were positive, with high variation among sampled families, but none of them was positive for M. gallisepticum and M. synoviae. The protected area closer to human settlements presented more resistant isolates to broad-spectrum antibiotics gentamicin (φ = 0.45) and tetracycline (φ = 0.37) and also presented the two positive samples to primary pathogenic Chlamydia psittaci. The birds in the sampled protected areas may host and spread potentially pathogenic microorganisms as C. psittaci and Citrobacter freundii in low frequency in balanced co-existence of host/parasite. However, antibiotic-resistant Enterobacterales in protected areas might represent an impact on its bird populations and on the conservation of the environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Populations of birds have been declining for decades, especially in the last 50 years (Rosenberg et al. 2019). Several anthropogenic factors are driven by the decline or extinction of the bird populations, which can include introduced, transmitted, or emerged pathogens (Daszak et al. 2000; Heard et al. 2013). Healthy wild migratory and resident birds are susceptible to several bacterial pathogens common to humans and domestic animals (Hubálek 2004; Benskin et al., 2009; Stenkat et al. 2014; Dias et al. 2019) and may present acute or chronic disease. They may die from the pathogen themselves, cure or become asymptomatic reservoirs, and act as biological carriers of several bacteria for prolonged periods (Hubálek 2004; Benskin et al., 2009). They can also be responsible for transporting vectors of bacteria (Hasle, 2013; Lugarini et al. 2015) or become pathogen reservoirs of these vector-borne bacteria (Hornok et al. 2014). Despite its relevance for pathogen transmission, gastrointestinal microbial structure in wild birds remains understudied. Therefore, descriptive epidemiological studies are needed to increase understanding of baseline microbial diversity within and among the avian group (Grond et al. 2018).
If birds can be contaminated by environmental bacteria from humans and domestic animals, they can be used as a sentinel of the contamination of soil, water, or plant matter with potential avian and/or human pathogens (Hamer et al. 2012). Understanding whether the different level of sharing human environment affect the infectious organism is important to access the risk of spreading these pathogens in stressful situations, such as climate changes, human contact, and overcrowding, which might unbalance the host/parasite state (Corrêa et al. 2013). However, several other factors as the life history of the hosts (i.e., diet, nesting environments, social interactions), water, and ground can influence the bird’s bacterial exposure, susceptibility, and spread (Benskin 2009; Grond et al. 2018). In this instance, the feeding ecology appears to be the main factor influencing the occurrence of different bacteria in healthy free-living birds (Brittingham et al. 1988; Stenkat et al. 2014). Terrestrial-foraging and water-dependent birds tend to increase the exposure to bacterial pathogens, because they may ingest contaminated food or water by bird droppings, nasal discharges, and respiratory exudates with Chlamydia psittaci, Salmonella, Escherichia coli, Enterococcus faecalis, Clostridium, among others (Hubálek 2004). The body size may also influence susceptibility to pathogens as the bacterial acquisition occurs predominantly through foraging, and larger individuals should eat more, and increase the exposure to infected food (Benskin et al., 2009). Some groups of birds such as raptors are also more exposed to the potential pathogens from the intestines of the prey they ingest or scavengers, which are exposed to bacteria from carcasses. Birds inhabiting opened areas, which also can be benefited by human activities or adapted to perianthropic habitats, would have more exposure to the bacterial pathogens than birds that inhabits forest habitats, because they may share foraging areas with sewage, carrion, water, and contaminated food by the excreta of other animals or humans (Fenlon 1983; Williams et al. 1976; Silva et al. 2010). The shedding rate of an agent or duration and level of its bacteremia in infected migrating birds might increase due to the stress and reduce the resistance to infections during migration. In this sense, the number of pathogenic agents associated with migratory birds is probably greater than with resident species (Hubálek 2004).
Therefore, we tested whether the bacteria occurrence varies according to the level of the shared habitat human or the type of vegetation the birds usually occupy (opened or forested areas), hosts’ group, and different history as migration and foraging behavior (foraging stratum and main trophic category), body mass, and sensitivity to human impacts. We were also interested in whether the protected area is preserving the wildlife from antibiotic-resistant bacteria. We focused on opportunistic and primary bacterial pathogens reported in birds, associated with humans and domestic animals shared environment, as Enterobacterales, Mycoplasma gallisepticum, M. synoviae, and Chlamydia psittaci (Brittingham et al. 1988; Andersen and Vanrompay 2000; Nascimento et al. 2005; Silva et al. 2010; Vilela et al. 2012; Dias et al. 2014, 2019; Afema and Sischo 2016; Silva et al. 2018).
Methods
Sampling
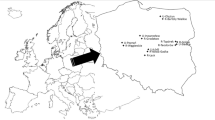
Sample collection was carried out at Guaribas Biological Reserve (GBR) in northern coastal Atlantic Forest of Paraíba state, Brazil, with a territorial extent of 4051.62 ha, divided into three small fragmented areas, surrounded by sugar cane plantations, villages, and indigenous communities in the municipalities of Mamanguape and Rio Tinto, and Raso da Catarina Ecological Station (RCES), a large continuous protected area with 104,842.84 ha in the municipalities of Jeremoabo, Rodelas, and Paulo Afonso, situated in Brazilian semiarid region, specifically in the Caatinga domain (Fig. 1). GBR birds would have a higher risk of acquiring and shedding pathogenic bacteria, considering the proximity of the urban areas, while remoteness habitats of RCES, far from urban influence, could have a lower occurrence of pathogenic bacterial pathogens.
Sample collection was carried out at Guaribas Biological Reserve (GBR) in northern coastal Atlantic Forest of Paraíba state, Brazil, with the territorial extent of 4051.62 ha, divided into three small fragmented areas, surrounded by sugar cane plantations, villages, and indigenous communities in the municipalities of Mamanguape and Rio Tinto, and Raso da Catarina Ecological Station (RCES), a large continuous protected area with 104,842.84 ha in the municipalities of Jeremoabo, Rodelas, and Paulo Afonso, situated in Brazilian semiarid region, specifically in the Caatinga domain
Birds were captured with mist nets, individually identified with aluminum leg bands, examined for body condition, body mass, sampled, and released. At the sampling moment, all birds were apparently healthy and samples were collected for culture-based methods and PCR. For culture and isolation, cloacal swabs were collected and stored in a Stuart medium at 4 °C and processed in 48 h. For molecular analysis, oropharyngeal and cloacal swabs were combined in the same vial with virus transport media (VTM): PBS-balanced salt solution supplemented with 0.5% bovine albumin, antimicrobial agents (200 U/mL penicillin G, 200 U/mL streptomycin, 25 μg/mL fungisone, and 6 μg/mL gentamycin), and 10% glycerol (Araujo et al. 2014). Samples were immediately stored in liquid nitrogen and then sent to the laboratory, stored at – 20 °C until processed.
