Abstract
A single-center retrospective study of G-band karyotyping and chromosomal microarray analysis (CMA) for the invasive prenatal diagnosis of 6159 fetuses with ultrasound abnormalities was conducted. This study aimed to investigate the incidence rates of chromosomal abnormalities and pregnancy outcomes and postpartum clinical manifestations by long-term follow-up and to explore the correlation between different types of prenatal ultrasound abnormalities and pathogenic chromosomal abnormalities. The overall incidence of pathogenic chromosomal aberrations in fetuses with ultrasound abnormalities was 7.58% (467/6159), which comprised 41.7% (195/467) with chromosome number abnormalities, 57.6% (269/467) with pathogenic copy-number variations (pCNVs), and 0.64% (3/467) with uniparental disomy (UPD). In addition, 1.72% (106/6159) with likely pathogenic copy-number variations (lpCNVs) and 3.04% (187/6159) with variants of unknown significance (VOUS) were detected by CMA. Ultrasound abnormalities were categorized into structural anomalies and soft marker anomalies. The incidence rate of pathogenic and likely pathogenic chromosomal abnormalities was significantly higher among fetuses with structural anomalies than soft markers (11.13% vs 7.59%, p < 0.01). We retrospectively analyzed the prenatal genetic outcomes for a large cohort of fetuses with different types of ultrasound abnormalities. The present study showed that the chromosomal abnormality rate and clinical outcomes of fetuses with different types of ultrasound abnormalities varied greatly. Our data have important implications for prenatal genetic counseling for fetuses with different types of ultrasound abnormalities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ultrasonic examination plays an important role in the discovery and diagnosis of fetal abnormalities, including soft markers and structural anomalies. Fetal structural abnormalities are found in up to 3% of all pregnancies [1], and these fetuses are at increased risk of chromosomal abnormalities. In addition, with the increasing capabilities and experience of technicians, an increasing number of ultrasound abnormalities are being detected, especially the abnormalities in various soft markers. Soft marker abnormalities can be easily detected by ultrasound examination during the second trimester. Previous studies have shown that soft marker abnormalities have a minor impact on fetal development and usually resolve in the third trimester, but they are considered a potential risk factor for chromosomal abnormalities [2, 3]. The incidence of chromosomal abnormalities varies among fetuses with different types of ultrasound abnormalities. It is particularly important to use suitable methods for genetic prenatal diagnosis of ultrasound abnormalities in fetuses.
Assessment of genome-wide copy-number variations (CNVs) is recommended as the first level of testing for the cytogenetic assessment of these fetuses with ultrasound abnormalities [4, 5]. Chromosomal microarray analysis (CMA) is already widely utilized in invasive prenatal diagnostics for fetuses with ultrasound abnormalities, advanced maternal age, aberrant first trimester screening, and other situations [6]. .CMA has been found to be useful to identify prenatal clinically relevant CNVs. In addition to chromosomal aneuploidy and CNVs, CMA with single nucleotide polymorphism (SNP) probes is also effective in detecting uniparental disomy (UPD), loss of heterozygosity (LOH), triploidy, and chimerism [7, 8]. Several previous studies showed that the application of CMA is valuable for fetuses with ultrasound abnormalities [9, 10], but focused on specific cases of pregnancy outcomes, and the sample sizes were limited. The large-scale studies on the association between chromosomal abnormality rates and ultrasound abnormalities in different groups are still lacking. Therefore, when encountering a certain type of ultrasound abnormality, clinicians may experience difficulties in choosing the most appropriate test. So further large sample studies are still necessary to clarify the correlation between different types of ultrasound abnormalities and chromosomal abnormalities, and provide more data for clinicians.
