Abstract
Cervical cancer (CC) is the most serious gynecological malignancy among women worldwide. As a subtype of noncoding RNAs (ncRNAs), circular RNAs (circRNAs) play important roles in the regulation of gene expression and cancer progression. It was discovered from the cancer-specific circRNA database (CSCD) that circ_0019435 was mainly distributed in the nucleus of HeLa-S3 cells. However, few researches have mentioned circ_0019435 with its function in cancers. The present study uncovered that circ_0019435 was upregulated in CC cells by qRT-PCR. Moreover, circ_0019435 was more stable than its linear isoform-ABCC2. Besides, no regulation of circ_0019435 on ABCC2 and the chemoresistance of CC cells were found. Then, it was unveiled by a series of functional assays including colony formation, trypan blue staining, and transwell invasion assays in that circ_0019435 ablation induced the suppression of proliferation, invasion, and EMT of HeLa and SiHa cells. The subcellular distribution of circ_0019435 was assessed by subcellular fractionation and FISH assay. Furthermore, it was disclosed that circ_0019435 binds to EZH2 to silence DKK1 and PTEN. Finally, rescue assays corroborated that DKK1 and PTEN were involved in circ_0019435-mediated CC cell progression. In conclusion, circ_0019435 regulates DKK1 and PTEN expression at the epigenetic level, thereby influencing the progression of CC cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer (CC) poses great threat to women’s lives, notably in the developing world [1]. Among various risk factors, human papillomavirus infection is directly related with this malignancy [2]. Encouraging achievements have been made in cervical cytology and biopsy, which facilitates the detection of early-stage and locally advanced CC cases [3, 4]. For instance, cervical stromal invasion and presence of GC at the preoperative cervical smear might predict the incidence of local recurrence in endometrial cancer [5]. Preoperative conization plays a role in protecting CC patients [6]. Furthermore, specific biomarkers, such as indocyanine green, CEA, SCC-Ag, and CD44, were used for the diagnosis and treatment of CC [7, 8]. However, CC still ranks the fourth in inducing death among female as the third most common gynecological cancer [9]. Thus, it is of substantial necessity to uncover more molecular mechanism underlying the progression of CC cells and develop new biomarkers, which have potential of elevating the cure ratio of CC.
Circular RNA (circRNA) belongs to noncoding RNAs (ncRNAs) and has been studied extensively in recent years [10]. Featuring the covalently closed loop and non-protein-coding ability, circRNA has been elucidated to engage in a variety of physiological and pathological processes [11,12,13]. In addition, it was revealed by genomewide analyses that circRNAs were dysregulated in several cancer tissues [14] and could serve as biomarkers in multiple cancers [15]. It has been well-established that through the interaction between circRNAs and RNA-binding proteins (RBPs), circRNAs could regulate the expression of downstream genes [16]. Moreover, the microRNA (miRNA) binding sites within the sequences of circRNAs enable circRNAs to sponge miRNAs [17]. Similarly, as another subtype of ncRNAs, long non-coding RNAs (lncRNAs) have also been elucidated to interact with RBPs to mediate the stability of target genes or act as miRNA sponges [18, 19]. Besides, lncRNAs could recruit enhancer of zeste homolog 2 (EZH2), which is the enzyme module in PcG repressor complex 2 (PRC2) trimethylating at lysine 27 on histone 3 (H3K27me3) to epigenetically silence downstream messenger RNAs (mRNAs) [20, 21]. Thereafter, we wondered whether circRNAs could epigenetically downregulate mRNAs with the same mechanism in cancers.
Circ_0019435, derived from ATP-binding cassette subfamily C member 2 (ABCC2), is chiefly occupied in the nucleus of HeLa-S3 cells as identified in the CSCD (http://gb.whu.edu.cn/CSCD/). Nevertheless, there are few researches concerning circ_0019435. The regulatory mechanism of circ_0019435 in CC remains unknown. Therefore, this study explored the role of circ_0019435 in CC, with the hypothesis that circ_0019435 might exert its function through recruiting EZH2.
