Abstract
The named entity recognition (NER) is a method for locating references to rigid designators in text that fall into well-established semantic categories like person, place, organisation, etc., Many natural language e applications, like summarization of text, question–answer models and machine translation, always include NER at their core. Early NER systems were quite successful in reaching high performance at the expense of using human engineers to create features and rules that were particular to a certain domain. Currently, the biomedical data is soaring expeditiously and extracting the useful information can help to facilitate the appropriate diagnosis. Therefore, these systems are widely adopted in biomedical domain. However, the traditional rule-based, dictionary based and machine learning based methods suffer from computational complexity and out-of-vocabulary (OOV)issues Deep learning has recently been used in NER systems, achieving the state-of-the-art outcome. The present work proposes a novel deep learning based approach which uses Bidirectional Long Short Term (BiLSTM), Bidirectional Encoder Representation (BERT) and Conditional Random Field mode (CRF) model along with transfer learning and multi-tasking model to solve the OOV problem in biomedical domain. The transfer learning architecture uses shared and task specific layers to achieve the multi-task transfer learning task. The shared layer consists of lexicon encoder and transformer encoder followed by embedding vectors. Finally, we define a training loss function based on the BERT model. The proposed Multi-task TLBBC approach is compared with numerous prevailing methods. The proposed Multi-task TLBBC approach realizes average accuracy as 97.30%, 97.20%, 96.80% and 97.50% for NCBI, BC5CDR, JNLPBA, and s800 dataset, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction and Background
We have noticed a tremendous growth in technology field which has led to produce huge amount of data e.g. the huge portion of population is dependent on Internet based applications which is also considered as one of the prime reason of generating the data [1]. This technological growth has proliferated the use and advancement in the biomedical domain such as health monitoring based on ECG signal, iSheet for remote diagnosis of isolated individuals, wearable device for cancer diagnosis etc. [2, 3]. However, extracting the useful information from the huge data always plays important role in analysing the historical data and in facilitating the appropriate diagnosis. For example, the MEDLINE database consist of more than 24 million abstracts of biomedical journals and it is still increasing [4]. This task of extracting the information and analysing the historical data can be accomplished with the help of data mining and Natural Language Processing (NLP) [5].
Because of its ability to extract and recognise entities from any text, entity recognition has attracted a lot of attention in this area of NLP. The Named Entity Recognition (NER) technique is used to identify and label a variety of entities, including people, organisations, places, and numerical properties. This process of NER is also adopted in biomedical domain where these entities include different types of diseases, genes, proteins and chemicals, etc. [6]. However, the BioMedical entities have several characteristics which lead to increase the complexities in entity recognition. For example, the biomedical entities include several characteristics such as descriptive entity names (e.g. ‘normal thymic epithelial cells’) leads to ambiguous term boundaries and same entity with same spelling such as N-acetylcysteine’, ‘N-acetyl-cysteine’ and ‘NAcetylCysteine’ [7]. Therefore, development of an efficient model for Biomedical NER becomes an essential part of NLP in biomedical domain. The BioNER models helps in several applications related to drug-to-drug interaction, and disease-treatment relationship. Moreover, the BioNER is also used in various biomedical entity search tools which helps to provide the solution for complex queries in biomedical texts. Generally, the traditional biomedical NER methods are classified as rule based, dictionary based and machine learning based BioNER [8].
-
Rule based approach: The data patterns and rules found in sentences serve as the foundation for the rule-based approach. However, the rule based methods can utilize the context information to overcome the issues of multiple named entity however, these rules must be written manually before employing.
-
Dictionary based approach: these methods follow the easy structure but suffer from issue of handling the unknown items and words with several meaning.
-
Machine learning methods: current advancements in machine learning schemes have led to develop the deep learning methods for biomedical text mining applications. LSTM (Long Short-Term Memory) and Conditional Random Field (CRF) are the two widely adopted methods in this domain. However, training of these machine learning methods on general non-specific domains improves the performance of system achieving the performance that is desired for bio-medical data will be a challenging task.