The present study retrospectively investigated the clinical ultrasound manifestations and outcomes of 6159 fetuses with ultrasound abnormalities by CMA and karyotyping with long-term follow-up. The present study further evaluated the clinical application of CMA in the prenatal diagnosis of CNVs. In particular, the potential diagnostic rates of CMA for different subgroups of ultrasound abnormalities were also evaluated. In addition, subgroup analyses were performed to better understand the genetic causes of ultrasound abnormalities and to make recommendations for prenatal genetic testing for each type of ultrasound abnormality.
Materials and Methods
Subjects
A total of 6159 fetuses with ultrasound abnormalities detected by fetal ultrasound or echocardiography and that underwent CMA and karyotyping were retrospectively reviewed. Invasive prenatal diagnosis was performed between January 2015 and December 2021 at the First Affiliated Hospital of the Fourth Military Medical University (Shanxi Province, Northwest China). All parents received prenatal counseling from a clinical geneticist about the risks associated with an invasive prenatal diagnosis, the advantages and limitations of CMA, and the risks of variants of unknown significance (VOUS) and incidental findings. The CMA results and following outcomes were analyzed: incidence rates of chromosomal anomalies in different group ultrasound abnormalities and the prognosis of fetuses. All pregnant women routinely provide written informed consents. In the present study, the amniotic fluid samples of fetuses were collected at 18 to 35 weeks of gestation, chorionic villus samples were collected from 26 fetuses at 11 to 13 weeks of gestation, and umbilical cord blood samples were collected from 22 fetuses at 24 to 28 weeks due to oligohydramnios. The pregnancy outcomes were obtained by telephone follow-up.
Karyotype Analysis
The amniotic fluid, chorionic villus, and umbilical cord blood samples were cultured and karyotyped according to standard cytogenetic protocols. The Giemsa-banding technique (450-550-band resolution) was used to analyze the cultured amniocytes or lymphocytes.
Chromosomal Microarray Analysis (CMA)
A QIAamp DNA Blood Mini Kit (Qiagen, Venlo, the Netherlands) was used to extract genomic DNA from amniotic fluid, chorionic villus, and umbilical cord blood samples. An Affymetrix CytoScan 750K array (Affymetrix, Santa Clara, CA, USA) was used and the procedure was performed according to the standard manufacturer’s protocol as described in our previous publication [11]. The Chromosome Analysis Suite v4.2 software was used to analyze the CEL files, based on data from the genome version GRCh37 (hg19). CNVs larger than 100 kb or those that affected more than 50 contiguous probes were considered, and regions of homozygosity larger than 10 Mb were analyzed.
Public databases such as DGV (http://www.ncbi.nlm.nih.gov/dbvar/), ClinGen(https://search.clinicalgenome.org/kb/gene-dosage), OMIM (http://www.ncbi.nlm.nih.gov/omim), DECIPHER (http://decipher.sanger.ac.uk/), ISCA (https://www.iscaconsortium.org/), UCSC (http://genome.ucsc.edu), and PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) were used for the interpretation of the results and to analyze genotype-phenotype correlations. According to the American College of Medical Genetics (ACMG) guidelines [12], the CNVs were classified into five categories: pathogenic copy-number variations (pCNVs),likely pathogenic copy-number variations (lpCNVs), benign, likely benign, and variants of unknown significance (VOUS). In the present study, to determine whether the pCNVs, lpCNVs and VOUS detected by CMA are de novo or inherited, some parents were tested, but benign and likely benign CNVs were not considered for the present study.
Clinical Follow-up Assessments and Statistical Analysis
Clinical follow-up assessments about the pregnancy outcomes, the detail data on postnatal conditions, and prenatal and postnatal development were performed regularly by telephone from 6 months to 3 years. SPSS 24.0 statistical software was used for statistical analysis of the data. Comparisons between groups were conducted using the chi-square test or the Fisher exact test. A p-value < 0.05 was considered statistically significant in the tests.
Results
Study Subjects
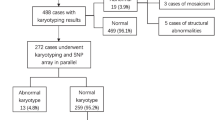
CMA detection was performed on 9141 pregnancies during the 7-year study period in our center. Among them, 6159 pregnant women underwent genetic CMA testing due to ultrasound abnormalities. The detection rates of pCNVs, lpCNVs, VOUS, and other findings in fetuses with different subgroups of ultrasonographic structural anomalies and soft markers are summarized in Tables 1 and 2.
