Abstract
Light is a key environmental cue that fundamentally regulates all aspects of plant growth and development, which is mediated by the multiple photoreceptors including the blue light photoreceptors cryptochromes (CRYs). In Arabidopsis, there are two well-characterized homologous CRYs, CRY1 and CRY2. Whereas CRYs are flavoproteins, they lack photolyase activity and are characterized by an N-terminal photolyase-homologous region (PHR) domain and a C-terminal extension domain. It has been established that the C-terminal extension domain of CRYs is involved in mediating light signaling through direct interactions with the master negative regulator of photomorphogenesis, COP1. Recent studies have revealed that the N-terminal PHR domain of CRYs is also involved in mediating light signaling. In this review, we mainly summarize and discuss the recent advances in CRYs signaling mediated by the N-terminal PHR domain, which involves the N-terminal PHR domain-mediated dimerization/oligomerization of CRYs and physical interactions with the pivotal transcription regulators in light and phytohormone signaling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptochromes (CRYs) are blue light photoreceptors that were first discovered in Arabidopsis. They act to regulate a broad spectrum of physiological processes, including seedling photomorphogenesis, photoperiodic flowering, circadian rhythm, and stomatal opening and development in plants (Ahmad et al. 1993; Guo et al. 1998; Kang et al. 2009; Liu et al. 2008b; Mao et al. 2005; Somers et al. 1998; Toth et al. 2001; Yang et al. 2000; Yu et al. 2010). Cryptochromes not only exist in plants, but in a variety of other organisms from bacterium to human as well. In Drosophila melanogaster, cryptochrome serves as the photoreceptor to entrain the circadian clock, and in mammals, they act as integral components of the circadian clock (Emery et al. 1998; Kume et al. 1999). In migratory butterfly and birds, cryptochrome is responsible for sensing the Earth’s magnetic field and providing precise navigation during their long-distance migration (Gegear et al. 2010). Arabidopsis has two well-characterized homologous cryptochromes, CRY1 and CRY2. They possess an N-terminal photolyase-homologous region (PHR), also known as CNT1 (N Terminus of CRY1) and CNT2 (N Terminus of CRY2), and a C-terminal extension (CCE), also known as CCT1 (C Terminus of CRY1) and CCT2 (C Terminus of CRY2) (Yang et al. 2000; Yu et al. 2010). The signaling mechanisms of CRYs concerning their regulation of transcription or stability of photoresponsive proteins and regulation of CRYs activity by photo-oligomerization and phosphorylation have been well documented in other recent reviews (Wang et al. 2018a,2020; Yang et al. 2017). In this review, we first summarize the role for the C-terminal domain of Arabidopsis CRYs in mediating light signaling, and then introduce the involvement of the N-terminal PHR domain of CRYs in mediating light signaling reported in recent years.
The C-terminal domain of Arabidopsis cryptochromes in mediating light signaling
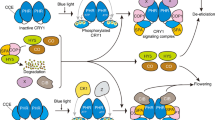
CRY1 is the first blue light photoreceptor identified in plants in 1993 (Ahmad et al. 1993). The insight into the signaling pathway of CRYs was obtained in 2000 through the demonstration that transgenic plants expressing the C-terminal domain of either CRY1 (CCT1) or CRY2 (CCT2) fused to β-glucuronidase (GUS) display a constitutive photomorphogenic phenotype shown by shortened hypocotyls, enhanced anthocyanin production and chloroplast development in the dark, and an early flowering phenotype in both long days and short days (Yang et al. 2000). These phenotypes are similar to the loss-of-function mutant of COP1, a RING-finger E3 ubiquitin ligase acting as the master negative regulator of photomorphogenesis and flowering (Deng et al. 1991; McNellis et al. 1994). COP1 interacts with its substrates such as HY5, a bZIP transcription factor acting as a key positive regulator of photomorphogenesis (Oyama et al. 1997), and CONSTANS (CO), a B-Box type Zn finger-containing transcription regulator acting as a master activator of photoperiodic flowering (Koornneef et al. 1991; Putterill et al. 1995), to ubiquitinate them and promote their degradation through the 26S proteasome (Fig. 1a, b) (Jang et al. 2008; Liu et al. 2008b; Osterlund et al. 2000). These findings suggest that CRY1 and CRY2 signaling in response to blue light activation is mediated through their C-terminal domain. Consistent with this proposition, both CRY1 and CRY2 directly interact with COP1 through CCT1 and CCT2 in blue light-independent manner in heterologous systems (Wang et al. 2001; Yang et al. 2001). Interestingly, Holtkotte et al. showed that the interactions of CRY1 and CRY2 with COP1 are blue light-dependent (Holtkotte et al. 2017), which may require SPAs, as SPAs interact with COP1 (Saijo et al. 2003; Seo et al. 2003), and CRY1 and CRY2 interact with SPAs in a blue light-dependent manner (see below) (Lian et al. 2011; Liu et al. 2011; Zuo et al. 2011). The outcome of the light activation of CRY1 and CRY2, through their CCT1- and CCT2-mediated physical interactions with COP1, is likely the disruption of the negative regulation of COP1 exerted on its substrates such as HY5 and CO. In this manner, HY5 and CO are relieved from COP1-dependent proteolysis and are able to perform their roles in photomorphogenesis and photoperiodic flowering (Fig. 1a, b). Since the establishment of the direct interactions of CRYs with COP1, it has been demonstrated that the red/far-red light photoreceptors phyB/phyA and the UV-B light photoreceptor UVR8 also physically interact with COP1 to mediate light signaling (Seo et al. 2004; Jang et al. 2010; Rizzini et al. 2011).