Bacterial isolation
We used a culture-based method, involving general and selective media. The advantages of culture-based study are multifold and include reproducible results with minimal error; enable isolation of specific target organisms; and provide key data on multiple antibiotic resistance (McLain et al. 2016). The samples were plated on Petri dishes containing blood agar base supplemented with sheep blood 8% and Levine agar, incubated at 37 °C for 24–48 h. Initial identification was performed by morphology and color of the bacterial colonies. An aliquot of the isolated colonies was submitted to the Gram stain. The Gram-positive cocci were submitted to catalase, and Staphylococcus colonies were submitted to the coagulase test. Samples that were characteristic of Gram-negative coccobacillus were subjected to simplified biochemical profile: capacity for decarboxylation of lysine; citrate used as a source of carbon; mobility; production of hydrogen sulfide (H2S); Simmons’ citrate agar; Voges-Proskauer (VP); and sulfide indole motility (SIM) agar. For the Salmonella isolation, the samples were first subjected to pre-enrichment in peptone water and incubated for 24 h at 37 °C. Then, an aliquot from the peptone water was enriched in tetrathionate and Rapapport-Vassiliadis selective mediums broths at 37 °C for 24 h under agitation. After these steps, the samples were plated in xylose lysine deoxycholate (XLD) agar and bright green bile agar and confirmed with biochemical tests using triple sugar iron (TSI), lysine iron agar (LIA), and urea broth, following Litchfield (1973).
Four samples, randomly chosen, of Escherichia coli isolates were stored at – 20 °C and submitted to PCR with the aim of amplification of attaching and effacing (eae) and bundle-forming pili structural (bfpA) genes of enteropathogenic (EPEC); aerobactin (iucD), cytotoxic necrotizing factor (cnf1), S fimbrial adhesion (sfa), and P fimbrial adhesin (papEF) genes of avian pathogenic (APEC); alpha-hemolysin (HlyA) for uropathogenic (UPEC); and additional APEC genes for serum resistance (iss) and temperature-sensitive hemagglutinin (tsh) following Saidenberg et al. (2012).
Fifty-nine isolates were selected to be submitted to an antibiogram sensitivity test in disc diffusion assay in Muller-Hilton Agar (Bauer et al., 1966). Isolates were classified as resistant or susceptible to each antibiotic using and the zones of inhibition according to the standards of the Clinical and Laboratory Standards Institute-CLSI (2019). The chosen antibiotics were based on Guo et al. (2016) that detected the highest frequencies and concentrations of tetracyclines, fluoroquinolones and sulphonamides as typical veterinary antibiotics in manure and soil of livestock farms soil. Therefore, one representative of the antimicrobial classes was used: tetracycline (30 μg), fluoroquinolone (norfloxacin, 10 μg), and sulfonamide (sulfazotrin 25 μg). Other classes of former studies considering bird bacteria were also considered as aminoglycoside (gentamicin, 10 μg) and beta-lactam (penicillin G, 10 U, ampicillin, 10 μg, amoxicillin, 10 μg) (e.g., Afema and Sischo 2016; Matias et al. 2016).
Mycoplasma spp. and Chlamydia psittaci DNA detection
The suspension of VTM was thawed and centrifuged for 5 min at 14,000 g; 500 μL was submitted to DNA extraction. DNA was extracted from swab samples with Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) or Qiagen DNA Easy Blood and Tissues Kit (Qiagen, Valencia CA, USA), according to manufactures.
For Mycoplasma spp., a set of primers based on the amplification of mycoplasmal 16S rRNA sequences (van Kuppeveld et al. 1992, 1994) was used: GPO-3 (5′-GGGAGCAAACAGGATrAGATACCCT-3′) and MGSO (5′-TGCACCATCTGTCACTCTGTTAACCTC-3′) that amplify a product of 280 bp with the thermal cycles, following and reaction according to Santos et al. (2013). Positive screened samples were submitted to Mycoplasma gallisepticum (MG)/Mycoplasma synoviae (MS) amplification.
For MG, the pair of primers was used: MG-14F (5′-GAGCTAATCTGTAAAGTTGGTC-3′) and MG-13R (5′-GCTTCCTTGCGGTTAGCAAC-3′) that produces an amplicon with 481 bp (OIE 2008). For MS, the pair of primers MS–F (5′-GAGAAGCAAAATAGTGATATCA-3′) and MS–R (5′-CAGTCGTCTCCGAAGTTAACAA-3′) was used, which generate products with 207 bp (Lauerman et al. 1993). The final volume of the reaction was 50 μL, including 2.5 μL of 10 × PCR buffer, 2 μL of MgCl2, 1 μL of dNTP mixture (l0 mM), 0.5 μL of each primer (20 pmol/μL), 0.5 μL of Taq (2.5 U/μL), 8.5 μL of ultra-pure water, and 5 μL of template DNA. As a positive control, we used the standard strains of “American Type Culture Collection” (ATCC) MG (MGR-13610) and MS (WVU-1853). PCR cycle was performed with 40 cycles at 94 °C for 30 s, at 55 °C for 30 s and at 72 °C for 1 min, and final extension at 72 °C for 5 min. Amplification products were subjected to 30-min electrophoresis in 1.5% agarose gel, under 80-Volt. Electrophoresis gels were carried out in 1 × TAE buffer (EDTA Tris acetic acid), exposed to ultra-violet light, and photographed by photo documentation.
For Chlamydia psittaci, PCR based on the conserved region of the major outer membrane protein (MOMP) gene was performed using the pairs of primers Cpsi A (5′-ATGAAACATCCAGTCTACTGG-3′) and Cpsi B (5′-TTGTGTAGTAATATTATCAAA-3′), which amplify a product of 300 bp. The final volume of the reaction was 25 μL, including 10 mM of Tris–HCl (pH 8.3), 4 mM of MgCl2, 0.2 mM of dNTP mixture, 5 pmol of each primer, 0.5 U of Taq polymerase, 5 μL of template DNA, and ultra-pure water. PCR cycle was performed with the first cycle at 94 °C for 5 min, followed by 40 cycles at 94 °C for 1 min, at 50 °C for 1 min and at 72 °C for 2 min, and final extension at 72 °C for 10 min. Brazilian strain Cpsi/Mm/BR01 (GenBank number JQ926183.1) and ultra-pure water were used as positive and negative controls, respectively. Amplification products were subjected to 40-min electrophoresis in 1.5% agarose gel colored with 0.5 μg/10 mL of GelRed (Uniscence do Brasil), under 100-Volt. Electrophoresis gels were carried out in 1 × TAE buffer (EDTA Tris acetic acid), exposed to ultra-violet light, and photographed by photo documentation.
Statistical analysis
We tested by the chi-square test whether there was the association between Enterobacterales and Mycoplasma occurrence and (1) the protected areas (GBR or RCES) and (2) type of vegetation (forested—arboreal Caatinga and semideciduous forest—vs. opened areas—Cerrado enclaves and shrubby Caatinga). Chlamydia psittaci occurrence was excluded due to the low number of positive samples. We tested by the G-test on the DescTools package test whether there was an association between Enterobacterales occurrence: (1) hosts’ traits as foraging stratum, and main trophic category, and (2) sensitive to anthropogenic activities. For the relationship between migratory status and occurrence of Enterobacterales, we used Fisher’s exact test, as the expected cell values fell below 5. The occurrence was calculated by the proportion of the positive samples for each bacterium divided by the number of examined samples * 100. The main trophic category of bird species and foraging strata was based in Wilman et al. (2014) when available or Silva et al. (2003), Donatelli et al. (2004), Telino-Júnior et al. (2005), and Del Hoyo et al. (2020). Sensitivity of species to anthropogenic impacts was based on Stotz et al. (1996) and migration status followed Somenzari et al. (2018). We used logistic regression to see if the presence or absence of Enterobacterales (dependent variable) of positive samples was associated with the body mass of the sampled birds (independent variable).