Prevalence of Chromosomal Abnormalities
The overall incidence rate of pathogenic chromosomal abnormalities in fetuses with ultrasound abnormalities was 7.58% (467/6159). Among these cases, 57.6% (269/467) were with pCNVs, 41.7% (195/467) with numerical chromosomal abnormalities, and 0.64% (3/467) with UPD.
The 195 fetuses with chromosomal number abnormalities were comprised of 92 trisomy 21, forty-four trisomy 18, five trisomy 13, 26 monosomy X, five XXX, seven XXY, five XYY, ten mosaicisms (including one mosaic trisomy 8, one mosaic trisomy 18, one mosaic trisomy 22, one mosaic trisomy 16, one mosaic trisomy 21, and five mosaic sex chromosome), and one triploid (69,XXX).
Two hundred sixty-nine (4.37%, 269/6159) cases with pCNVs were detected. The fragment size of chromosomal pCNVs detected ranged from 73 to 80.1 Mb. There were 190 (3.08%; 190/6159) with microdeletions, 46 (0.75%; 46/6159) with microduplications, and 33 (0.54%; 33/6159) cases with both deletions and duplications. A total of 35 microdeletion or microduplication syndromes were found in 187 cases, including 22q11.2 microdeletion syndrome, 22q11.2 microduplication syndrome, 1p36 microdeletion syndrome, 15q11.2 microdeletion syndrome, 15q13.3 microdeletion syndrome, 16p11.2 microdeletion syndrome, 16p11.2 microduplication syndrome, 16p13.11 microdeletion syndrome, 17q12 microdeletion syndrome, 17q12 microduplication syndrome, 1q21.1 microdeletion syndrome, 1q21.1 microduplication syndrome, 1q44 deletion syndrome, 2q13 microdeletion syndrome, 3q29 microdeletion syndrome, 6q25.1 microdeletion syndrome, 7q11.23 microduplication syndrome, 8p23.1 deletion syndrome, 8p23.1 microduplication syndrome, alpha-thalassemia/mental retardation syndrome, type 1, cri du chat syndrome, hereditary stress susceptibility neurosis, Jacobsen syndrome, KBG syndrome, Miller-Dieker syndrome, Pallister-Killian syndrome, Phelan–McDermid syndrome, Smith-Magenis syndrome, tetrasomy 18p syndrome, Williams-Beuren syndrome, Wolf–Hirschhorn syndrome, MECP2 duplication syndrome, and X-linked ichthyosis. In the remaining cases, some rare CNVs such as 21q22.12q22.3 microdeletion, 6q27 1 Mb microdeletion, and 2p16.1p14 microduplication were included; the details are summarized in Table 3.
In addition, 1.72% (106/6159) with lpCNVs and 3.04% (187/6159) with VOUS were detected by CMA; the details of lpCNVs are summarized in Table 4 and VOUS are summarized in Supplementary table 1.
Subgroup Analysis of the Different Types of Ultrasound Abnormalities
There were 2982 fetuses with ultrasonographic structural anomalies, 9.32% (278/2982) with pathogenic chromosomal abnormalities including pCNVs, aneuploid, and UPD. 1.81% (54/2982) with lpCNVs and 3.42% (95/2982) with VOUS were detected by CMA. Congenital heart diseases (CHDs) were the most common ultrasound abnormalities presented in 1795 fetuses; the detection rate of pathogenic and likely pathogenic chromosomal abnormalities for fetuses with CHDs was 7.47% (134/1795) and 1.62% (29/1795) respectively. The incidence of pathogenic chromosomal abnormalities for fetuses with skeletal and central nervous system abnormalities was respectively 6.03% and 10.98%. Three thousand one hundred seventy-seven fetuses with ultrasonographic soft markers, 5.95% (189/3171) with pathogenic anomaly. 1.64% (52/3177) with lpCNVs, and 3.08% (98/3177) with VOUS were detected by CMA. The detection rate of pathogenic and likely pathogenic chromosomal abnormalities in fetuses with ultrasonographic structural anomalies (332/2982, 11.13%) was significantly higher than that in fetuses with ultrasonographic soft markers (241/3177, 7.59%) (p < 0.001).