CRY1 and CRY2 C and N termini-mediated interactions with COP1 and SPAs in light control of plant development. Upon blue light (BL) irradiation, the C terminus of CRY1 and CRY2 (CCT1 and CCT2)-mediated interactions with COP1 and SPAs inhibit COP1 activity and stabilize HY5 and CO proteins, and promote photomorphogenesis (a) and photoperiodic flowering (b), respectively. Moreover, CRY2 also interacts with CIBs through its N terminus (CNT2) and TOEs through both N and C termini to regulate floral initiation (b)
SUPPRESSORS OF PHYA (SPAs, SPA1–4) share a similar nuclear-localized WD repeats with COP1 that mediates COP1′s interactions with CRY1 and CRY2 (Wang et al. 2001; Yang et al. 2001), and the loss-of-function mutants of SPAs show a constitutive photomorphogenic phenotype, similar to cop1 mutant (Laubinger 2004). The COP1 activity is dependent on SPAs, as SPAs interact with COP1 and act as enhancers of the COP1 E3 ligase activity (Hoecker et al. 2001; Seo et al. 2003). Two studies have shown that CRY1 interacts with SPAs through its C terminus in a blue light-dependent manner, to promote the dissociations of COP1 from SPAs (Lian et al. 2011; Liu et al. 2011). These studies suggest that, on one hand, CRY1 and CRY2 interact with COP1 to directly inhibit its activity, and on the other hand, they also interact with SPAs in a blue light-dependent manner, to indirectly attenuate COP1 activity by disrupting the COP1–SPAs core complex, leading to efficient promotion of HY5 and CO stability (Fig. 1a, b). Interestingly, CRY2 interacts with SPA1 through its N terminus but not its C terminus (Zuo et al. 2011). Recent studies have demonstrated that CRY2 inhibits the E3 ubiquitin ligase activity of COP1 on its substrates by physically displacing the substrates from COP1 (Ponnu et al. 2019; Lau et al. 2019) which suggests that CRY1 may act in the same manner.
The N-terminal domain mediates oligomerization of Arabidopsis cryptochromes
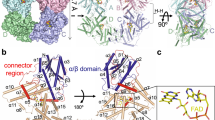
Although plant and mammal CRYs share amino acid sequence similarity to photolyases, mammal CRYs and photolyases function in monomers (Brautigam et al. 2004; Czarna et al. 2013), whereas plant CRYs act in dimers/oligomers (Ma et al. 2020a, b; Sang et al. 2005; Shao et al. 2020; Yu et al. 2007). The insight into the CNT1-mediated homodimerization of CRY1 was first obtained in 2005 through the demonstration that transgenic lines expressing CNT1 in the wild-type background show a dominant-negative phenotype under blue light, similar to cry1 mutant (Sang et al. 2005). In these lines, CNT1 physically interacts with the endogenous CRY1 to interfere with its homodimerization. CNT1-mediated homodimerization is essential for CCT1 function, and the substitution of CNT1 with GUS can activate CCT1 and lead to a constitutive light response (Sang et al. 2005; Yang et al. 2000). CNT1 and GUS with mutations or deletions that compromise their ability to dimerize or oligomerize fail to activate CCT1. CRY2 N terminus also mediates homodimerization of CRY2, which is required for CRY2 function (Yu et al. 2007). These studies therefore demonstrate that Arabidopsis CRY1 and CRY2 homodimerize/homooligomerize through their N terminus, which is necessary for their photoreceptor activity. These findings are supported by the following recent exciting studies. CRY2 undergoes blue light-dependent oligomerization, which can be suppressed or inactivated by the interactions of CRY2 with two closely related cryptochrome inhibitory proteins, Blue-light inhibitor of Cryptochrome 1 and 2 (BIC1 and BIC2) (Wang et al. 2016a). BIC1 and BIC2 repression of CRY2 oligomerization compromises all the known photobiochemical and photophysiological activities of