Materials and Methods
Cell Culture and Treatment
Human normal cervical epithelial cell line (HcerEpic) and CC cell lines (HeLa, SiHa, C4-1, and C-33A) were purchased from ATCC (Manassas, VA). RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA) with the addition of 10% FBS (Thermo Fisher Scientific) and 1% penicillin/streptomycin was applied for cell incubation in the humidified environment in 5% CO2 at 37°C. Actinomycin D (ActD; Sigma-Aldrich, St. Louis, MO) and RNase R (Epicentre, Madison, WI) were separately bought and applied for detecting the RNA stability of linear ABCC2 and circ_0019435 in SiHa and HeLa cells.
Total RNA Extraction and Quantitative Real-time PCR (qRT-PCR)
Total RNA extraction was achieved via TRIzol reagent (Invitrogen, Carlsbad, CA) for reverse transcription. SYBR Green PCR Master Mix (Invitrogen) was utilized for qRT-PCR, with GAPDH and U6 as internal controls. Relative gene expressions were calculated via 2−ΔΔCt formula.
Nucleic Acid Electrophoresis
Agarose gels with TE buffer from Thermo Fisher Scientific were employed for cDNA and gDNA PCR production of circ_0019435 with DL600 (KeyGen, Nanjing, China) as DNA markers. DNAs were separated by electrophoresis and treated with UV irradiation successively.
Cell Transfection
The specific shRNAs targeting RNAs (circ_0019435, EZH2, DKK1, and PTEN) and nonspecific shRNAs as negative control (NC), as well as the pcDNA3.1/EZH2 and empty pcDNA3.1 vector, were synthesized by Genechem (Shanghai, China). CC cells were transfected with plasmids for 48 h by the application of Lipofectamine 3000 (Invitrogen).
Colony Formation
Clonogenic CC cell samples were put and cultured in 6-well plates for 14 days. After being fixed in methanol, cells were dyed with 0.5% crystal violet for observation and analysis. At last, colonies (> 50 cells) were counted.
Trypan Blue Staining
Cell samples were seeded and cultured in the 24-well plates for 48 h at 37°C. After being washed and trypsinized, cells were colored with trypan blue staining (Beyotime, Shanghai, China). Cell counting chambers were applied to count stained cells for analysis.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Staining
Chemosensitivity was evaluated through MTT assays. CC cells were cultured with cisplatin in the 96-well plates firstly. After 24 h, MTT solution was added to cells, followed by the incubation for 4 h. Then, the absorbance at a wavelength of 490 nm was evaluated via microplate reader after CC cells were treated with DMSO. Finally, IC50 (half maximal inhibitory concentration) of cisplatin was analyzed.
JC-1 Assay
CC cells were mixed with 10 mM of JC-1 (Beyotime) for 30 min at 37°C before being labeled with fluorescence. After cells were washed in PBS, EnSpire Reader (PerkinElmer, Waltham, MA) was applied for apoptosis analysis.
Flow Cytometer Apoptosis Analysis
CC cells were first treated with 70% cold ethanol on ice for fixation, then with Annexin-V fluorescein isothiocyanate (FITC)/propidium iodide (PI) dual staining kit in the dark. Cell apoptosis was monitored and analyzed by using flow cytometry (BD Biosciences, San Jose, CA).
Transwell Invasion Assay
Invasion of CC cells was analyzed by using Matrigel-coated transwell chambers (Corning Co, Corning, NY). Cells invading to the lower chambers were fixed in 4% paraformaldehyde, stained with crystal violet, and counted with the microscopy.
Western Blot
Total proteins were extracted from CC cells, subjected to 10% SDS-PAGE and transferred to PVDF membranes. Then, membranes were blocked by 5% non-fat milk for incubation with primary antibodies (Abcam, Cambridge, MA) against GAPDH (internal reference) and varied proteins (MMP9, MMP7, MMP2, Snail, Twist, Slug, ZEB1, Vimentin, N-cadherin, E-cadherin, DKK1, PTEN, EZH2, β-catenin, c-myc, cyclin D1, p-PI3K, PI3K, p-Akt, and Akt). Secondary antibodies were then added. ECL (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used to visualize and analyze band images finally.