Clinical Follow-up Assessments
In the current study, the average telephone follow-up time for these fetuses was 1 year, ranging from 3 months to 3 years. Among 194 aneuploid fetuses, 183 underwent termination of pregnancy, 2 were lost to follow-up, 9 were born without obvious clinical defects including six with XXY, one with XXX, and two with 45,X. Among 269 fetuses with pCNVs, 205 underwent termination of pregnancy, 24 were lost to follow-up, and 35 were born. However, there were 5 fetuses who had postnatal death and 8 fetuses showed developmental delay, hypotonia, and feeding difficulties after birth, others without obvious clinical defects at follow-up. Among the 106 fetuses of lpCNV, 35 underwent termination of pregnancy, 12 were lost to follow-up, 53 were born apparently normal, 1 had postnatal death, and 5 showed developmental delay after birth. Among 193 cases of VOUS, 33 underwent termination of pregnancy due to the chromosomal abnormalities, 40 were lost to follow-up, 111 were born apparently normal, 5 showed developmental delay after birth, and 4 had postnatal death. In the 5393 fetuses with normal results, 41 died after birth, 651 underwent termination of pregnancy, 5352 fetuses were apparently normal at birth, and 565 were lost to follow-up. In the present study, the detail clinical follow-up evaluation for different types of CMA results after prenatal diagnosis are summarized in Table 5.
Discussion
Most of the existing studies have focused on soft markers or structural anomalies, but large-scale studies are lacking. Currently, limited data on the clinical outcomes of pregnancies with specific types of ultrasound abnormalities are available. Previous studies have shown that CMA detects 6 to 18.7% of chromosomal abnormalities in fetuses with ultrasound abnormalities [9, 13] and may identify 1.5 to 7.4% of pathogenic CNVs in fetuses with ultrasound abnormalities and normal karyotypes [10, 14]. Our study indicated that the overall frequency of pathogenic chromosomal abnormalities including aneuploidy and CNVs was 7.58% (467/6159); the rate of CNVs with normal karyotype detected by CMA was 3.75% (231/6159), which was in accord with some previous studies [15, 16]. The fetuses with ultrasonographic structural anomalies (9.32%, 278/2982) were significantly higher than fetuses with soft markers (5.95%, 189/3177). We report for the first time that the incidence of chromosomal abnormalities in fetuses with ultrasound structural abnormalities was 9.32% (278/2982) and ranged from 2.53 to 34.62% in groups with different structural anomalies. The incidence of pathogenic chromosomal abnormalities was highest among fetuses with two or more ultrasound structural abnormalities (34.62%, 90/260). lpCNVs were 1.62% (29/1795). The incidence of pathogenic chromosomal abnormalities among fetuses with different structural anomalies is as follows: central nervous system malformations were 10.98% (19/173), congenital heart defects were 7.47% (134/1795), skeletal system malformations were 6.03% (14/232), gastrointestinal system malformations were 4.76% (4/84), and genitourinary system malformations were 4.39% (12/273). Our study findings are slightly different from those of previous studies, in which cardiovascular system, central nervous system, and musculoskeletal system malformations were mostly associated with pathogenic CNVs, but the incidence of chromosomal abnormalities was not completely uniform among fetuses with the same type of ultrasound abnormalities in different studies [17,18,19]; this may be caused by selection bias, different populational factors, and different sample sizes. However, the incidence rate of pathogenic chromosomal abnormalities among fetuses with respiratory system and facial malformations was relatively low, with rates of 3.49% (3/86) and 2.53% (2/79) respectively. Among the fetuses with soft markers, the overall frequency of pathogenic chromosomal abnormalities including aneuploidy and CNVs was 5.95% (189/3177); aneuploidy was 3.02% (96/3177), which accounts for 51.06%; pCNVs was 