CRY2, strongly demonstrating that the oligomerization of CRY2 is crucial for their photoreceptor activities. Most recently, Ma et al. and Shao et al. have successfully obtained the 3D structure of the blue light-perceiving PHR domain of dimeric and tetrameric bioactive Arabidopsis CRYs at high resolution using cryo-EM and X-ray crystallography (Ma et al. 2020b; Shao et al. 2020). The structural analysis indicates that two molecules of the blue-light-activated PHR domain form a head-to-head dimer, and two dimmers form a tetramer, supporting the notion that photo-oligomerization is necessary for CRYs' function. Moreover, Ma et al. have revealed that BIC2 displays a waist belt-like structure, which interacts with photoexcited CRY2 and wraps around the groove between the α/β- and α-domains of PHR domain to prevent CRY2 oligomerization (Ma et al. 2020a). Comparison of the structures between the CRY2 tetramer and the BIC2-CRY2N demonstrates that the interface 2 in the CRY2 tetramer clashes with BIC2 in the BIC2–CRY2N complex, implying that BICs may inhibit CRY2 oligomerization by occupying CRY2’s oligomeric surface. Furthermore, they show that BIC2 not only restrains electron and proton transfer during FAD photoreduction, but also inhibits the blue light-dependent oligomerization of CRY2 and prevents CRY2 from interacting with its signaling partners, as well.
The N-terminal domain of Arabidopsis cryptochromes is involved in mediating light response
It had been thought that the C terminus of CRYs is responsible for mediating light signaling by interacting with downstream proteins, while the N terminus is responsible for absorbing light signal through the non-covalently bound FAD and mediating dimerization of CRYs (Lin et al. 1995; Sang et al. 2005; Wang et al. 2001; Yang et al. 2001, 2000; Yu et al. 2007). However, several studies suggest that PHR domain of CRYs is implicated in regulating CRY’s activity. Many missense mutations within CNT1 compromised CRY1 ability to mediate blue light inhibition of hypocotyl elongation and undergo light-dependent phosphorylation (Ahmad et al. 1995; Bouly et al. 2003; Kanai et al. 1997; Shalitin et al. 2002, 2003). The direct evidence supporting the involvement of CRY1 N terminus in mediating light signaling was obtained in 2015 with the transgenic lines expressing CNT1 fused to the nuclear localization signal (NLS) in the cry1 mutant background that show enhanced blue light inhibition of hypocotyl elongation (He et al. 2015). These lines display a wild-type phenotype in red and far-red light, respectively. Importantly, expression of CNT1 with the missense mutations (Ahmad et al. 1995) compromised its ability to promote blue light responsiveness. These findings suggest that CNT1 can mediate CRY1 signaling. This speculation is supported by recent studies in which many CRYs N-terminus-interacting proteins have been identified. The first one is cryptochrome-interacting bHLH 1 (CIB1), with which CRY2 interacts through its N terminus in a blue light-dependent manner (Liu et al. 2008a) (Fig. 1b). The formation of CRY2–CIB1–CO complex in response to blue light promotes the expression of the florigen gene, FLOWERING LOCUST T (FT) at dusk, and induces floral initiation (Liu et al. 2018). Du et al. carried out yeast two-hybrid screening using CNT1 as a bait, and identified an AP2-like transcriptional factor, TOE1, as an interacting protein (Du et al. 2020), which interacts with CO to inhibit its transcriptional activation of FT to repress flowering (Zhai et al. 2015; Zhang et al. 2015). Both N and C termini of CRY2 are involved in mediating the blue light-dependent interactions of CRY2 with TOE1 and TOE2 to regulate flowering under long days (Fig. 1b).