The rule and dictionary based approaches are easily scalable but it has to be ‘fitted’ manually with dataset. Moreover, these systems fail to achieve the desired performance because of not including the word in the training set and it causes out-of-vocabulary problem (OOV). This problem affects the performance of medical domain analysis because of frequent addition of new drugs, medicines and perhaps new viruses [9]. Currently, deep learning based approaches have gained attention in this domain of NLP. The deep learning based methods include LSTM, BILSTM, and CRF based models to overcome the issues of traditional NER based methods. In this work, we present deep learning based solution to achieve the efficient performance for bioNER. The proposed model uses BilSTM, CRF, BERT and multi-tasking model with transfer learning approach.
Main contributions of research work as follows:
-
(1)
Focus on aforementioned challenges in biomedical Named Entity Recognition and
-
(2)
Focus on developing a new method using deep learning—Multi-tasking Bidirectional Long Short Term (BiLSTM), Bidirectional Encoder Representation (BERT) and Conditional Random Field mode (CRF) (TLBBC) and
-
(3)
Implementation of Transfer Learning to improve the performance of the model.
Literature Survey
The text mining and NLP methods in this bio-medical domain includes biology with computer science to address several problems of data collection, processing and data analysis for healthcare applications. As discussed before, the traditional NER methods are categorized into three divisions: rule based methods, dictionary based methods and machine learning based methods. This section briefly describes the existing methods of bioNER by considering aforementioned methods.
Eftimov et al. [10] presented a rule-based method for diet recommendation. This method is divided into the two phases: the phases include detection and determination of the entities mentioned in the dataset and the other performs selection and extraction. This model uses sentence segmentation as pre-processing step and each segmented sentence is processed through the chunking and splitting the sentence. Further, this model uses POS tagging and chunking in the first step of detection of entities. These chunks are used to define the matrices of dictionaries and entities. In extraction phase, the sentence segmentation is presented as graph and syntactic parser are used for entity extraction. Asghari et al. [11] reported the growth of unstructured data over web which increased the demand of NER to understand the text data. Authors used the BERT based model for Arabic NER. This model is pre-trained on monolingual data.
Cho et al. [12] reported that the existing methods fail to handle the new and unseen entities therefore development of an advanced NER method is required to overcome the issues of NER. The deep learning methods have gained attention due to their efficient pattern learning. Moreover, some researchers have suggested to incorporate high-level attributes in the embedding layer. Based on these assumptions, we introduce a deep learning-based model that uses combinatorial feature embedding to represent the biomedical word tokens. The model proposed uses combination of Bi-LSTM and CRF. Further, the outcome of these models is improved by integrating the character level representation obtained from CNN and bi-LSTM. Furthermore, this model includes an attention mechanism that directs attention to the tokens in the phrase that are important. This attention mechanism helps to mitigates the long-term dependency difficulty.
Naseem et al. [13] reported several challenges in bio-medical NER such as limited amount of training data, multiple instances of same entity and effect of acronyms. The traditional methods neglect these issues and directly train the deep learning models on general corpora. To overcome the issues of existing methods, authors introduced ALBERT (A Lite Bidirectional Encoder Representations from Transformers for Biomedical Text Mining)—bioALBERT. This model has an attention mechanism that focuses attention on the crucial tokens in the phrase. The main aim of this trained model is to capture the context dependent NER.
Hong et al. [14] reported that the BioNER is accomplished with the help of CRF and it is labelled as sequence labelling problem. The CRF based methods focus on connectivity between labels to obtain the yield structure. The deep learning based CRF models. The deep learning models focus on the estimating the individual labels. The connected labels are described in the form of static number. Despite this, it might be difficult to accurately segregate entity mentions in biological literature because the phrases are sometimes lengthy and specific when compared to generic terms. As a result, restricting the label-label transitions to static values is a barrier to BioNER's performance growth.