For estimation of the association between the Enterobacterales occurrence and well-sampled hosts’ families and orders, we used a minimum sample size of ~ 8 positive samples, considering accurate estimates of prevalence depends on adequate sampling (Jovani and Tella 2006): Columbiformes (Columbidae), Apodiformes (Trochilidae), and Passeriformes (Thamnophilidae, Dendrocolaptidae, Pipridae, Rhynchocyclidae, Tyrannidae, Polioptilidae, Passerellidae, and Thraupidae). Turdidae was excluded as presented no Enterobacterales. For Mycoplasma, we compared the occurrence of well-sampled families: Columbiformes (Columbidae) and Passeriformes (Thamnophilidae, Furnariidae, Pipridae, Tyrannidae, Passerellidae, Parulidae, Thraupidae, and Cardinalidae). Rhynchocyclidae and Turdidae were not considered, because there were no positive samples for these families. For the association, we used the G-test on the DescTools package. Significance was established at P < 0.05. Phi coefficient (φ) was used to measure the strength of association between two categorical variables and was calculated with the Psych package. All statistical tests were performed in R v3.6.3 (R Core Team 2020).
Results
We sampled 507 individuals from 91 species, 30 families of 10 orders of birds, 245 from GBR, and 262 from RCES, from March 2012 to December 2013. The most common sampled bird species belonged to Passeriformes order (88.4%) with 20 families, followed by Apodiformes (4.7%) and Columbiformes (3.6%); the other seven orders have had less than 10 samples each (Suppl. Material, Table S1). Most (97.2%) of the species were residents and only six (2.8%) species were considered partially migratory, according to Somenzari et al. (2018).
Bacterial isolation
For bacterial isolation, samples from 292 individuals were collected (92 from RCES and 200 from GBR). Overall, 136 samples (43.5%) were positive for at least one bacterium colony in the culture-dependent method, representing 161 isolates. Most of them (57.1% of the total) were Gram-negative strains that belonged to Enterobacterales. The occurrence of Enterobacterales was higher for Klebsiella aerogenes (11.3%) and Escherichia coli (10.7%), followed by Gram-positive bacteria Staphylococcus spp. (6.8%) and Bacillus spp. (6.5%). Only one strain of Staphylococcus was positive coagulase (S. aureus; 0.3% occurrence). No sample was positive to Salmonella spp. Escherichia coli isolates submitted to PCR did not amplify fragments of the tested genes of pathotypes EPEC, APEC, and UPEC.
The occurrence of Enterobacterales was significantly higher (χ2 = 8.8, df = 1, P < 0.01; φ = 0.18) in RCES (41.3%) than GBR (23.5%) (Suppl. Material, Tables S2). We did not find significant differences in the prevalence of Enterobacterales between hosts’ orders and families, type of vegetation, migratory status, forage strata, main trophic category, and sensitivity to anthropogenic activities (Suppl. Material, Tables S1 and S3). The logistic regression did not show an association of the occurrence of aerobically bacteria and the body mass of the sampled birds (Z = − 1.539; P = 0.12).
From isolates submitted to antibiogram, 20.3% was susceptible to all seven tested antibiotics (Suppl. Material, Table S4). Klebsiella aerogenes have more isolates resistant to tetracycline; however, it was not significant when compared with other tested bacteria. GBR presented more resistant isolates to gentamicin (χ2 = 9.5, df = 1, P = 0.002; φ = 0.45) and tetracycline (χ2 = 4.8, df = 1, P = 0.03; φ = 0.37) than RCES. No difference was detected between the types of vegetation.
Mycoplasma spp. and Chlamydia psittaci DNA detection
We tested 287 samples (176 from RCES and 111 from GBR) oropharyngeal/cloacal samples of healthy birds for Mycoplasma spp. and obtained 10.8% amplicons of 280 bp (Suppl. Material, Tables S1 and S2). From 31 Mycoplasma spp.-positive samples, none presented amplified products for MG and MS. For Chlamydia psittaci, we tested 292 samples (169 from RCES and 123 from GBR) and only 0.7% yielded C. psittaci DNA from Gray-fronted Dove (Leptotila verreauxi) and Striped Cuckoo (Tapera naevia), both from GBR (Suppl. Material, Tables S1–S3). Because we obtained only two positive samples, we did not perform any statistical analysis.
There was a significant difference in the occurrence of Mycoplasma spp. (G-test = 22.2, df = 9, P < 0.01, φ = 0.37) according to the family Cardinalidae (50%), Furnariidae (27.3%), Columbidae (22.2%), Pipridae (16.7%), Parulidae (12.5%), Tyrannidae (10.5%), Passerellidae (7.1%), Thamnophilidae (3.7%), and Thraupidae (3.03%) (Suppl. Material, Table S1). We did not find significant differences in the frequency of Mycoplasma between hosts’ orders, sampled protected area, and the type of vegetation.
Discussion
This study reported the occurrence of Enterobacterales, Mycoplasma spp., and C. psittaci in swabs of free-living birds from protected areas of Caatinga and Atlantic Forest in northeastern Brazil. Formerly, it was demonstrated that the microbiota of passerine and woodpeckers (Brittingham et al. 1988) was mostly composed of Gram-positive microorganisms. However, current studies showed frequent isolation of Enterobacterales bacteria in healthy free-living populations by culture-dependent methods (Saidenberg et al. 2012; Vilela et al. 2012; Stenkat et al. 2014; Serafini et al. 2015; Vaz et al. 2017; Machado et al. 2018). Therefore, Gram-negative bacteria may represent some part of the total aerobically cloacal bacteria of free-living wild birds, not representing an uncommon finding (Grond et al. 2018). Our sample units included understory passerines and near-passerine birds, and high occurrence of Enterobacterales was common as previously reported to asymptomatic passerines (Horn et al. 2015; Matias et al. 2016; Beleza et al. 2019), birds of Caatinga (Saidenberg et al. 2012; Machado et al. 2018), and Atlantic Forest, including birds in protected areas (Serafini et al. 2015; Vaz et al. 2017). These bacteria are often related to secondary infections, but can function as the primary pathogen in certain circumstances, depending on the virulence and the host response to the infections (Benskin et al., 2009). Escherichia coli represents one the most commonly isolated Gram-negative bacterial species of the normal gastrointestinal microbiota of many passerines (Horn et al. 2015; Matias et al. 2016) and several non-passerine avian species (Silva et al. 2009; Marietto-Gonçalves et al. 2010; Santos et al. 2010; Saidenberg et al. 2012; Corrêa et al. 2013; Lopes et al. 2015; Serafini et al. 2015; Matias et al. 2016; Vaz et al. 2017; Machado et al. 2018). Our results showed that E. coli was common in passerines and non-passerines species of Caprimulgiformes, Columbiformes, Galbuliformes, and Piciformes. Escherichia coli is often implicated in primary or secondary avian diseases (Gerlach 1994) and mortality in birds (Marietto-Gonçalves et al. 2007). Therefore, access to the degree of pathogenicity is important, due to the risk of the spread of pathogens in the environment, contributing to the epidemiological chain of several enteric diseases. However, the four tested samples used in this study did not show genes related to pathogenicity.