The phytochrome-interacting factors (PIFs) act as the key downstream proteins of phytochromes to negatively regulate photomorphogenesis (Leivar et al. 2011; Ni et al. 1998, 1999). Two independent studies have demonstrated that CRY1 and CRY2 can also physically interact with PIF4 or PIF5 via their N termini to regulate hypocotyl elongation under high temperature and canopy shade low blue light, respectively, to regulate their transcription activity (Fig. 2a) (Ma et al. 2016; Pedmale et al. 2016). It is shown that CRY1 physically interacts with the G-protein β subunit, AGB1, in a blue light-dependent manner, through its N terminus (Lian et al. 2018) (Fig. 2a). AGB1 interacts directly with HY5 to inhibit its DNA-binding activity, and blue light-triggered interaction of CRY1 with AGB1 inhibits the association of AGB1 with HY5, and promotes HY5 DNA-binding activity and photomorphogenesis. This study suggests that CRY1 utilizes its C terminus to stabilize HY5 protein via interactions with COP1 and SPAs (Lian et al. 2011; Liu et al. 2011), and its N terminus to promote HY5 DNA-binding activity via interaction with AGB1 (Lian et al. 2018). CRY1 also interacts with CIB1-related proteins HBI1 through its N terminus in a blue light-dependent manner (Fig. 2a), to inhibit the DNA-binding ability of HBI1 and hypocotyl elongation (Wang et al. 2018b). Moreover, the N termini of CRYs mediate the direct interactions of CRYs with the key transcription regulators in the phytohormone signaling pathways to regulate phytohormone signaling (see below).
CRY1 and CRY2 C and N termini-mediated interactions with transcription regulators in light control of plant development. Upon blue light irradiation, the N termini of CRY1 and CRY2 (CNT1 and CNT2)-mediated interactions with PIF4/5 and HBI1 to directly regulate their DNA-binding activity, and with AGB1 to indirectly regulate HY5–DNA-binding activity, to mediate light inhibition of hypocotyl elongation (a). CNT1 and CNT2 also mediate the interactions of CRY1 and CRY2 with Aux/IAA and ARF proteins (Aux/IAAs and ARFs), and BES1, to regulate Aux/IAAs stability, and ARFs and BES1 DNA-binding activity, which leads to repression of auxin and brassinosteroid (BR) signaling, respectively, and inhibition of hypocotyl cell elongation (b)
The N-terminal domain mediates the interactions of cryptochromes with the key transcription regulators in phytohormone signaling pathways
Phytohormones such as auxin (IAA) and brassinosteroid (BR) act to regulate the same physiological processes regulated by light, which include photomorphogenesis, flowering time, and stomatal development (Kim et al. 2012; Lincoln et al. 1990; Mockaitis et al. 2008; Vert et al. 2005; Zhang et al. 2014; Zhu et al. 2013). The IAA and BR biosynthesis-deficient mutants, as well as the loss-of-function mutants of their receptors TIR1/AFBs and BRI1 show a reduced hypocotyl elongation phenotype during early seedlings photomorphogenic development (Chory 1991; Clouse et al. 1996; Dharmasiri et al. 2005; Kim et al. 1996; Richards et al. 2001; Szekeres et al. 1996; Ueguchi-Tanaka et al. 2005). It has been established that auxin promotes the assembly of its co-receptor complex comprising F-box proteins TIR1/AFBs and transcription regulators’ Aux/IAA proteins (Aux/IAAs), and subsequent ubiquitination and degradation of Aux/IAAs, thus releasing the inhibitory effects of AUX/IAAs on a family of transcription factors, Auxin Response Factors (ARFs), to activate auxin-responsive gene expression (Calderon et al. 2012; Dharmasiri et al. 2005; Gray et al. 2001; Kepinski et al. 2005; Kim et al. 1997). There are 29 AUX/IAAs and 23 ARFs in Arabidopsis. Aux/IAAs are small short-lived proteins with four domains (DI to DIV), of which DII mediates the interactions of Aux/IAAs with TIR1/AFBs, and thus is required for auxin-triggered degradation of Aux/IAAs (Calderon et al. 2012; Kepinski et al. 2005). The DIII/IV-containing C-terminal region of Aux/IAAs mediates their own homodimerization and heterodimerization with ARFs (Kim et al. 1997; Korasick et al. 2014; Nanao et al. 2014; Ulmasov et al. 1997). Typical ARF proteins have a conserved N-terminal DNA-binding domain, followed by a non-conserved middle region, and a conserved C-terminal dimerization domain (Liscum et al. 2002). The DNA-binding domain is responsible for ARFs binding to the promoters of target genes (Ulmasov et al. 1999). Recently, it has been reported that on one hand, CRY1 interacts with Aux/IAAs in a blue light-dependent manner through its N terminus, to suppress the auxin-induced TIR1-Aux/IAAs interactions and subsequent degradation of Aux/IAAs (Fig. 2b) (Xu et al. 2018). On the other hand, CRY1 physically interacts with ARF6 and ARF8 in a blue light-dependent manner through its N terminus (Mao et al. 2020) (Fig. 2b). The N-terminal DNA-binding domain of ARF6 mediates the interaction of ARF6 with CRY1, and the CRY1–ARF6 interaction leads to the repression of the DNA-binding activity of ARF6 and its target gene expression (Fig. 2b). Altogether, these studies suggest that the direct repression of auxin-responsive genes expression mediated by CNT1-mediated interactions of CRY1 with Aux/IAAs and ARFs constitutes two layers of the regulatory mechanisms by which light inhibits auxin signaling and hypocotyl elongation.