2.9% (92/3177), which accounts for 48.94%. The incidence of chromosomal aneuploidy in fetuses with ultrasonographic soft markers is high. The incidence of pathogenic chromosomal abnormalities was highest in fetuses with thickened nuchal fold (16.36%, 45/275), aneuploidy accounts for 86.7% (39/45), and pCNVs account for 13.3% (6/45). Hu et al.’s [2] previous study also showed that the incidence of aneuploidy was highest in thickened nuchal fold fetuses compared with other soft markers. The incidence of pathogenic chromosomal abnormalities for fetuses with thickened nuchal translucency in our study was higher than that in Hu et al.’s report, but the sample size is larger and more representative. Karyotype analysis can detect most abnormalities in fetuses with nuchal translucency (NT) abnormalities, but there was still 2.18% fetuses with pCNVs; CMA is more meaningful. Our study indicated that the use of CMA in prenatal diagnosis is necessary and can significantly improve the detection rate of pathogenic CNVs. In addition, for fetuses with an absent/hypoplastic nasal bone, the incidence rate of aneuploidies and pCNVs was 3.85% (7/182) and 2.2% (4/182) respectively, so the incidence of aneuploidies especially trisomy 21 was also higher in this group. Huang et al. [20] also showed a strong correlation between chromosomal abnormalities and fetal nasal bone hypoplasia. Excluding fetuses with NT thickening and absent/hypoplastic nasal bones, the incidence of pCNVs was high among fetuses with other ultrasonographic soft marker abnormalities. For fetuses with mild ventriculomegaly, the incidence rate of chromosome aneuploidies and pCNVs was 1.16% (3/158) and 5.81% (15/158) respectively, and the incidence of pCNVs was significantly higher than that of aneuploidies. Previous studies have reported that the incidence of chromosomal abnormalities ranges from 5.7 to 12.1% in different cohorts [21,22,23,24]. The overall incidence of pathogenic chromosomal abnormalities was 6.98%, and this difference may be attributed to selection bias. CMA is a promising prenatal diagnosis tool that can provide valuable data for accurately assessing fetal prognosis and deciding whether to continue pregnancy during prenatal clinical consultation.
In the present study, a total of 35 types of microdeletion/microduplication syndromes were detected in 187 fetuses, such as 22q11.2 microdeletion syndrome, 1p36 microdeletion syndrome, 15q11.2 microdeletion syndrome, 3q29 microdeletion syndrome, and Williams-Beuren syndrome. The 22q11.2 microdeletion syndrome was the most common chromosomal microdeletion syndrome, showing a wide phenotypic spectrum include congenital heart disease, gastrointestinal symptoms, and psychiatric illnesses and has an estimated incidence of 1/4000–6000 livebirths [25,26,27]. 22q11.2 microdeletion syndrome is closely related to congenital heart diseases (CHDs), which prompts genetic counseling, especially for complex heart abnormalities associated with other malformations, for which it is suggested that CMA detection be conducted to prevent the birth of children with birth defects. High-resolution CMA allowing for the detection of submicroscopic imbalances, except for the usual microdeletion syndromes, was also helpful to detect single-gene diseases caused by deletions. In the present study, 7 male fetuses with DMD gene deletion without any family history of dystrophinopathy were incidentally detected using CMA, and karyotyping of the fetuses showed normal 46,XY. The deletion was further verified by denaturing high-performance liquid chromatography, and parental study revealed maternal inheritance or de novo inheritance. The deletion or disruption of the DMD gene may result in Duchenne muscular dystrophy (DMD) or Becker muscular dystrophy (BMD). DMD seriously affects the quality of life and survival of the patients and currently, there is no effective treatment. Prenatal diagnosis is necessary to provide accurate prognostic information for genetic counseling and potential options for the family regarding clinical management.