It was stated that Enterobacterales in carnivorous or omnivorous birds was a consequence of its growing motivation by animal protein diet (Glunder 2002). Brittingham et al. (1988) and Gerlach (1994) suggested that this bacteria family usually does not belong to the normal gastrointestinal microbiota of granivorous and herbivorous birds. However, Stenkat et al. (2014) isolated Enterobacterales regularly from all trophic categories sampled. In fact, there was no evidence of the influence of the main trophic category in the Enterobacterales occurrence in our study, showing that other factors are also involved in the occurrence of the Gram-negative in the gastrointestinal tract. We discarded the hypothesis that the ground-foraging birds could host higher enterobacterial occurrence. However, the sampled birds here were passerines and near passerines that are supposed to have less contact with fecal material, as they perch in branches and are not concentrated in the ground roosting. Waterbirds tend to be in contact with feces on the ground or in water and are more likely to become infected in case of environmental contamination (Silva et al. 2010, 2018; Stenkat et al. 2014); birds exposed to large aggregations for feeding or breeding may also spread gastrointestinal microbiota among conspecifics (Grond et al. 2018), which are not the case of our sampled birds. We also discarded the initial hypothesis that these birds were more exposed to human and domestic waste (less sensitive to anthropogenic impacts), being more affected by Enterobacterales. In fact, we could find a higher occurrence of Enterobacterales in the RCES than GBR. These results have to be carefully interpreted as there was a low association between the two variables (φ = 0.18), showing that other variables are hidden and influencing results.
The environment can be the main factor affecting passerine gut bacteria (Hird et al. 2014), but the host taxonomy might be the strongest determinant of the gut microbial community (Hird et al. 2015). Then, the susceptibility among bird groups is highly variable (Matias et al. 2016), and the influence of the environment in the gastrointestinal bacteria can be less expressive for some bird groups (Stenkat et al. 2014). The susceptibility of the bird depends on the specificity of each strain of the bacteria (Afema and Sischo 2016), which can justify the difference in the occurrence of Mycoplasma in well-sampled families. Nineteen host species have described the hosting DNA of Mycoplasma spp. There are several species of Mycoplasma, but only MG, MS, and M. meleagridis are considered economically important to the poultry and turkeys and obligatory notifiable according to the Brazilian National Avian Sanitary Program. Several other mollicutes, including M. iowa, M. iners, M. gallinarum, M. pullorum, M. gallopavonis, M. gallinaceum, M. columbinasale, M. columbinum, M. columborale, M. lipofaciens, M. glycophilum, M. cloacale, M. anseris, Uraaplasma galorale, and Acholeplasma laidlawii are not pathogens of major concern with very low or even lack of pathogenicity (Nascimento et al. 2005).
We found similar positivity (10.6%) as found by Silva et al. (2016) in captive psittacine birds (16.5%) in northeastern Brazil, showing that it is possible to find Mycoplasma spp. in health birds in captivity or wildlife. The Mycoplasma prevalence might be higher in healthy captivity birds, and also comprehend pathogenic MG and MS (Carvalho et al. 2017; Magalhães et al. 2020), which was not the case in this pioneering study in healthy free Brazilian birds, probably due to low exposition to competent hosts, as birds of prey, Galliformes, Piciformes (Magalhães et al. 2020) or Psittaciformes (Carvalho et al. 2017). The differences found between the well-sampled families in the prevalence of Mycoplasma reaching 50% for Cardinalidae are probably related to the high specificity to their hosts and must be focused on in future studies. Some wild passerines as House Finches (Carpodacus mexicanus) are affected by conjunctivitis caused by MG (Farmer et al. 2005), as a result of shifts and mutations (Staley et al. 2018), which might also represent an important factor to control the bird populations in Brazil in the future.
Chlamydia psittaci is widespread in Brazil and has been detected in different avian species, especially in psittacines maintained in captivity (Raso et al. 2002; Santos et al. 2014) or in healthy free-living populations (Raso et al. 2006; Ribas et al. 2014), and pigeons (Leal et al. 2015; Ferreira et al. 2016). The prevalence of C. psittaci in the sampled protected areas was low, in agreement with other rural sampled sites. Even in more susceptible bird’s groups, the prevalence tends to be low or null in cloacal and tracheal or oropharyngeal samples (Raso et al. 2006; Ribas et al. 2014; Vaz et al. 2017), while in urban habitats (Leal et al., 2015) and captive psittacine birds the prevalence is higher, with 10–72% of positivity for C. psittaci (Raso et al. 2002; Santos et al. 2014; Vilela et al. 2019), especially when birds are submitted to the stress of the illegal trade, high-density facilities and poor management conditions (Santos et al. 2014; Vilela et al. 2019). The abundance of susceptible hosts may be linked with the high prevalence, while less impacted habitats present low occurrence of the bacteria, due to the diversity of birds (Lima et al. 2011). The presence of C. psittaci in GBR was related especially with Columbidae, a common reservoir family for the pathogen (Kaleta and Taday 2003), but it is the first time it is found in L. verreauxi. On the other hand, Cuculiformes is one of the families with less effort spent in surveys and is new host species for C. psittaci.
The absence of Salmonella, MG, MS, pathogenic E. coli and other bird primary bacteria in this study, as Klebsiella pneumoniae (Gerlach 1994) and also viruses in these sampled populations (Lugarini et al. 2018) can be justified by the low contamination and spread bacteria and other pathogens in the protected relicts. These protected areas showed low risk for emergence of bacteria in birds, different of previously searched high disturbed areas (e.g., Silva et al. 2009; Afema and Sischo 2016; Dias et al. 2019), showing their importance to safeguard birds healthy. Low primary bacteria occurrence across sampled sites may be directly related to high species richness and low migrant density that is responsible to spread pathogens (Afema and Sischo 2016). Most of the sampled species are resident and the occurrence of these bacteria represents the local microbiota of the birds. Compared to migratory birds, residents may be less exposed to diverse microbiota (Grond et al. 2018). Therefore, the “dilution effect” of bird richness, especially non-competent hosts, can account for the low capacity of transmission of pathogens for humans and animals (Swaddle and Calos 2008; Keesing et al. 2010).