Ning et al. [15] reported the different complexities in entity recognition such as large number of entities, non-uniform name of rules, complexity in entity word formation. Moreover, the traditional machine learning based methods focus on manual feature extraction. The feature quality directly affects the accuracy. Therefore, authors adopted the DL based method for efficient feature extraction. In this article, authors introduced a hybrid DL model which uses a combination of Bi LSTM and CRF with Glove for NER. In first phase, uses Glove model which uses semantic features to train the word vector, similarly, the BLSTM model is used to train the word vector along with the morphological features. This process generates the final representation of word and these models are processed through the BilSTM–CRF model to recognize the entities.
Wei et al. [16] reported that the traditional machine learning suffers from the computational complexity issues in feature extraction process. Moreover, the advanced neural network processes improve the overall performance but these methods do not pay attention to significant areas while extracting the features. Therefore, authors have presented attention based model by combining BiLSTM and CRF mode. The BiLSTM model helps to obtain the contextual information whereas the attention mechanism helps to improve the vector representation in BiLSTM. This combined model of BiLSTM and CRF solves the problem of strong dependence of tags. Çelikmasat et al. [17] also used the combined model of BiLSTM and CRF. However, authors this methodology suggested to use transformer based model such as BERT and BioBERT in embedding layer to overcome the issues of CRF models.
Zhang et al. [18] introduced fully-shared multi-task learning model which uses pre-trained bioBERT model with attention mechanism to incorporate the syntactic data. This model is carried out into two parts: one part of this model considers single BioNER where the model is trained on single dataset whereas in another part focus on multi-task method where all datasets are trained together. In single-task model, attention mechanism is also incorporated which mainly integrates the syntactic data into BioBERT encoder. Further, open source NLP toolkit is also employed to extract the different types of syntactic information. Later, the attention mechanism is used to assign the weights for each token and its features. Khan et al. [19] discussed that including new domain in existing NER models leads to several complexities such as labelled data, limited memory devices, and relying of single training data which can cover different slot types. In order to alleviate these issue, authors introduced neural network based multi-task model. In this work, the slot tagging using multiple dataset is considered as multi-task learning problem. Harnoune et al. [20] proposed a knowledge extraction method by using BERT and CRF model. This approach is based on the knowledge based graphs. The knowledge graph helps to represent the knowledge retrieval problem in a simple manner where the problem is transformed into different perspectives.
Proposed Multi-Tasking TLBBC Model
This section presents the proposed Multi-tasking Transfer Learning BiLSTM, BERT and CRF (TLBBC) deep learning based solution for biomedical NER. However, the Deep learning model performance depends on labelled and annotated data. Several researches have reported the issue of unknown words in the data which affects the overall performance. To overcome this issue, transfer learning is considered as promising technique.
Let us consider that variable \(\phi\) represents a set of labels indicating whether the word belongs to the specific entity or not. We consider that a sequence of word is represented as \(w=\left\{{w}_{1},{w}_{2},\dots ,{w}_{n}\right\}\) and the output containing the labels is presented as\(y=\left\{{y}_{1},{y}_{2},\dots {y}_{n}\right\} ,{y}_{i}\in \phi\).
Long Short-Term Memory (LSTM) and Bidirectional Long Short-Term Memory (BiLSTM) Model
The LSTM model is known as specific type of recurrent neural network that is used for modelling the dependencies between elements and input sequence with the help of recurrent connections. Figure 1 shows a basic representation of recurrent network.