CMA exhibits a high efficiency for the diagnosis of fetal chromosomal abnormalities and unavoidable and multiple VOUS with unclear relevance to the detected clinical phenotypes [14]. The identification of VOUS during prenatal diagnosis continues to be a challenging issue prenatally, which may lead to difficulty in clinical genetic counseling and stress for pregnant women and their families and even result in excessive induction of labor. VOUS has been identified in less than 5% of all prenatal samples [28, 29]. In the current study, a 1.72% prevalence rate of lpCNVs and a 3.04% prevalence rate of VOUS were detected in the 6159 fetuses, which was consistent with previous reports [9, 30], but higher than those of a previous study [10]. These differences may be caused by different interpretation biases and reporting standards. 15q11.2 duplication, including the BP1-BP2 region, encompasses four highly conserved genes: TUBGCP5, NIPA1, NIPA2, and CYFIP1, which are the most common lpCNVs (31.1%, 33/106). CNVs involving this region present a major challenge in prenatal testing because they have been reported in affected individuals with healthy family members of affected probands. Most CNVs in this area are inherited without significant clinical manifestations from parents. The phenotypes associated with CNVs are known for their variability, incomplete penetrance, and wide phenotypical spectrum, even among members of the same family [31]. The microduplication of 15q11.2 had a low penetrance, but increased the risk of developmental delays and mental retardation [32, 33]. CMA with SNP probes can also detect loss of heterozygosity (LOH) and uniparental disomy (UPD). Excluding clearly imprinted genes, the clinical significance of LOH and UPD is unclear, and the recessive disease-causing genes contained in the regions increase the risk of hereditary diseases, making genetic counseling difficult. Therefore, further studies are needed to accurately assess fetuses with VOUS.
Conclusions
Fetuses with ultrasound abnormalities are at increased risk of chromosomal abnormalities including CNVs and aneuploidy. Our present study aimed to investigate the incidence rates of chromosomal abnormalities and pregnancy outcome and postpartum clinical manifestations by long-term follow-up. Detection by CMA with SNP probes can be used as an effective method for the prenatal genetic diagnosis of fetal ultrasound abnormalities and can enhance the detection rate of chromosomal abnormalities. Prenatal CMA should be recommended for fetuses with ultrasound abnormalities. The present study also provides important data including the prevalence and distribution of chromosomal abnormalities among fetuses with different types of ultrasound aberrations and pregnancy outcomes that may assist physicians and geneticists in proper genetic counseling.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Mone F, McMullan DJ, Williams D, et al. Evidence to support the clinical utility of prenatal exome sequencing in evaluation of the fetus with congenital anomalies: scientific impact paper No. 64 [February] 2021. BJOG. 2021;128(9):e39–50. https://doi.org/10.1111/1471-0528.16616.
Hu T, Tian T, Zhang Z, et al. Prenatal chromosomal microarray analysis in 2466 fetuses with ultrasonographic soft markers: a prospective cohort study. Am J Obstet Gynecol. 2021;224(5):516. https://doi.org/10.1016/j.ajog.2020.10.039.
Hu ZM, Li LL, Zhang H, et al. Clinical application of chromosomal microarray analysis in pregnant women with advanced maternal age and fetuses with ultrasonographic soft markers. Med Sci Monit. 2021;27:e929074. https://doi.org/10.12659/MSM.929074.
American College of O and Gynecologists Committee on G. Committee Opinion No. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol. 2013;122(6):1374–7. https://doi.org/10.1097/01.AOG.0000438962.16108.d1.
Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–64. https://doi.org/10.1016/j.ajhg.2010.04.006.
Stosic M, Levy B, Wapner R. The use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol Clin North Am. 2018;45(1):55–68. https://doi.org/10.1016/j.ogc.2017.10.002.
Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril. 2018;109(2):201–12. https://doi.org/10.1016/j.fertnstert.2018.01.005.
Brady PD, Vermeesch JR. Genomic microarrays: a technology overview. Prenat Diagn. 2012;32(4):336–43. https://doi.org/10.1002/pd.2933.
Huang H, Cai M, Xue H, et al. Single nucleotide polymorphism array in genetic evaluation of fetal ultrasound abnormalities: a retrospective follow-up study. Am J Transl Res. 2022;14(5):3516–24.
Xiang J, Ding Y, Song X, et al. Clinical utility of SNP array analysis in prenatal diagnosis: a cohort study of 5000 pregnancies. Front Genet. 2020;11:571219. https://doi.org/10.3389/fgene.2020.571219.
Song T, Xu Y, Li Y, et al. Detection of submicroscopic chromosomal aberrations by chromosomal microarray analysis for the prenatal diagnosis of central nervous system abnormalities. J Clin Lab Anal. 2020;34(10):e23434. https://doi.org/10.1002/jcla.23434.
Riggs ER, Andersen EF, Cherry AM, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22(2):245–57. https://doi.org/10.1038/s41436-019-0686-8.
Cai M, Huang H, Su L, et al. Fetal congenital heart disease: associated anomalies, identification of genetic anomalies by single-nucleotide polymorphism array analysis, and postnatal outcome. Medicine. 2018;97(50):e13617. https://doi.org/10.1097/MD.0000000000013617.
Ganapathi M, Nahum O, Levy B. Prenatal diagnosis using chromosomal SNP microarrays. Methods Mol Biol. 2019;1885:187–205. https://doi.org/10.1007/978-1-4939-8889-1_13.
Cai M, Lin N, Su L, et al. Copy number variations in ultrasonically abnormal late pregnancy fetuses with normal karyotypes. Sci Rep. 2020;10(1):15094. https://doi.org/10.1038/s41598-020-72157-6.
Cai M, Lin N, Guo N, et al. Using single nucleotide polymorphism array for prenatal diagnosis in a large multicenter study in Southern China. Sci Rep. 2023;13(1):7242. https://doi.org/10.1038/s41598-023-33668-0.
Di Pasquo E, Kuleva M, Arthuis C, et al. Prenatal diagnosis and outcome of fetuses with isolated agenesis of septum pellucidum: cohort study and meta-analysis. Ultrasound Obstet Gynecol. 2022;59(2):153–61. https://doi.org/10.1002/uog.23759.
Qiao F, Wang Y, Zhang C, et al. Comprehensive evaluation of genetic variants using chromosomal microarray analysis and exome sequencing in fetuses with congenital heart defect. Ultrasound Obstet Gynecol. 2021;58(3):377–87. https://doi.org/10.1002/uog.23532.
Su J, Qin Z, Fu H, et al. Association of prenatal renal ultrasound abnormalities with pathogenic copy number variants in a large Chinese cohort. Ultrasound Obstet Gynecol. 2022;59(2):226–33. https://doi.org/10.1002/uog.23702.
Huang H, Cai M, Ma W, et al. Chromosomal microarray analysis for the prenatal diagnosis in fetuses with nasal bone hypoplasia: a retrospective cohort study. Risk Manag Healthc Policy. 2021;14:1533–40. https://doi.org/10.2147/RMHP.S286038.
Wang J, Zhang Z, Li Q, et al. Prenatal diagnosis of chromosomal aberrations by chromosomal microarray analysis in foetuses with ventriculomegaly. Sci Rep. 2020;10(1):20765. https://doi.org/10.1038/s41598-020-77400-8.
Huang RN, Chen JY, Pan H, et al. Correlation between mild fetal ventriculomegaly, chromosomal abnormalities, and copy number variations. J Matern Fetal Neonatal Med. 2020;35(24):4788–96. https://doi.org/10.1080/14767058.2020.1863941.