Overall, 5.1% of tested isolates have been considered resistant to three tested antibiotics or more. These results have to be carefully interpreted, as few antimicrobial classes were opportunistic tested (Magiorakos et al. 2012). Klebsiella aerogenes, Corynebacterium spp., and Staphylococcus spp. presented resistant isolates to all the tested antimicrobial drugs. However, Enterobacterales are intrinsic resistant to penicillin (Magiorakos et al. 2012), and it is known that K. aerogenes and Citrobacter freundii are intrinsic resistant to penicillin, ampicillins, and cephalosporin (CLSI 2019). The fact of tetracycline showed resistance for more than half of K. aerogenes isolates; the potential pathogenic bacterium E. coli showed resistance to a broad-spectrum of antimicrobial drugs such as tetracycline, gentamicin, and norfloxacin, and S. aureus and Shigella isolates which showed high resistance to sulfazotrin antibiotic resistant are important concerns since even in low disturbance regions we can observe the resistance of bacteria against antibiotics. Amoxicillin, ampicillin, tetracycline, gentamicin, and sulfonamide are commonly used human and veterinary drugs, and the emergence of resistance may indicate a widespread of resistant bacteria in different environments (Machado et al. 2018), with the excessive and inadequate use of antibiotics (Nascimento et al. 2003). The spread and elevated use of antibiotics in songbirds for example, at home, can be the cause of the spread of multi-resistant drug isolates into wild birds (Horn et al. 2015). Livestock is the main reservoir of resistant bacteria for environmental contamination (Sayah et al. 2005) and the contact between poultry, backyard chickens, and captive birds is common (Scherer et al. 2011). In questionnaires applied for subsistence farmers around GBR (data not showed), most of them are related to the use a broad range of antibiotics, including tetracycline, which are commonly found in pet shops in the region and are sold without veterinary prescription. The backyard chickens created surrounding GBR, which might be responsible for the resistant strains (Guo et al. 2016), were seen in strict contact with the wild birds since they forage inside boundaries of the protected area, especially in the fragment near to the urban area of Rio Tinto municipality. Notably, GBR presented a higher frequency of resistant isolates. Not surprised, the only C. psittaci-positive samples were from GBR. For C. psittaci, birds can develop acute, subacute, chronic, and unapparent diseases; the last form of the disease is more common, responsible for the intermittent spread of the pathogen, shedding the organism over long periods, contributing to the dissemination, and representing a significant source of infection for other birds (Fudge 1996). Another potentially primary pathogenic enterobacteria found only in GBR was C. freundii, isolated from one apparently healthy individual, indicating that wild birds at GBR host it in low frequency, and may be transient reservoirs (Glunder 2002). This bacterium was also isolated previously from black cormorants Phalacrocorax carbo (Stenkat et al. 2014) and red-tailed parrots Amazona brasiliensis (Vaz et al. 2017), and here from one black-cheeked gnateater (Conopophaga melanops), a threatened species. No bird sampled presented signals of any diseases and the presence of bacteria, suggesting a balanced co-existence of host/parasite. However, the lack of clinical signs can be also attributed to the fast removal of ill and dead birds from predators and scavengers, which do not allow us to notice mobility or mortality (Brand 1989). The occurrence of a primary pathogen in a free-living population without apparent infection can alert to the potential risk of outbreaks if stressful episodes, such as abrupt variation in weather conditions and environmental changes, including habitat loss (Ribas et al. 2014). Moreover, C. psittaci represents important zoonosis (Petrovay and Balla 2008) and the contact of humans with free-living birds is common in northeast Brazil, driven by the trapping and traffic (Alves et al. 2013), which can also impact public health (Raso et al. 2014).
Brazil has a high diversity of resident and migratory birds and several habitats that are crucial for their conservation. Here, it was demonstrated the presence of Enterobacterales as the usual constituents of the aerobically cloacal bacteria of wild birds in protected areas and the low prevalence of Mycoplasma spp. and C. psittaci. In addition, in Guaribas Biological Reserve closer to human settlement, the samples presented resistant isolates to broad-spectrum antibiotics like gentamicin and tetracycline and also presented few samples to primary pathogenic bacteria as C. psittaci and C. freundii. Then, the birds in the sampled protected areas may host and spread potentially pathogenic microorganisms in low-frequency balanced co-existence of host/parasite; however, primary pathogenic bacteria and drug-resistant bacteria in free-living birds associated with human and domestic animal shared environment can represent an impact to its bird populations and conservation of the environment.
Data availability
All the material has been submitted.
Code availability
Not applicable.
References
Afema JA, Sischo WM (2016) Salmonella in wild birds utilizing protected and human impacted habitats, Uganda. EcoHealth 3:558–569. https://doi.org/10.1007/s10393-016-1149-1
Andersen AA, Vanrompay D (2000) Avian chlamydiosis. Rev Sci Tech 19:396–404
Alves RRN, Lima JRF, Araujo HFP (2013) The live bird trade in Brazil and its conservation implications: an overview. Bird Conserv Int 23:53–65. https://doi.org/10.1017/S095927091200010X
Araujo J, Azevedo-Júnior SM, Gaidet N, Hurtado RF, Walker D, Thomazelli LM, Ometto T, Seixas MMM, Rodrigues R, Galindo DB, Silva ACS, Rodrigues AMM, Bomfim LL, Mota MA, Larrazábal ME, Branco JO, Serafini P, Simão-Neto I, Franks J, Webby RJ, Webster RG, Durigon EL (2014) Avian Influenza Virus (H11N9) in migratory shorebirds wintering in the Amazon region. Brazil PLoS ONE 9:e110141. https://doi.org/10.1371/journal.pone.0110141
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496. https://doi.org/10.1093/AJCP/45.4_TS.493
Beleza AJF, Maciel WC, Carreira AS, Bezerra WGA, Carmo CC, Havt A, Gaio FC, Teixeira RSC (2019) Detection of Enterobacterales, antimicrobial susceptibility, and virulence genes of Escherichia coli in Canaries (Serinus canaria) in northeastern Brazil. Pesq Vet Bras 39:201–208. https://doi.org/10.1590/1678-5150-pvb-5829