A genetic screen for Arabidopsis photomorphogenic development mutants identified det2 (de-etiolated 2), a BR biosynthesis-deficient mutant with a shortened hypocotyl and expanded cotyledon phenotype in the dark (Chory 1991; Li et al. 1996), indicating a link between BR and light signaling. Extensive studies of the BR signal transduction pathway have led to the discovery of the key signaling components such as BES1 and BZR1, and bHLH transcription factors that activate the transcription of BR-responsive genes (He et al. 2005; Yin et al. 2005). When BR level is high in plant cells, BR signal is perceived by BR receptor BRI1, a membrane-localized receptor kinase (Kim et al. 2011; Li et al. 1997). This releases the inhibitor BKI1 (Jaillais et al. 2011; Wang et al. 2011; Wang 2006), activates the intracellular kinase domain of BRI1, and enables BRI1 to bind to its co-receptor BAK1 (Li et al. 2002; Nam et al. 2002). Subsequently, the negative regulator BIN2 is dephosphorylated by BSU1 and inactivated through a multistep cascade of phosphorylation events (Kim et al. 2011; Tang et al. 2008), and BES1 and BZR1 are eventually released from BIN2-induced phosphorylation or dephosphorylated by PP2A (Tang et al. 2011; Wang et al. 2002; Yin et al. 2002; Zhao et al. 2002). Dephosphorylated BES1 and BZR1 can then bind to their target genes and regulate their expression, leading to BR responses (Sun et al. 2010; Vert et al. 2006; Yin et al. 2005). When BR level is low, BIN2 is activated, and BZR1 and BES1 are phosphorylated by BIN2 and thus unable to bind to their target genes (Vert et al. 2006; Zhao et al. 2002). Therefore, the BR signaling mechanism involves BR-induced formation of the physiologically active, dephosphorylated forms of BES1 and BZR1. By screening for CNT1-interacting proteins via yeast two-hybrid system using CNT1 as a bait, Wang et al. identified BES1-INTERACTING MYC-LIKE 1 (BIM1) (Wang et al. 2018c), a bHLH protein that interacts with BES1 to enhance its activity (Yin et al. 2005). Interestingly, blue light-activated CRY1 interacts specifically with the dephosphorylated BES1 through its N terminus (Fig. 2b), leading to the inhibition of BES1’s DNA-binding activity and repression of its target gene expression. This study suggests that blue light-dependent and BR-induced interaction of CRY1 with BES1 may constitute a strictly regulated mechanism by which plants optimize photomorphogenesis according to the availability of external light and internal BR signals (Fig. 2b) (Wang et al. 2018c). Another study by He et al. has also demonstrated that CRY1 interacts with BIN2 and BZR1 to enhance BIN2-dependent phosphorylation and cytoplasmic retention of BZR1 (He et al. 2019). Because these BR signaling regulators interact with each other, cryptochromes may form a dynamic complex with them to directly modulate the BR-responsive transcriptional regulons in response to the dynamic changes in light and BR signals.
Conclusions and future perspectives
To well get adapted to the environment and maintain an optional status of growth and development, plants must not only efficiently utilize the ambient light signals, but also coordinate light and auxin/brassinosteroid signals as well. We now understand that CRY1-mediated light signaling integrates with auxin and BR signaling by its N terminus-mediated interactions with Aux/IAAs, ARFs and BES1, respectively. Like auxin and BR, gibberellin (GA) is also an essential phytohormone promoting plant growth and development. The GA biosynthesis-deficient mutant and the loss-of-function mutant of GA receptor GID1 also show a reduced hypocotyl elongation phenotype (Griffiths et al. 2006), and it is shown that both the N and C termini of CRY1 are involved in mediating CRY1 repression of GA-promoted hypocotyl elongation through inhibition of GA-responsive genes expression in blue light (Wang et al. 2016b). It will be interesting to explore how CRY1’s N and C termini may mediate CRY1 regulation of GA signaling. To date, the studies on CRY1 function and signaling mechanism have primarily been carried out during the early seedlings photomorphogenic development stage, whether and how CRY1 might regulate vegetative development are not well understood, which will be worth investigating in future studies. Moreover, great efforts should be made to screen for more potential CRY1 N and C terminus-interacting proteins, which may lead to the elucidation of the new functions of CRY1 throughout the whole life cycle of plants and the related signaling mechanisms.