The LSTM network considers the input sequence in the form of vector as \(X=\left\{{x}_{1},{x}_{2},\ldots ,{x}_{T}\right\}\) where \({x}_{i}\) represents the vector of words. Similarly, the output is also represented in the form of sequence of vector as \(H=\left\{{h}_{1},{h}_{2},\ldots ,{h}_{T}\right\}\) where \({h}_{i}\) represents the hidden state of vector. At any given step \(t\), the input are \({x}_{t}\), \({c}_{t-1}\), and \({h}_{t-1}\); and, it produces the output as \({c}_{t}\) and \({h}_{t}\) as:
where \(\sigma \left(.\right)\) is the element wise sigmoid operation \({\text{tanh}}\left(.\right)\) Represents the hyperbolic tangent function and \(\odot\) is the element wise multiplication. Similarly, the \({i}_{t}\), \({f}_{t}\) and \({o}_{t}\) denotes the input, forget and output gates, respectively. The \({g}_{t}\) and \({c}_{t}\) represents the intermediate computation steps. The \({W}^{j},{U}^{j}\) and \({b}^{j}\) are the trainable parameters where \(j\in \left\{i,f,o,g\right\}\). However, the aforementioned LSTM model can process the data in one direction. Therefore, researchers have introduced a bidirectional LSTM model which can improve the performance by feeding the input to LSTM twice. Once this process is done in original direction and later it processes the input in the reverse direction. Both outputs are concatenated to obtain the final output. The main advantage of this bidirectional model is that it allows to detect the dependencies from previous and subsequent words in sequence. Current advancements in BioNER have suggested to incorporate CRF model with BiLSTM which takes input sequence \(X\) to predict output \(y\). The prediction score \(s\) can be defined as:
where \(P\) is the output of BiLSTM layer as \(n\times k\) form of matrix, \(n\) denotes length of the sequence and \(k\) is the label count. \(A\) denotes the transition matrix and \({A}_{i,j}\) is the probability of transition from label \(i\) to label \(j\). Further, the possible sequence label is defined as \({Y}_{X}\) for a given input sequence \(X\). The training process helps to maximize the likelihood of label sequence \(y\) in the given sequence \(X\):
Transfer Learning
Different biomedical text mining tasks have seen a rise in popularity of its technique based on the pre-trained language models. As an illustration, authors in [20] employed a transfer learning-based strategy which is used train the weights of LSTM model in forward and backward direction. This task is accomplished with the help of Word2Vec embedding model which is trained on a sizable collection of biological data. However, the Word2vec model in such methods has to be adjusted in accordance with the fluctuations in the biological data.
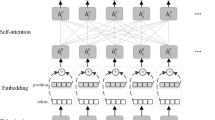
In this work, we have used combination of pre-trained language model where BERT is used and transfer learning approach where we have used task defined output layers. Below given Fig. 2 depicts the architecture of proposed DL based model for BioNER.
According to proposed model, the given input sequence is processed through the shared layer module where lexicon encoder, embedding vectors, transformer encoder and contextual embedding vector modules are used. Further, task specific layers are used for different datasets. Initially, a series of embedding vectors (X) is used to represent the vector which consist of token and word and segment embeddings. Later, the transformer encoder module generates the shared contextual embedding vectors by capturing the contextual data for each token. The generation of task-specific representations for each task/dataset is accomplished last, and then the operations required for entity recognition are performed. The generation of task-specific representations for each task/dataset is done last, and then the operations required for entity recognition are performed. Below given Fig. 3 shows the complete process from input word sequence to output.
-
Lexicon layer: a sentence \(s=\left\{{w}_{1},{w}_{2},{w}_{3},\dots ,{w}_{n}\right\}\) denotes the sequence of tokens of length \(n\).Along with this, the \(\left[CLS\right]\) and \(\left[SEP\right]\) are also employed to denote the start and end of the sentence where \(\left[CLS\right]\) is the first token and \([SEP]\) is the last token. This layer helps to map the sentence \(s\) into embedding vectors \(X=\left\{{x}_{1},\dots {x}_{n}\right\}\) which is obtained with the help of word, segment and position embeddings.