Benskin CMH, Wilson K, Jones K, Hartley IR (2009) Bacterial pathogens in wild birds: a review of the frequency and effects on infection. Biol Rev Camb Philos Soc 84:349–373. https://doi.org/10.1111/j.1469-185X.2008.00076
Brand CJ (1989) Chlamydial infections in free-living birds. J Am Vet Med Assoc 195:15311535
Brittingham MC, Temple SA, Duncan RM (1988) A survey of the prevalence of selected bacteria in wild birds. J Wildl Dis 24:299–307
Carvalho AM, Andrade MA, Linhares GF, Jaime VS (2017) Pesquisa de Mycoplasma em aves da família Psittacidae mantidas em diferentes cativeiros no Brasil Central. Pesq Vet Bras 37:1159–1164. https://doi.org/10.1590/s0100-736x2017001000019
Clinical and Laboratory Standards Institute (2019) Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI M100-S29, Wayne, Pennsylvania
Corrêa IM, Flores F, Schneiders GH, Pereira LQ, Brito BG, Lovato M (2013) Detecção de fatores de virulência de Escherichia coli e análise de Salmonella spp. em psitacídeos. Pesq Vet Bras 33:241–246. https://doi.org/10.1590/S0100-736X2013000200017
Daszak P, Cunningham AA, Hyatt AD (2000) Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287:443–449. https://doi.org/10.1126/science.287.5452.443
Del Hoyo J, Elliott A, Sargatal J, Christie DA (2020) Birds of the World. Cornell Lab of Ornithology, New York
Dias PA, Moraes TP, Wilsmann DE, Ferrasso MM, Marinheiro MF, Heinen JG, Calabuig CIP, Timm CD (2019) Ocorrência de Campylobacter e Enterobacterales em aves silvestres e frangos de corte. Arq Bras Med Vet Zootec 71:225–231. https://doi.org/10.1590/1678-4162-10289
Dias PA, Wilsmann DE, Heinen JG, Corsini CD, Calabuig C, Timm CD (2014) Primeiro relato de Salmonella enterica e Campylobacter spp. isolados de Garibaldi (Chrysomus ruficapillus) e Canário-da-terra (Sicalis flaveola) silvestres. Rev Inst Adolfo Lutz 73:368–371. https://doi.org/10.18241/0073-98552014731629
Donatelli RJ, Costa TVV, Ferreira CD (2004) Dinâmica da avifauna em fragmento de mata na Fazenda Rio Claro, Lençóis Paulista, São Paulo, Brasil. Rev Bras Zool 21:97–114. https://doi.org/10.1590/S0101-81752004000100017
Farmer KL, Hill GE, Roberts SR (2005) Susceptibility of wild songbirds to the House Finch strain of Mycoplasma gallisepticum. J Wildl Dis 41:317–325. https://doi.org/10.7589/0090-3558-41.2.317
Fenlon DR (1983) A comparison of Salmonella serotypes found in the faeces of gulls feeding at a sewage works with serotypes present in the sewage. J Hyg 91:47–52. https://doi.org/10.1017/s0022172400060010
Ferreira VL, Dias RA, Raso TP (2016) Screening of feral pigeon (Columba livia) for pathogens of veterinary and medical importance. Rev Bras Cienc Avi 18:701–704. https://doi.org/10.1590/1806-9061-2016-0296
Fudge AM (1996) Avian chlamydiosis. In: Rosskopf –Jr.WJ, Woerpel RW (Eds) Diseases of caged and aviary birds. 3rd edn. Williams & Wilkins Company, Malvern, pp 572–585
Gerlach H (1994) Bacteria. In: Ritchie BW, Harrison GJ, Harrison LR (eds) Avian medicine: principles and application. Wingers Publishing, Lake Worth, pp 949–983
Glunder G (2002) Influence of diet on the occurrence of some bacteria in the intestinal flora of wild and pet birds. Dtsch Tierarztl Wchschr 109:266–270
Grond K, Sandercock BK, Jumpponen A, Zeglin LH (2018) The avian gut microbiota: community, physiology and function in wild birds. J Avian Biol 49:e01788. https://doi.org/10.1111/jav.01788
Guo XY, Hao LJ, Qiu PZ, Chen R, Xu J, Kong XJ (2016) Pollution characteristics of 23 veterinary antibiotics in livestock manure and manure-amended soils in Jiangsu province. China J Environ Sci Heal B 51:383–392. https://doi.org/10.1080/03601234.2016.1142743
Hamer SA, Lehrer E, Magle SB (2012) Wild birds as sentinels for multiple zoonotic pathogens along an urban to rural gradient in greater Chicago, Illinois. Zoonoses Public Health 59:355–364. https://doi.org/10.1111/j.1863-2378.2012.01462
Hasle G (2013) Transport of ixodid ticks and tick-borne pathogens by migratory birds. Front Cell Infect Microbiol 3:48. https://doi.org/10.3389/fcimb.2013.00048
Heard MJ, Smith KF, Ripp KJ, Berger M, Chen J, Dittmeier J, Goter M, McGarvey ST, Ryan E (2013) The threat of disease increases as species move toward extinction. Conserv Biol 27:1378–1388. https://doi.org/10.1111/cobi.12143
Hird SM, Carstens BC, Cardiff SW, Dittmann DL, Brumfield RT (2014) Sampling locality is more detectable than taxonomy or ecology in the gut microbiota of the brood-parasitic Brown-headed Cowbird (Molothrus ater). PeerJ 2:e321. https://doi.org/10.7717/peerj.321
Hird SM, Sánchez C, Carstens BC, Brumfield RT (2015) Comparative gut microbiota of 59 Neotropical bird species. Front Microbiol 6:1403. https://doi.org/10.3389/fmicb.2015.01403
Horn RV, Cardoso WM, Lopes ES, Teixeira RSC, Albuquerque AH, Rocha-e-Silva RC, Machado DN, Bezerra WGA (2015) Identification and antimicrobial resistance of members from the Enterobacterales family isolated from Canaries (Serinus canaria). Pesq Vet Bras 35:552–556. https://doi.org/10.1590/S0100-736X2015000600011
Hornok S, Kováts D, Csörgő T, Meli ML, Gönczi E, Hadnagy Z, Takács N, Farkas R, Hofmann-Lehmann R (2014) Birds as potential reservoirs of tick-borne pathogens: first evidence of bacteraemia with Rickettsia helvetica. Parasit Vectors 7:128. https://doi.org/10.1186/1756-3305-7-128
Hubálek Z (2004) An annotated checklist of pathogenic microorganisms associated with migratory birds. J Wildl Dis 40:639–659. https://doi.org/10.7589/0090-3558-40.4.639
Jovani R, Tella JL (2006) Parasite prevalence and sample size: misconceptions and solutions. Trends Parasitol 22:214–218. https://doi.org/10.1016/j.pt.2006.02.011
Kaleta EF, Taday EMA (2003) Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol 32:435–461. https://doi.org/10.1080/03079450310001593613
Keesing F, Belden LK, Dobson P, Harvell CD, Holt RD, Hudson P, Jolles A, Jones KE, Mitchell CE, Myers SS, Bogich T, Ostfeld RS (2010) Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468:647–652. https://doi.org/10.1038/nature09575
Lauerman LH, Hoerr FJ, Sharpton SM, van Santen VL (1993) Development and application of a polymerase chain reaction assay for Mycoplasma synoviae. Avian Dis 37:829–834
Leal DC, Negrão VB, Santos F, Raso TF, Barrouin-Melo SM, Franke CR (2015) Ocorrência de Chlamydophila psittaci em Pombos (Columba livia) na cidade de Salvador, Bahia. Arq Bras Med Vet Zootec 67:771–776
Lima VY, Langoni H, Silva AV, Pezerico SB, Castro AP, Silva RC, Araújo-Jr JP (2011) Chlamydia psittaci and Toxoplasma gondii infection in Pigeons (Columba livia) from São Paulo state, Brazil. Vet Parasitol 175:9–14