Subcellular Fractionation
Cytoplasmic and nuclear RNAs were isolated by PARIS™ Kit (Invitrogen) as instructed by the supplier to analyze the distribution of circ_0019435 via qRT-PCR.
Fluorescence In Situ Hybridization Analysis (FISH)
RNA FISH probe designed for circ_0019435 were synthesized by RiboBio (Guangzhou, China) and transfected into CC cells. Nuclei were counterstained with Hoechst. Cells were observed under the confocal microscope (ZEISS, Jena, Germany) to locate circ_0019435.
RNA-Binding Protein Immunoprecipitation (RIP)
RIP assay was implemented with specific antibodies against EZH2 and control IgG via Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA).
Dual-Luciferase Reporter Assay and TOP/FOP Luciferase Reporter Assay
DKK1 promoter was cloned into the pGL3-Basic vector (Promega, Madison, WI) for the co-transfection with sh-circ_0019435#1 or sh-NC into CC cells. For TOP/FOP luciferase reporter assay, the TOP Flash/FOP Flash reporter vectors were procured from Addgene (Cambridge, MA) and co-transfected into cells with sh-circ_0019435#1 or sh-NC. Forty-eight hours later, Dual-Luciferase Reporter Assay System (Promega) was used to analyze luciferase activities.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was carried out by applying the antibodies against EZH2, H3K27me3, and control IgG by the use of Magna ChIP Kit (Millipore) as required by the supplier before the analysis of precipitated chromatin via qRT-PCR.
Statistical Analyses
Data were shown as mean ± SD, representing three independently conducted assays. Student’s t-test or one-way/two-way ANOVA was undertaken for statistical analysis with GraphPad Prism 5 (GraphPad, San Diego, CA), with p < 0.05 as significant cutoff. Dunnett or Tukey was applied for the post hoc test.
Results
Upregulated Expression of Circ_0019435 in CC Cells
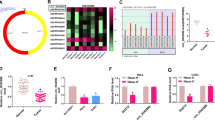
According to the CSCD (http://gb.whu.edu.cn/CSCD/) website, it was found that the nuclei of HeLa-S3 cells contained more circ_0019435 than the cytoplasm (Fig. 1A). Therefore, we decided to investigate the expression of circ_0019435 in CC cell lines (HeLa, SiHa, C4-1, and C-33A) and the normal cervical cell line (HcerEpic). The results indicated that circ_0019435 exhibited higher expression in CC cell lines than that in normal cervical cell line, especially in HeLa and SiHa cell lines (Fig. 1B), suggesting the potential involvement of circ_0019435 in the progression of CC cells. Therefore, we selected HeLa and SiHa cells for the follow-up experiments.
Upregulated expression of circ_0019435 in CC cells. (A) The circ_0019435 was spotted in CSCD (http://gb.whu.edu.cn/CSCD/). (B) qRT-PCR was used to detect circ_0019435 expression in CC cell lines (HeLa, SiHa, C4-1, and C-33A) and the human normal cervical cell line (HcerEpic). (C) qRT-PCR was used to evaluate the expression of circ_0019435 and ABCC2 after the treatment of actinomycin D (ActD). (D) The loop structure of circ_0019435 was verified in CC cells by nucleic acid electrophoresis. Solid triangle represents divergent primers, and hollow triangle represents convergent primers. (E) qRT-PCR was used to assess the expression of circ_0019435 and ABCC2 in HeLa and SiHa cells treated with RNase R. (F) qRT-PCR was used to analyze the knockdown efficiency of sh-circ_0019435. (G) qRT-PCR was utilized to detect ABCC2 relative expression in sh-circ_0019435#1/2-transfected CC cells. (H) MTT staining assays were used to test cisplatin sensitivity of HeLa and SiHa cells after the ablation of circ_0019435. *P<0.05, **P<0.01
In subsequence, ActD treatment and RNase R treatment were employed to verify the circularization of circ_0019435. It was observed that after the treatment of ActD, circ_0019435 was more stable than its linear isoform, ABCC2 (Fig. 1C). Moreover, circ_0019435 was revealed to be only amplified in cDNA group by divergent primers rather than in gDNA (Fig. 1D). Furthermore, after HeLa and SiHa cells were treated with RNase R, circ_0019435 expression showed no remarkable change, while ABBC2 mRNA level was reduced significantly (Fig. 1E). All the above evidence verified the circular structure of circ_0019435.