-
Transformer layer: this layer helps to map the input vectors and contextual embedding vectors. In this work, we have used BERT encoder model as shared layer. Further, the BERT model is fine tuned in training stage with the help of multi-task objective function.
-
Task specific layer: in this stage, each dataset is processed through a shallow liner layer. Each dataset is considered as a spate slot tagging task. In this work, we have used softmax layer
Training
The multi-task model is the foundation of the suggested strategy. The following is a definition of the multi-task model configuration: let us consider that total \(m\) datasets are present and each dataset \({D}_{i}\) from \(m\) consists of \({n}_{i}\) training samples as \({D}_{i}={\left\{\left({s}_{j}^{i},{y}_{j}^{i}\right)\right\}}_{j=1}^{ni}\). The training set of each dataset is denoted as \({X}_{i}=\left\{{X}_{1}^{i},\dots ,{X}_{ni}^{i}\right\}\) and their corresponding labels are denoted as \({Y}_{i}=\left\{{y}_{1}^{i},\dots ,{y}_{ni}^{i}\right\}\). The loss function for this multi-task model is given as follows:
Results and Discussion
The suggested method is based on supervised learning scheme which requires labelled data to train the model. In the proposed work, we have used supervised deep learning approach on the following benchmark datasets: NCBI-disease corpus, BC5CDR, JNLPBA, Species-800 and BC2GM dataset. The proposed deep learning model is executed on VSCode tool. For Neural networks, we have used Keras 2.4.0 with tensor flow running on the top of keras applications. Keras is a neural network library. it is widely used in designing the customized neural network model. The complete coding is done by using Python 3.8.
Dataset Details
The NCBI Disease dataset: There are 793 PubMed abstracts in this dataset, and subsets have been created for training (593), development (100), and testing (100). Utilizing concept IDs from either MeSH or OMIM, disease mentions are added to the NCBI Disease corpus. We only take into account mapping illness references to a regulated vocabulary of diseases due to the limited chemical vocabulary.
The BC5CDR corpus: This dataset consists of overall 1500 PubMed abstracts. These abstracts are equally divided to obtain the training, development and testing sets.
JNLPBA: This biomedical dataset was derived from the GENIA 3.02 corpus. In order to create this dataset, a controlled search was applied on MEDLINE. During this search, total 2000 abstracts were collected and annotated to form the 48 classes based on the chemical properties.
Species-800: A corpus for species entities called Species-800 was created based on personally annotated abstracts. There are 800 PubMed abstracts are mentioned in the dataset. In this process, total 800 abstracts were collected from eight different categories. From each category, 100 abstracts are finalized to include in the final list. These categories are bacteriology, botany, entomology, medicine, mycology, protistology, virology, and zoology. This increased the taxonomic mention diversity in the corpus. 800 has been annotated with a concentration on species, although higher taxa (including genera, families, and orders) have also been taken into account.
The BioCreative II Gene Mention Recognition (BC2GM): Dataset was developed by Smith et al. in 2008 and comprises information where contestants are requested to recognize a gene mention in a phrase by providing its start and end characters. With the use of a set of phrases and a list of gene mentions for each sentence, the training set is created (GENE annotations). English only; registration is necessary to access. There are 20 files in the n/a file type.
Parameter Setting
Keras plays a major role in development of deep learning architectures. The traditional deep learning approaches consist of four layers, which are Dense layer, activation layer, drop-out layer and Lambda layer. Point-wise multiplication is performed by the dense layer, learning is started by the activation layer, overfitting is prevented by the drop out layer, and random fluctuations in the model are lessened by the lambda layer.
To solve the overfitting issue, the initial dropout value in this experiment was taken into account as 0.6 at the input layer and 0.5 at the LSTM and SoftMax layers. Due to insufficient training data, fewer neurons often result in under-fitting of the data. Consequently, each hidden layer uses 120 hidden neurons. For an optimizer to prevent a large loss function value, the weights must be changed. Here, the learning settings are adjusted at each epoch. Epsilon and learning rate are set to 0.01 and 1e−08, respectively. The batch size is 40, and there are 250 epochs, each of which displays the loss/error relative to the training set.