References
Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166. https://doi.org/10.1038/366162a0
Ahmad M, Lin C, Cashmore AR (1995) Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J 8:653–658. https://doi.org/10.1046/j.1365-313x.1995.08050653.x
Bouly JP et al (2003) Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur J Biochem 270:2921–2928. https://doi.org/10.1046/j.1432-1033.2003.03691.x
Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci U S A 101:12142–12147. https://doi.org/10.1073/pnas.0404851101
Calderon VL et al (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8:477–485. https://doi.org/10.1038/nchembio.926
Chory J (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3:445–459. https://doi.org/10.1105/tpc.3.5.445
Clouse SD, Langford M, Mcmorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111:671–678. https://doi.org/10.1104/pp.111.3.671
Czarna A et al (2013) Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153:1394–1405. https://doi.org/10.1016/j.cell.2013.05.011
Deng XW, Caspar T, Quail PH (1991) cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev 5:1172–1182. https://doi.org/10.1101/gad.5.7.1172
Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445. https://doi.org/10.1038/nature03543
Du SS et al (2020) Photoexcited cryptochrome2 interacts directly with TOE1 and TOE2 in flowering regulation. Plant Physiol 184:487–505. https://doi.org/10.1104/pp.20.00486
Emery P, So WV, Kaneko M, Hall JC, Rosbash M (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95:669–679. https://doi.org/10.1016/s0092-8674(00)81637-2
Gegear RJ, Foley LE, Casselman A, Reppert SM (2010) Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463:804–807. https://doi.org/10.1038/nature08719
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF (TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276. https://doi.org/10.1038/35104500
Griffiths J et al (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18:3399–3414. https://doi.org/10.1105/tpc.106.047415
Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363. https://doi.org/10.1126/science.279.5355.1360
He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307:1634–1638. https://doi.org/10.1126/science.1107580
He SB, Wang WX, Zhang JY, Xu F, Lian HL, Li L, Yang HQ (2015) The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of Hypocotyl elongation. Mol Plant 8:822–825. https://doi.org/10.1016/j.molp.2015.02.008
He G, Liu J, Dong H, Sun J (2019) The Blue-light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol Plant 12:689–703. https://doi.org/10.1016/j.molp.2019.02.001
Hoecker U, Quail PH (2001) The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276:38173–38178. https://doi.org/10.1074/jbc.M103140200
Holtkotte X, Ponnu J, Ahmad M, Hoecker U (2017) The blue light-induced interaction of cryptochrome 1 with COP1 requires SPA proteins during Arabidopsis light signaling. Plos Genet 13:e1007044. https://doi.org/10.1371/journal.pgen.1007044
Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 25:232–237. https://doi.org/10.1101/gad.2001911
Jang S et al (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. Embo J 27:1277–1288. https://doi.org/10.1038/emboj.2008.68
Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22:2370–2383. https://doi.org/10.1105/tpc.109.072520
Kanai S, Kikuno R, Toh H, Ryo H, Todo T (1997) Molecular evolution of the photolyase-blue-light photoreceptor family. J Mol Evol 45:535–548. https://doi.org/10.1007/pl00006258
Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21:2624–2641. https://doi.org/10.1105/tpc.109.069765
Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–451. https://doi.org/10.1038/nature03542
Kim BC, Soh MC, Kang BJ, Furuya M, Nam HG (1996) Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J 9:441–456. https://doi.org/10.1046/j.1365-313x.1996.09040441.x
Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci U S A 94:11786–11791. https://doi.org/10.1073/pnas.94.22.11786
Kim T, Guan S, Burlingame AL, Wang Z (2011) The CDG1 kinase mediates Brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like Kinase BIN2. Mol Cell 43:561–571. https://doi.org/10.1016/j.molcel.2011.05.037
Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482:419–422. https://doi.org/10.1038/nature10794
Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229:57–66. https://doi.org/10.1007/BF00264213
Korasick DA et al (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A 111:5427–5432. https://doi.org/10.1073/pnas.1400074111
Kume K et al (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193–205. https://doi.org/10.1016/s0092-8674(00)81014-4
Lau K, Podolec R, Chappuis R, Ulm R, Hothorn M (2019) Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. Embo J 38:e102140
Laubinger S (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16:2293–2306. https://doi.org/10.1105/tpc.104.024216
Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16:19–28. https://doi.org/10.1016/j.tplants.2010.08.003
Li J, Chory J (1997) A putative Leucine-Rich repeat receptor kinase involved in Brassinosteroid signal transduction. Cell 90:929–938. https://doi.org/10.1016/s0092-8674(00)80357-8
Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272:398–401. https://doi.org/10.1126/science.272.5260.398
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110:213–222. https://doi.org/10.1016/s0092-8674(02)00812-7
Lian HL et al (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25:1023–1028. https://doi.org/10.1101/gad.2025111
Lian H et al (2018) Photoexcited CRYPTOCHROME 1 interacts directly with G-Protein beta subunit AGB1 to regulate the DNA-binding activity of HY5 and photomorphogenesis in Arabidopsis. Mol Plant 11:1248–1263. https://doi.org/10.1016/j.molp.2018.08.004
Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR (1995) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269:968–970. https://doi.org/10.1126/science.7638620
Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2:1071–1080. https://doi.org/10.1105/tpc.2.11.1071
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49:387–400
Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008a) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322:1535–1539. https://doi.org/10.1126/science.1163927
Liu LJ et al (2008b) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20:292–306. https://doi.org/10.1105/tpc.107.057281
Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25:1029–1034. https://doi.org/10.1101/gad.2025011
Liu Y, Li X, Ma D, Chen Z, Wang JW, Liu H (2018) CIB1 and CO interact to mediate CRY2-dependent regulation of flowering. Embo Rep 19:e45762. https://doi.org/10.15252/embr.201845762
Ma D et al (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci U S A 113:224–229. https://doi.org/10.1073/pnas.1511437113
Ma L et al (2020a) Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2. Nat Struct Mol Biol 27:472–479. https://doi.org/10.1038/s41594-020-0410-z
Ma L et al (2020b) Structural insights into the photoactivation of Arabidopsis CRY2. Nat Plants 6:1432–1438. https://doi.org/10.1038/s41477-020-00800-1
Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) From the cover: a role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci U S A 102:12270–12275. https://doi.org/10.1073/pnas.0501011102
Mao Z et al (2020) Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225:848–865. https://doi.org/10.1111/nph.16194
McNellis TW, von Arnim AG, Araki T, Komeda Y, Misera S, Deng XW (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6:487–500. https://doi.org/10.1105/tpc.6.4.487
Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24:55–80. https://doi.org/10.1146/annurev.cellbio.23.090506.123214
Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110:203–212. https://doi.org/10.1016/s0092-8674(02)00814-0
Nanao MH et al (2014) Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun 5:3617. https://doi.org/10.1038/ncomms4617
Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95:657–667. https://doi.org/10.1016/s0092-8674(00)81636-0
Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400:781–784. https://doi.org/10.1038/23500
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466. https://doi.org/10.1038/35013076
Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11:2983–2995. https://doi.org/10.1101/gad.11.22.2983
Pedmale UV et al (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164:233–245. https://doi.org/10.1016/j.cell.2015.12.018
Ponnu J, Riedel T, Penner E, Schrader A, Hoecker U (2019) Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1909181116
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857. https://doi.org/10.1016/0092-8674(95)90288-0
Richards DE, King KE, Ait-ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of Gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52:67–88. https://doi.org/10.1146/annurev.arplant.52.1.67
Rizzini L et al (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332:103–106. https://doi.org/10.1126/science.1200660
Saijo Y (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Gene Dev 17:2642–2647. https://doi.org/10.1101/gad.1122903
Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17:1569–1584. https://doi.org/10.1105/tpc.104.029645
Seo HS, Yang J, Ishikawa M, Bolle C, Ballesteros ML, Chua N (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995–999. https://doi.org/10.1038/nature01696
Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18:617–622. https://doi.org/10.1101/gad.1187804
Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417:763–767. https://doi.org/10.1038/nature00815
Shalitin D, Yu X, Maymon M, Mockler T, Lin C (2003) Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15:2421–2429. https://doi.org/10.1105/tpc013011
Shao K et al (2020) The oligomeric structures of plant cryptochromes. Nat Struct Mol Biol 27:480–488. https://doi.org/10.1038/s41594-020-0420-x
Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282:1488–1490. https://doi.org/10.1126/science.282.5393.1488
Sun Y et al (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19:765–777. https://doi.org/10.1016/j.devcel.2010.10.010
Szekeres M et al (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85:171–182. https://doi.org/10.1016/s0092-8674(00)81094-6
Tang W et al (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321:557–560. https://doi.org/10.1126/science.1156973