Next, we studied whether circ_0019435 affects the chemosensitivity of CC cells. Before that, we transfected HeLa and SiHa cells with sh-circ_0019435#1/#2 to knock down circ_0019435. As depicted in Fig. 1F, the expression of circ_0019435 was inhibited conspicuously, verifying the knockdown efficiency of circ_0019435#1/#2. Afterward, qRT-PCR analysis revealed that ABBC2 mRNA expression was not significantly influenced by circ_0019435 silencing (Fig. 1G). Then, considering the important role of ABCC2 in the chemosensitivity of cancer cells [22], we then detected the influence of circ_0019435 on the resistance to cisplatin of HeLa and SiHa cells. As revealed in Fig. 1H, the shortage of circ_0019435 posed little influence on the chemoresistance of CC cells. The abovementioned results proved the circularization of circ_0019435 and indicated that circ_0019435 has no effect on the chemoresistance of CC cells.
Circ_0019435 Accelerates the Proliferation, Invasion, and EMT of CC Cells
Given that circ_0019435 could not mediate the chemoresistance of CC cells, we went on investigating the effect of circ_0019435 on the proliferation, apoptosis, invasion, and EMT of CC cells via colony formation, trypan blue staining, JC-1, flow cytometry, and transwell and western blot assays. Firstly, colony formation assay demonstrated that circ_0019435 depletion remarkably hindered the clonogenic ability of HeLa and SiHa cells (Fig. 2A). Moreover, result of trypan blue staining reflected that the proliferation of CC cells was significantly blocked by circ_0019435 repression (Fig. 2B). Additionally, JC-1 assay uncovered that the ratio of viable cells was overtly decreased after circ_0019435 knockdown (Fig. 2C). Consistently, it was suggested that the apoptosis was obviously prompted in sh-circ_0019435#1/#2-transfected CC cells as shown in flow cytometry analysis (Fig. 2D). Hence, circ_0019435 was positively related to the proliferation of CC cells while being negatively to the apoptosis. Then, the invasive ability of CC cells was tested by transwell assays (Fig. 2E). Result showed that the number of the invaded Hela and SiHa cells into the lower chambers was significantly decreased from 166 (or 138) to 112 and 98 (or 76 and 74) when circ_0019435 was silenced, implying circ_0019435 promotes the invasion of CC cells. Subsequently, western blot assay was conducted to evaluate protein levels of mesenchymal markers (N-cadherin, vimentin, ZEB1, Slug, Twist, Snail, MMP2, MMP7, and MMP9) and the epithelial marker (E-cadherin) in CC cells to evaluate the influence of circ_0019435 on EMT (Fig. 2F). It was revealed through the result that the protein levels of mesenchymal biomarkers were decreased significantly, while that of E-cadherin was increased. Taken all together, circ_0019435 accelerates the proliferation, invasion, and EMT of CC cells.