Chang Q, Yang Y, Peng Y, et al. Prenatal detection of chromosomal abnormalities and copy number variants in fetuses with ventriculomegaly. Eur J Paediatr Neurol. 2020;25:106–12. https://doi.org/10.1016/j.ejpn.2020.01.016.
Duan HL, Zhu XY, Zhu YJ, et al. The application of chromosomal microarray analysis to the prenatal diagnosis of isolated mild ventriculomegaly. Taiwan J Obstet Gynecol. 2019;58(2):251–4. https://doi.org/10.1016/j.tjog.2019.01.015.
Cirillo A, Lioncino M, Maratea A, et al. Clinical manifestations of 22q11.2 deletion syndrome. Heart Fail Clin. 2022;18(1):155–64. https://doi.org/10.1016/j.hfc.2021.07.009.
Kotcher RE, Chait DB, Heckert JM, et al. Gastrointestinal features of 22q11.2 deletion syndrome include chronic motility problems from childhood to adulthood. J Pediatr Gastroenterol Nutr. 2022;75(2):e8–e14. https://doi.org/10.1097/MPG.0000000000003491.
Xue J, Shen R, Xie M, et al. 22q11.2 recurrent copy number variation-related syndrome: a retrospective analysis of our own microarray cohort and a systematic clinical overview of ClinGen curation. Translational pediatrics. 2021;10(12):3273–81. https://doi.org/10.21037/tp-21-560.
Shaffer LG, Rosenfeld JA, Dabell MP, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn. 2012;32(10):986–95. https://doi.org/10.1002/pd.3943.
Papoulidis I, Sotiriadis A, Siomou E, et al. Routine use of array comparative genomic hybridization (aCGH) as standard approach for prenatal diagnosis of chromosomal abnormalities. Clinical experience of 1763 prenatal cases. Prenat Diagn. 2015;35(13):1269–77. https://doi.org/10.1002/pd.4685.
Wang JC, Radcliff J, Coe SJ, et al. Effects of platforms, size filter cutoffs, and targeted regions of cytogenomic microarray on detection of copy number variants and uniparental disomy in prenatal diagnosis: Results from 5026 pregnancies. Prenat Diagn. 2019;39(3):137–56. https://doi.org/10.1002/pd.5375.
Maya I, Perlman S, Shohat M, et al. Should we report 15q11.2 BP1-BP2 deletions and duplications in the prenatal setting? J Clin Med. 2020;9(8):2602. https://doi.org/10.3390/jcm9082602.
Picinelli C, Lintas C, Piras IS, et al. Recurrent 15q11.2 BP1-BP2 microdeletions and microduplications in the etiology of neurodevelopmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2016;171(8):1088–98. https://doi.org/10.1002/ajmg.b.32480.
Benitez-Burraco A, Barcos-Martinez M, Espejo-Portero I, et al. Variable penetrance of the 15q11.2 BP1-BP2 microduplication in a family with cognitive and language impairment. Mol Syndro Mol. 2017;8(3):139–47. https://doi.org/10.1159/000468192.
Acknowledgements
We thank all the family members for participating in the research and the support of financial fundings.
Funding
This work was supported by the Xi’an City Innovation Ability strong base plan-medical research project under Grant 21YXYJ0106 and the National Natural Science Foundation of China under Grant 82172993.
Author information
Authors and Affiliations
Contributions
JZ, LJ, and HY conceived and designed the study. TS drafted the manuscript. YX revised the manuscript. YL and JZ performed the statistical analysis and participated in its design. FG revised the grammar of the manuscript. All authors read and approved the final manuscript submitted for publication.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 51 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Song, T., Xu, Y., Li, Y. et al. Clinical Experience of Prenatal Chromosomal Microarray Analysis in 6159 Ultrasonically Abnormal Fetuses. Reprod. Sci. 31, 1089–1107 (2024). https://doi.org/10.1007/s43032-023-01399-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-023-01399-2