Tang W et al (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13:124–131. https://doi.org/10.1038/ncb2151
Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127:1607–1616. https://doi.org/10.1104/pp.010467
Ueguchi-Tanaka M et al (2005) Gibberellin insensitive dWARF1 encodes a soluble receptor for gibberellin. Nature 437:693–698. https://doi.org/10.1038/nature04028
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971. https://doi.org/10.1105/tpc.9.11.1963
Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19:309–319. https://doi.org/10.1046/j.1365-313x.1999.00538.x
Vert G, Chory J (2006) Downstream nuclear events in brassinosteroid signalling. Nature 441:96–100. https://doi.org/10.1038/nature04681
Vert G, Nemhauser JL, Geldner N, Hong F, Chory J (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol 21:177–201. https://doi.org/10.1146/annurev.cellbio.21.090704.151241
Wang X (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313:1118–1122. https://doi.org/10.1126/science.1127593
Wang Q, Lin C (2020) Mechanisms of cryptochrome-mediated photoresponses in plants. Annu Rev Plant Biol 71:103–129. https://doi.org/10.1146/annurev-arplant-050718-100300
Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294:154–158. https://doi.org/10.1126/science.1063630
Wang ZY et al (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2:505–513. https://doi.org/10.1016/s1534-5807(02)00153-3
Wang H et al (2011) Dual role of BKI1 and 14–3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev Cell 21:825–834. https://doi.org/10.1016/j.devcel.2011.08.018
Wang Q et al (2016a) Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354:343–347. https://doi.org/10.1126/science.aaf9030
Wang W et al (2016b) Transcriptome analyses reveal the involvement of both C and N Termini of Cryptochrome 1 in its regulation of phytohormone-responsive gene expression in Arabidopsis. Front Plant Sci 7:294. https://doi.org/10.3389/fpls.2016.00294
Wang Q, Zuo Z, Wang X, Liu Q, Gu L, Oka Y, Lin C (2018a) Beyond the photocycle-how cryptochromes regulate photoresponses in plants? Curr Opin Plant Biol 45:120–126. https://doi.org/10.1016/j.pbi.2018.05.014
Wang S et al (2018b) CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J Exp Bot 69:3867–3881. https://doi.org/10.1093/jxb/ery209
Wang W et al (2018c) Photoexcited CRYPTOCHROME1 interacts with Dephosphorylated BES1 to regulate Brassinosteroid signaling and Photomorphogenesis in Arabidopsis. Plant Cell 30:1989–2005. https://doi.org/10.1105/tpc.17.00994
Xu F et al (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit Auxin Signaling in Arabidopsis. Mol Plant 11:523–541. https://doi.org/10.1016/j.molp.2017.12.003
Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103:815–827. https://doi.org/10.1016/s0092-8674(00)00184-7
Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13:2573–2587. https://doi.org/10.1105/tpc.010367
Yang Z, Liu B, Su J, Liao J, Lin C, Oka Y (2017) Cryptochromes Orchestrate transcription regulation of diverse blue light responses in plants. Photochem Photobiol 93:112–127. https://doi.org/10.1111/php.12663
Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191. https://doi.org/10.1016/s0092-8674(02)00721-3
Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120:249–259. https://doi.org/10.1016/j.cell.2004.11.044
Yu X et al (2007) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci U S A 104:7289–7294. https://doi.org/10.1073/pnas.0701912104
Yu X, Liu H, Klejnot J, Lin C (2010) The Cryptochrome blue light receptors. Arabidopsis Book 8:e0135. https://doi.org/10.1199/tab.0135
Zhai Q et al (2015) Transcriptional mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 27:2814–2828. https://doi.org/10.1105/tpc.15.00619
Zhang JY, He SB, Li L, Yang HQ (2014) Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc Natl Acad Sci U S A 111:E3015–E3023. https://doi.org/10.1073/pnas.1400542111
Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29:975–987. https://doi.org/10.1101/gad.251520.114
Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE (2002) Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol 130:1221–1229. https://doi.org/10.1104/pp.102.010918
Zhu JY, Sae-Seaw J, Wang ZY (2013) Brassinosteroid signalling. Development 140:1615–1620. https://doi.org/10.1242/dev.060590
Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21:841–847. https://doi.org/10.1016/j.cub.2011.03.048
Acknowledgements
We apologize to colleagues whose work may not be cited. This work was supported by The National Key Research and Development Program of China grant (2017YFA0503802), The National Natural Science Foundation of China grants (31530085, 31900609, 31900207, and 32000183), and The Science and Technology Commission of Shanghai Municipality grant (18DZ2260500).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflicts of interest to this work.
Rights and permissions
About this article
Cite this article
Wang, W., Mao, Z., Guo, T. et al. The involvement of the N-terminal PHR domain of Arabidopsis cryptochromes in mediating light signaling. aBIOTECH 2, 146–155 (2021). https://doi.org/10.1007/s42994-021-00044-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-021-00044-3