Circ_0019435 accelerates the proliferation, invasion, and EMT of CC cells. (A–B) Colony formation and trypan blue staining assays were applied to evaluate CC cell proliferation. (C–D) JC-1 and flow cytometry analysis were performed to evaluate the apoptosis of CC cells. (E) Transwell assay was implemented to detect the invasion of CC cells. The number of invaded HeLa and SiHa cells into the lower chambers changed from 166 to 112 and 98 (or from 138 to 76 and 74) in the NC and sh-circ_0019435 groups. (F) Western blot was used to measure the protein level of mesenchymal markers (N-cadherin, vimentin, ZEB1, Slug, Twist, Snail, MMP2, MMP7, and MMP9), and the epithelial marker (E-cadherin) in CC cells after circ_0019435 was knocked down. *P<0.05, **P<0.01
Circ_0019435 Downregulates DKK1 and PTEN Expression via Recruiting EZH2
To further the study of circ_0019435, we detected the localization of circ_0019435 in HeLa and SiHa cells. Both subcellular fractionation and FISH assay manifested that circ_0019435 was mostly localized in the nucleus, implying that circ_0019435 might take part in the transcriptional regulation of genes (Fig. 3A–B). With the aim of exploring the mechanism between circ_0019435 and EZH2, RIP assay was conducted to prove the combination between circ_0019435 and EZH2 (Fig. 3C). Further, the expression of several mRNAs targeted by EZH2 was analyzed in sh-circ_0019435#1-transfected cells. qRT-PCR results disclosed that circ_0019435 negatively mediated the expression of dickkopf WNT signaling pathway inhibitor 1 (DKK1) and phosphatase and tensin homolog (PTEN) for the downregulated circ_0019435 notably enhanced the mRNA expression of DKK1 and PTEN relative to other mRNAs (Fig. 3D). Subsequently, luciferase reporter assay showed that the shortage of circ_0019435 enhanced the luciferase activity of both DKK1 and PTEN promoter vectors (Fig. 3E), suggesting that circ_0019435 might negatively influence the transcription of DKK1 and PTEN.
Circ_0019435 downregulates DKK1 and PTEN expression via recruiting EZH2. (A–B) Subcellular fractionation and FISH assays explored the localization of circ_0019435 in HeLa and SiHa cells. (C) RIP assay verified the binding between EZH2 and circ_0019435. (D) qRT-PCR was conducted to measure the relative expression of the downstream mRNAs of EZH2 in sh-circ_0019435#1/2-transfected cells. (E) Luciferase reporter assay was implemented to test the interaction between circ_0019435 and DKK1 or PTEN promoters. (F) qRT-PCR and western blot assays aimed to examine the knockdown efficiency of EZH2. (G) qRT-PCR and western blot assays were performed to measure DKK1 and PTEN expression in sh-NC or sh-EZH2#1-transfected cells. (H) ChIP assay was utilized to examine the combination between EZH2 and DKK1 or PTEN promoter. (I) ChIP assay was adopted for measuring the enrichment of DKK1 and PTEN promoter in IgG, EZH2, and H3K27me3 antibody precipitates. (J) qRT-PCR and western blot analyses were performed to attest the overexpression efficiency of pcDNA3.1/EZH2. (K) qRT-PCR and western blot assays were conducted for the measurement of DKK1 and PTEN expression in different transfected cells. *P<0.05, **P<0.01
After testing the knockdown efficiency of sh-EZH2 in HeLa and SiHa cells (Fig. 3F), we preliminarily found that EZH2 negatively regulated DKK1 and PTEN with the remarkably enhanced mRNA and protein levels of DKK1 and PTEN in sh-EZH2 group as shown in qRT-PCR and western blot results (Fig. 3G). Furthermore, ChIP assay uncovered that the promoters of DKK1 and PTEN were significantly enriched in sh-EZH2#1 group rather than in sh-NC group (Fig. 3H), revealing that EZH2 might inhibit DKK1 and PTEN expression in terms of transcription.
With the aim of probing the mechanism of circ_0019435, EZH2, DKK1, and PTEN, ChIP assay in Fig. 3I was implemented to unveil that circ_0019435 deficiency resulted in a suppression in the recruitment of EZH2 to DKK1 and PTEN promoter, as well as the trimethylation of H3K27me3 at promoter regions. To confirm whether circ_0019435 recruits EZH2 to repress the expression of DKK1 and PTEN, we conducted the rescue experiments after the overexpression efficiency of EZH2 was affirmed in Fig. 3J. It was analyzed that the elevated mRNA and protein levels of DKK1 and PTEN in sh-circ_0019435#1-transfected cells could be impaired by EZH2 overexpression (Fig. 3K). Taken together, circ_0019435 could downregulate the expression of DKK1 and PTEN via recruiting EZH2.