Litchfield JH (1973) Salmonella and the food industry: methods for isolation, identification and enumeration. Critical Rev Food Technol 3:415–456
Lopes ES, Maciel WC, Albuquerque AH, Machado DN, Bezerra WGA, Vasconcelos RH, Lima BP, Marietto Gonçalves GA, Teixeira RSC (2015) Prevalence and antimicrobial resistance profile of Enterobacteria isolated from Psittaciformes of illegal wildlife trade. Acta Sci Vet 43:1313
Lugarini C, Hurtado R, Araujo J, Ometto T, Thomazelli L, Seixas M, Durigon E, Silva JC (2018) Lack of detection of avian influenza, Newcastle disease, and West Nile viruses in wild birds of northeastern Brazil. J Wildl Dis 54:422–425. https://doi.org/10.7589/2017-09-218
Lugarini C, Martins TF, Ogrzewalska M, Vasconcelos NC, Ellis VA, Oliveira JB, Pinter A, Labruna MB, Silva JC (2015) Rickettsial agents in avian ixodid ticks in northeast Brazil. Ticks Tick Borne Dis 6:364–375. https://doi.org/10.1016/j.ttbdis.2015.02.011
Machado DN, Lopes ES, Albuquerque AH, Horn RV, Bezerra WGA, Siqueira RAS, Lopes IT, Nunes FP, Teixeira RSC, Cardoso WM (2018) Isolation and antimicrobial resistance profiles of Enterobacteria from nestling Grey-Breasted Parakeets (Pyrrhura griseipectus). Rev Bras Cienc Avic 20:103–110. https://doi.org/10.1590/1806-9061-2017-0551
Magalhães BSN, Pereira VLA, Dias TS, Machado LS, Silva MM, Nascimento ER, Mendes-de-Almeida F, Almosny NRP (2020) Investigação de Mycoplasma spp. em aves do Zoológico do Rio de Janeiro por isolamento e PCR. Pesq Vet Bras 40:220–225. https://doi.org/10.1590/1678-5150-PVB-6447
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Marietto-Gonçalves GA, Almeida SM, Lima ET, Andreatti-Filho RL (2010) Detecção de Escherichia coli e Salmonella sp. em microbiota intestinal de Psittaciformes em fase de reabilitação para soltura. Braz J Vet Res Anim Sci 47:185–189. https://doi.org/10.11606/issn.1678-4456.bjvras.2010.26853
Marietto-Gonçalves GA, Lima ET, Sequeira JL, Andreatti Filho RL (2007) Colisepticemia em Papagaio Verdadeiro (Amazona aestiva). Rev Bras Saúde Prod an 8:56–60
Matias CAR, Pereira IA, Reis EMF, Rodrigues DP, Siciliano S (2016) Frequency of zoonotic bacteria among illegally traded wild birds in Rio de Janeiro. Braz J Microbiol 47:882–888. https://doi.org/10.1016/j.bjm.2016.07.012
McLain JE, Cytryn E, Durso LM, Young S (2016) Culture-based methods for detection of antibiotic resistance in agroecosystems: advantages, challenges, and gaps in knowledge. J Environ Qual 45:432–440. https://doi.org/10.2134/jeq2015.06.0317
Nascimento AM, Cursino L, Gonçalves-Dornelas H, Reis A, Chartone-Souza E, Marini MÂ (2003) Antibiotic-resistant Gram-negative bacteria in birds from the Brazilian Atlantic Forest. Condor 105:358–361. https://doi.org/10.1093/condor/105.2.358
Nascimento ER, Pereira VLA, Nascimento MGF, Barreto ML (2005) Avian mycoplasmosis update. Rev Bras Cienc Avi 7:1–9. https://doi.org/10.1590/S1516-635X2005000100001
OIE Terrestrial Manual (2008) Avian mycoplasmosis (Mycoplasma gallisepticum, Mycoplasma synoviae) In: OIE. Manual of diagnostic tests and vaccines for terrestrial animals pp 525–541
Petrovay F, Balla E (2008) Two fatal cases of psittacosis caused by Chlamydia psittaci. J Med Microbiol 57:1296–1298. https://doi.org/10.1099/jmm.0.2008/001578-0
R Core Team (2020) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org/index.html. Accessed 11 August 2021
Raso TF, Berchieri-Júnior A, Pinto AA (2002) Evidence of Chlamydophila psittaci infection in captive Amazon Parrots in Brazil. J Zoo Wildl Med 33:118–121. https://doi.org/10.1638/1042-7260(2002)033[0118:EOCPII]2.0.CO;2
Raso TF, Ferreira VL, Timm LN, Abreu MFT (2014) Psittacosis domiciliary outbreak associated with Monk Parakeets (Myiopsitta monachus) in Brazil: need for surveillance and control. JMM Case Reports 1:1–4. https://doi.org/10.1099/jmmcr.0.003343
Raso TF, Seixas GHF, Guedes NMR, Pinto AA (2006) Chlamydia psittaci in free-living Blue-fronted Amazon Parrots (Amazona aestiva) and Hyacinth Macaws (Anodorhynchus hyacinthinus) in the Pantanal of Mato Grosso do Sul, Brazil. Vet Microbiol 117:235–241
Ribas JM, Sipinski EAB, Serafini PP, Ferreira VL, Raso TF, Pinto AA (2014) Chlamydia psittaci assessment in threatened Red-tailed Amazon (Amazona brasiliensis) parrots in Paraná, Brazil. Ornithologia 6:144–147
Rosenberg KV, Dokter AM, Blancher PJ, Sauer JR, Smith AC, Smith PA, Stanton JC, Panjabi A, Helft L, Parr M, Marra PP (2019) Decline of the North American avifauna. Science 366:120–124. https://doi.org/10.1126/science.aaw1313
Saidenberg AB, Teixeira RHF, Guedes NMR, Allgayer MC, Melvelville PA, Benites NR (2012) Molecular detection of enteropathogenic Escherichia coli in asymptomatic captive psittacines. Pesq Vet Bras 32:922–926. https://doi.org/10.1590/S0100-736X2012000900017
Santos HF, Flôres ML, Lara VM, Sá MF, Battisti L, Lovato LT (2010) Cloacal microbiota identification and evaluation of the antimicrobial resistance in captive cracids from Rio Grande do Sul, Brazil. Pesq Vet Bras 30:1077–1082. https://doi.org/10.1590/S0100-736X2010001200013
Santos F, Leal DC, Raso TF, Souza BMPS, Cunha RM, Martinez VHR, Barrouin-Melo SM, Franke CR (2014) Risk factors associated with Chlamydia psittaci infection in psittacine birds. J Med Microbiol 63:458–463. https://doi.org/10.1099/jmm.0.060632-0
Santos SB, Pinheiro-Júnior JW, Oliveira AAF, Mota AR, Oliveira-Júnior MB, Veras GA, Nascimento ER, Mota RA (2013) Ocorrência de Mollicutes e Ureaplasma spp. em surto de doença reprodutiva em rebanho bovino no estado da Paraíba. Pesq Vet Bras 33:315–318. https://doi.org/10.1590/S0100-736X2013000300007
Sayah RS, Kaneene JB, Johnson Y, Miller R (2005) Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl Environ Microbiol 71:1394–1404. https://doi.org/10.1128/AEM.71.3.1394-1404.2005
Scherer AL, Scherer JFM, Petry MV, Sander M (2011) Occurrence and interaction of wild birds at poultry houses in southern Brazil. Rev Bras Ornitol 19:74–79
Serafini PP, Meurer R, Biesdorf SM, Sipinski EAB (2015) O uso da microbiologia como ferramenta para a conservação de aves ameaçadas: dados preliminares para o Papagaio-de-cara-roxa, Amazona brasiliensis (Aves: Psittacidae) no Paraná. Arq Ciênc Vet Zool UNIPAR 18:65–69
Silva JMC, Souza MA, Bieber AGD, Carlos CJ (2003) Aves da Caatinga: status, uso do habitat e sensitividade. In: Leal IR, Tabarelli M, Silva JMC (eds) Ecologia e conservação da Caatinga. Universitária da UFPE, Recife, pp 237–274
Silva LTR, Santos SB, Rameh-de-Albuquerque LC, Siqueira DB, Amorim MMR, Almeida JC, Oliveira AAF, Mota RA (2016) Detecção molecular e isolamento de Mycoplasma spp. em psitacídeos no estado de Pernambuco. Brasil Arq Bras Med Vet Zootec 68:113–118. https://doi.org/10.1590/1678-4162-8387