Circ_0019435 Regulates CC Progression Dependent on DKK1 and PTEN
Considering that DKK1 is the inhibitor of Wnt/β-catenin pathway [23] and PTEN is one of the PI3K/Akt pathway–related proteins [24], we investigated the influences of circ_0019435 knockdown on these two pathways via western blot analysis of pathway-related proteins and TOP/FOP flash luciferase activity assays. As shown in Fig. 4A, the expression of β-catenin, c-myc, and cyclin D1 were suppressed by circ_0019435 inhibition. Besides, the luciferase activity of TOP/FOP flash was weakened by circ_0019435 depletion (Fig. 4B), confirming that Wnt/β-catenin pathway could be activated by circ_0019435. Moreover, the p-PI3K and p-Akt protein levels were also manifested to be reduced in sh-circ_0019435#1/2-transfected cells (Fig. 4C), hinting that circ_0019435 might regulate PI3K/Akt pathway as well. Next, to investigate whether circ_0019435 regulates the progression of CC via targeting DKK1 and PTEN, we silenced DKK1 and PTEN for follow-up assays. The mRNA and protein expression levels of DKK1 and PTEN were both downregulated dramatically after the transfection of sh-DKK1 or sh-PTEN (Fig. 4D–E). Cell proliferation was determined by colony formation and trypan blue staining assays. As reflected in Fig. 4F–G, the restrained proliferative ability induced by circ_0019435 silencing was partially compensated by DKK1 depletion while being nearly totally rescued by the co-transfection of sh-PTEN. Furthermore, cell apoptosis was assessed by JC-1 and flow cytometry analyses. The results showed that the promotion of cell apoptosis induced by circ_0019435 repression could be significantly countervailed by the shortage of DKK1 and PTEN (Fig. 4H–I). Finally, cell invasive capacity was determined by transwell invasion assays. The results showed that the concurrence of DKK1 and PTEN deficiency could successfully restore the circ_0019435 inhibition–induced reduced number of invaded cells indicated by the change of number from 171 to 78, 149, and 164 (or from 135 to 53, 116, and 130) (Fig. 4J). Collectively, circ_0019435 regulates Wnt/β-catenin pathway and PI3K/Akt pathway, facilitating cell proliferation and invasion but inhibiting cell apoptosis in a DKK1- and PTEN-dependent manner.
Circ_0019435 regulates CC progression dependent on DKK1 and PTEN. (A) Western blot was applied to analyze the expression of DKK1 and Wnt/β-catenin pathway–related proteins (β-catenin, c-myc, and cyclin D1) in the HeLa and SiHa cells transfected with sh-NC or sh-circ_0019435#1. (B) TOP/FOP flash luciferase reporter assay was performed to verify the effect of circ_0019435 on the activation of Wnt/β-catenin pathway in the transfected cells. (C) Western blot analysis of PI3K/Akt pathway–related proteins was conducted in different groups. (D–E) qRT-PCR and western blot analyses were employed to verify the knockdown efficiency of sh-DKK1 and sh-PTEN. (F–G) Colony formation and trypan blue staining assays detected the proliferation of the indicated transfected cells. (H–I) JC-1 and flow cytometry analyses were conducted to evaluate the apoptosis of the indicated transfected cells. (J) Transwell invasion assay was performed to assess the invasive capacity of the indicated transfected cells. *P<0.05, **P<0.01
Discussion
Previous literature elucidated that circRNAs such as circRNA_101996 [25], circRNA8924 [26], and circ_0000263 [27] could facilitate the malignancy of CC via constructing a ceRNA regulatory network. From the CSCD website, we figured out a cricRNA—circ_0019435, which is mainly distributed in the nucleus of HeLa-S3 cells. However, the role circ_0019435 plays in the cellular processes of CC cells remains obscure.
In this study, circ_0019435 exhibited an upregulated expression in CC cells. In addition, the circular structure of circ_0019435 was corroborated in HeLa and SiHa cells. Besides, the interaction between circ_0019435 and ABCC2 was also explored. The results indicated that circ_0019435 could not regulate ABCC2 in CC cells. More importantly, circ_0019435 was confirmed not to affect the chemosensitivity of HeLa and SiHa cells. However, cell proliferation, invasion, and EMT could be enhanced by circ_0019435.
Then, the nuclear localization of circ_0019435 suggested that circ_0019435 has the potential for processing transcriptional regulation. EZH2 has been reported to interact with lncRNA PVT1 to epigenetically inhibit miR-195 expression, thereby promoting the EMT of CC cells [28]. For EZH2, previous studies have reported that lncRNAs participate in the EZH2 oncogenic regulatory network in lung cancer [29], prostate cancer [30], and so on. Herein, we investigated whether circ_0019435 participates in the EZH2 regulatory network epigenetically in CC. Results disclosed that circ_0019435 could bind to EZH2. Furthermore, circ_0019435 suppressed the transcription of DKK1 and PTEN, so did EZH2. More importantly, ChIP assay affirmed that circ_0019435 transcriptionally downregulates DKK1 and PTEN via recruiting EZH2. Given that DKK1 could impair the activation of Wnt/β-catenin pathway [23] and PTEN could suppress the activation of PI3K/Akt pathway [24], the regulation of circ_0019435 on these two pathways in CC cells was also taken into exploration. Then, the effects of circ_0019435 on the activation of Wnt/β-catenin and PI3K/Akt pathways were revealed. In the end, rescue assays illustrated that the inhibited cellular process of CC cells induced by circ_0019435 depletion could be reversed by the downregulation of DKK1 and PTEN, meaning that DKK1 and PTEN are involved in circ_0019435-mediated CC cell proliferation, apoptosis, and invasion.
This study verified the mechanism of circ_0019435/EZH2/DKK1/PTEN axis in the progression of CC cells for the first time and demonstrated that circ_0019435 recruits EZH2 to suppress the transcription of DKK1 and PTEN, thereby posing the promoting effects on cell proliferation, invasion, and EMT in CC. However, whether circ_0019435 activates Wnt/β-catenin and PI3K/Akt pathways to expedite the occurrence and development of CC needs more experimental and clinically analytical supports, which would be implemented in further studies. Besides, we will further explore the underlying regulatory mechanisms of circ_0019435 via recruiting other protein-coding genes in the cellular process of CC progression.
In conclusion, the present study sheds a light into understanding the pathogenesis of CC.
Data Availability
Not applicable.
References
Siva S, Deb S, Young RJ, Hicks RJ, Callahan J, Bressel M, et al. (1)(8)F-FDG PET/CT following chemoradiation of uterine cervix cancer provides powerful prognostic stratification independent of HPV status: a prospective cohort of 105 women with mature survival data. Eur J Nucl Med Mol Imaging. 2015;42(12):1825–32. https://doi.org/10.1007/s00259-015-3112-8.
Sherman SM, Lane EL. Awareness of risk factors for breast, lung and cervical cancer in a UK student population. Journal of cancer education : the official journal of the American Association for Cancer Education. 2015;30(4):660–3. https://doi.org/10.1007/s13187-014-0770-3.
Zhang HH, Li AH. Long non-coding RNA FEZF1-AS1 is up-regulated and associated with poor prognosis in patients with cervical cancer. Eur Rev Med Pharmacol Sci. 2018;22(11):3357–62. https://doi.org/10.26355/eurrev_201806_15156.
Sakuragi N. Refining insight into cervical cancer progression. Lancet Oncol. 2014;15(4):371–2. https://doi.org/10.1016/s1470-2045(14)70085-3.
Casarin J, Bogani G, Serati M, Pinelli C, Laganà AS, Garzon S, et al. Presence of glandular cells at the preoperative cervical cytology and local recurrence in endometrial cancer. Int J Gynecol Pathol. 2020;39(6):522–8. https://doi.org/10.1097/pgp.0000000000000642.
Casarin J, Bogani G, Papadia A, Ditto A, Pinelli C, Garzon S, et al. Preoperative conization and risk of recurrence in patients undergoing laparoscopic radical hysterectomy for early stage cervical cancer: a multicenter study. J Minim Invasive Gynecol. 2021;28(1):117–23. https://doi.org/10.1016/j.jmig.2020.04.015.
Valenti G, Vitale SG, Tropea A, Biondi A, Laganà AS. Tumor markers of uterine cervical cancer: a new scenario to guide surgical practice? Updat Surg. 2017;69(4):441–9. https://doi.org/10.1007/s13304-017-0491-3.