Silva MA, Fernandes EFST, Santana SC, Marvulo MFV, Barros MR, Vilela SMO, Reis EMF, Mota RA, Silva JCR (2018) Isolation of Salmonella spp. in Cattle Egrets (Bubulcus ibis) from Fernando de Noronha Archipelago. Brazil Braz J Microbiol 9:559–563. https://doi.org/10.1016/j.bjm.2018.01.004
Silva MA, Marvulo MFV, Mota RA, Silva JCR (2010) A importância da ordem Ciconiiformes na cadeia epidemiológica de Salmonella spp. para a saúde pública e a conservação da diversidade biológica. Pesq Vet Bras 30:573–580. https://doi.org/10.1590/S0100-736X2010000700011
Silva VL, Nicoli JR, Nascimento TC, Diniz CG (2009) Diarrheagenic Escherichia coli strains recovered from urban Pigeons (Columba livia) in Brazil and their antimicrobial susceptibility patterns. Curr Microbiol 59:302–308. https://doi.org/10.1007/s00284-009-9434-7
Somenzari M, Amaral PP, Cueto VR, Guaraldo AC, Jahn AE, Lima DM, Lima PC, Lugarini C, Machado CG, Martinez J, Nascimento JLX, Pacheco JF, Paludo D, Prestes NP, Serafini PP, Silveira LF, Sousa AEBA, Sousa NA, Souza MA, Telino-Júnior WR, Whitney BM (2018) An overview of migratory birds in Brazil. Pap Avulsos Zool 58:e20185803. https://doi.org/10.11606/1807-0205/2018.58.03
Staley M, Hill GE, Josefson CC, Armbruster JW, Bonneaud C (2018) Bacterial pathogen emergence requires more than direct contact with a novel passerine host. Infect Immun 86:e00863-17. https://doi.org/10.1128/IAI.00863-17
Stenkat J, Krautwald-Junghanns ME, Schmitz Ornes A, Eilers A, Schmidt V (2014) Aerobic cloacal and pharyngeal bacterial flora in six species of free-living birds. J Appl Microbiol 117:1564–1571. https://doi.org/10.1111/jam.12636
Stotz DF, Fitzpatrick JW, Parker TA III, Moskovits DK (1996) Neotropical birds: ecology and conservation. University of Chicago Press, Chicago
Swaddle JP, Calos SE (2008) Increased avian diversity is associated with lower incidence of human West Nile infection: observation of the dilution effect. PLoS ONE 3:e2488. https://doi.org/10.1371/journal.pone.0002488
Telino-Júnior WR, Dias MM, Azevedo-Junior SM, Lyra-Neves RM, Larrazabal MEL (2005) Estrutura trófica da avifauna na Reserva Estadual de Gurjaú, Zona da Mata Sul, Pernambuco, Brasil. Rev Bras Zool 22:962–973. https://doi.org/10.1590/S0101-81752005000400024
van Kuppeveld FJM, van der Logt JT, Angulo AF, van Zoest MJ, Quint WG, Niesters HG, Galama JM, Melchers WJ (1992) Genus- and species-specific identification of Mycoplasmas by 16 rRNA amplification. Appl Environ Microbiol 58:2606–2615
van Kuppeveld FJM, Johansson KE, Galama JMD, Kissing J, Bölske G, van der Logt JT, Melchers WJ (1994) Detection of mycoplasma contamination in cell cultures by a mycoplasma group-specific PCR. Appl Environ Microbiol 60:149–152
Vaz FF, Serafini PP, Locatelli-Dittrich R, Meurer R, Durigon EL, Araújo J, Thomazelli LM, Ometto T, Sipinski EAB, Sezerban RM, Abbud MC, Raso TF (2017) Survey of pathogens in threatened wild Red-tailed Amazon Parrot (Amazona brasiliensis) nestlings in Rasa Island, Brazil. Braz J Microbiol 48:747–753. https://doi.org/10.1016/j.bjm.2017.03.004
Vilela DAR, Marín SY, Resende M, Coelho HLG, Resende JS, Ferreira-Júnior FC, Ortiz MC, Araújo AV, Raso TF, Martins NRS (2019) Phylogenetic analyses of Chlamydia psittaci ompA gene sequences from captive Amazona aestiva (Aves: Psittaciformes) with hepatic diseases. Rev Sch Tech Off Int Epiz 38:1–20
Vilela SMO, Pinheiro-Júnior JW, Silva JSA, Pace F, Silveira WD, Saukas TN, Reis EMF, Mota RA (2012) Research of Salmonella spp. and evaluation of pathogenicity, cytotoxicity of Escherichia coli isolates proceeding from Sparrows (Passer domesticus). Pesq Vet Bras 32:931–935. https://doi.org/10.1590/S0100-736X2012000900019
Williams BM, Richards DW, Lewis J (1976) Salmonella infection in the Herring Gull. Vet Rec 98:51
Wilman H, Belmaker J, Simpson J, De La Rosa C, Rivadeneira MM, Jetz W (2014) EltonTraits 1.0: species-level foraging attributes of the world’s birds and mammals. Ecol 95:2027. https://doi.org/10.1890/13-1917.1
Acknowledgements
We are thankful to the National Council for Scientific and Technological Development (CNPq 481663/2011-8) for its financial support, for the fellowships to RAM and JCRS, to the National Center for Bird Conservation Research (CEMAVE), and especially to the field team Adriana C. Dias, Ailton de Oliveira, Andressa N. Pessoa, Andressa C. Scabin, Antonio E.B. Sousa, Cristine Prates, Cayo L.G. Silva, Dillys Costa, Fabiane F. Dias, Fernanda P. Marques, Flor Maria Guedes Las Casas, Ingrid M.D. Torres, Jonathas L. Souza, José T.A. Santos, Leontina H.M. Andrade, Ludmila M. Magroski, Márcia S. Amorim, Marcus M.R. Amorim, Murilo Arantes, Nathália C. T. Vasconcelos, Rachel Lyra-Neves, Randson M. C. Paixão, Renata F. Hurtado, Roberta Rodrigues, Sebastião S. Santos, and Thayz R. Enedino.
Funding
This study was funded by the National Council for Scientific and Technological Development (CNPq 481663/2011–8).
Author information
Authors and Affiliations
Contributions
C.L. conceived of the presented idea. C.L. and M.M.R.A. performed the field expeditions. L.T.R.S., C.L., and S.B.S. performed the Mycoplasma PCRs. M.M.R.A. and D.C.V.L. performed the aerobic bacteria isolation. A.B.S. performed the PCR for virulent genes of Escherichia coli, T.F.R. performed the Chlamydia psittaci PCR. J.C.R.S. and R.A.M. supervised the project. C.L. performed the computations and analytical methods. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All captures and procedures were authorized by The Chico Mendes Institute for Biodiversity Conservation (SISBIO numbers 23405, 36538 and 36299, SNA numbers 3604, 3625) and by the Bioethics Committee of the “Universidade Federal Rural de Pernambuco” (number 040/2013).
Consent to participate
Not applicable.
Consent for publication
All the authors are in accordance to publish this article. We declare that it is an original article and it has not been published before; not under consideration for publication anywhere else. The final version has been approved by all co-authors.
Competing interests
The authors declare no competing interest.
Additional information
Communicated by Caio Graco Machado.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lugarini, C., Silva, L.T.R., de Amorim, M.M.R. et al. Free-living birds from Caatinga and Atlantic Forest of northeast Brazil as hosts of Enterobacterales, Mycoplasma spp., and Chlamydia psittaci. Ornithol. Res. 29, 149–159 (2021). https://doi.org/10.1007/s43388-021-00063-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43388-021-00063-0