Rossetti D, Vitale SG, Tropea A, Biondi A, Laganà AS. New procedures for the identification of sentinel lymph node: shaping the horizon of future management in early stage uterine cervical cancer. Updat Surg. 2017;69(3):383–8. https://doi.org/10.1007/s13304-017-0456-6.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Smolle E, Haybaeck J. Non-coding RNAs and lipid metabolism. Int J Mol Sci. 2014;15(8):13494–513. https://doi.org/10.3390/ijms150813494.
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–8. https://doi.org/10.1080/15476286.2015.1020271.
Zhou ZB, Huang GX, Fu Q, Han B, Lu JJ, Chen AM, et al. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther. 2019;27(3):531–41. https://doi.org/10.1016/j.ymthe.2019.01.006.
Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging. 2019;11(24):12412–27. https://doi.org/10.18632/aging.102580.
Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. https://doi.org/10.1186/s13059-014-0571-3.
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. https://doi.org/10.1186/s12943-017-0663-2.
Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2018. https://doi.org/10.1002/jnr.24356.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–58. https://doi.org/10.1093/nar/gkw027.
Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–16. https://doi.org/10.1093/bib/bbv031.
Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics, proteomics & bioinformatics. 2017;15(3):177–86. https://doi.org/10.1016/j.gpb.2016.12.005.
Ding J, Li J, Wang H, Tian Y, Xie M, He X, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8(8):e2997. https://doi.org/10.1038/cddis.2017.328.
Zhang E, He X, Yin D, Han L, Qiu M, Xu T, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. https://doi.org/10.1038/cddis.2015.356.
Comsa E, Nguyen KA, Loghin F, Boumendjel A, Peuchmaur M, Andrieu T, et al. Ovarian cancer cells cisplatin sensitization agents selected by mass cytometry target ABCC2 inhibition. Future Med Chem. 2018;10(11):1349–60. https://doi.org/10.4155/fmc-2017-0308.
Guo J, Liu M, Yang D, Bouxsein ML, Saito H, Galvin RJ, et al. Suppression of Wnt signaling by Dkk1 attenuates PTH-mediated stromal cell response and new bone formation. Cell Metab. 2010;11(2):161–71. https://doi.org/10.1016/j.cmet.2009.12.007.
Benhamou D, Labi V, Getahun A, Benchetrit E, Dowery R, Rajewsky K, et al. The c-Myc/miR17-92/PTEN axis tunes PI3K activity to control expression of recombination activating genes in early B cell development. Front Immunol. 2018;9:2715. https://doi.org/10.3389/fimmu.2018.02715.
Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen X, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234(8):14296–305. https://doi.org/10.1002/jcp.28128.
Liu J, Wang D, Long Z, Liu J, Li W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48(1):173–84. https://doi.org/10.1159/000491716.
Cai H, Zhang P, Xu M, Yan L, Liu N, Wu X. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol. 2019;234(7):11391–400. https://doi.org/10.1002/jcp.27796.
Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25(7):637–44. https://doi.org/10.1080/1061186x.2017.1307379.
Su M, Xiao Y, Tang J, Wu J, Ma J, Tian B, et al. Role of lncRNA and EZH2 interaction/regulatory network in lung cancer. J Cancer. 2018;9(22):4156–65. https://doi.org/10.7150/jca.27098.
Ling Z, Wang X, Tao T, Zhang L, Guan H, You Z, et al. Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J Exp Clin Cancer Res. 2017;36(1):159. https://doi.org/10.1186/s13046-017-0629-7.
Acknowledgements
We appreciate all the participants who provide supports for this research.
IRB Approval
Not applicable.
Funding
This study was supported by Ningbo Natural Science Foundation (Grant No. 2019A610305) and Medical Scientific Research Foundation of Zhejiang Province (Grant No. 2020KY832).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare that no competing interest exists in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Wang, Q., Zhuo, Z., Yu, H. et al. Circ_0019435 Exerts Its Functions in the Cellular Process of Cervical Cancer via Epigenetically Silencing DKK1 and PTEN. Reprod. Sci. 28, 2989–2999 (2021). https://doi.org/10.1007/s43032-021-00625-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-021-00625-z