Performance Measurement
To estimate the outcome of suggested approach, measure precision \((P)\), recall \(\left(R\right)\), and F1-score \(\left(F1\right)\) which are computed based on true positive, false positive and false negative as given below:
Here, TP denotes the true positive which shows the model correctly predicted the test class to its corresponding positive class, true negative (TN) is shows that the model has correctly predicted the corresponding negative class. False positive (FP) denotes that model has incorrectly predicted the positive class and false negative (FN) denotes the number of negative classes are predicted incorrectly.
Comparative Performance
A comparative analysis of proposed approach where outcomes are compared with several existing methods like GCN, CRF, combined BiLSTM–CRF, combined FastText–BiLSTM–CRF, combined BERT–BiLSTM–CRF and combined BioBERT–BiLSTM–CRF. The proposed approach achieves average performance as 97.30%, 89.50%, 88.50% and 87.60% in terms of Accuracy, Precision, Recall, and F1 Score for NCBI dataset, respectively. The performance which obtained is presented in below given Table 1.
The combined transfer learning and multi-task model are the main advantages of proposed multitask transfer learning based BiLSTM CRF model.
Similarly, we measure the performance of Proposed Multitask-TLBBC and compared with aforementioned existing methods. The proposed approach has reported the average performance as 97.20%, 85.30%, 89.50% and 84.50% in terms of Accuracy, Precision, Recall, and F1 Score for BC5CDR Disease Dataset (Table 2).
Table 3 shows the comparative analysis for JNLPBA dataset where proposed approach obtained the average performance as 96.80%, 89.40%, 88.70%, and 86.30% in terms of Accuracy, Precision, Recall, and F1 Score.
Finally, we measure the overall performance for s800 dataset. The obtained performance is presented in Table 4. The Proposed Multitask-TLBBC reported the average accuracy as 97.50%, average precision as 87.90%, average recall as 86.10% and average.
Figure 4 depicts the loss performances of the proposed approach for 250 epochs. As the number of epochs are increasing the training loss decreases but validation loss increases but the validation loss still remains negligible and it doesn’t impact the accuracy performance. Below given Table 5 shows the performance measurement for initial 10 epochs in terms of Training Loss, Validation Loss, Precision, Recall, F1, and Accuracy.
Conclusion
Due to current demand of NER in bio medical domain, several systems have been developed such as rule based, dictionary based and machine learning based NER systems. These systems achieve desired performance in the normal text data but fail to achieve the noteworthy performance for medical data due to OOV issues. Moreover, computational complexity also remains a challenging task. Recently, deep learning has reported the promising performance for medical data. Therefore, we focused on deep learning based system and presented a combined model which uses BiLSTM, CRF, BERT and transfer learning. The performance of proposed approach is measured in terms of accuracy, precision, recall and F1-score and compared with existing methods. This comparative analysis shows that the proposed approach achieves better performance in terms.
Data Availability
The dataset generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Dash S, Shakyawar SK, Sharma M, Kaushik S. Big data in healthcare: management, analysis and future prospects. J Big Data. 2019;6(1):1–25.
Lou Z, Wang L, Jiang K, Wei Z, Shen G. Reviews of wearable healthcare systems: materials, devices and system integration. Mater Sci Eng R Rep. 2020;140: 100523.
Pooja H, Jagadeesh MP. A collective study of data mining techniques for the big health data available from the electronic health records. In: 2019 1st International conference on advanced technologies in intelligent control, environment, computing & communication engineering (ICATIECE), Bangalore, India; 2019. p. 51–55. https://doi.org/10.1109/ICATIECE45860.2019.9063623.
Zilbermint M. Diabetes and climate change. J Commun Hosp Intern Med Perspect. 2020;10(5):409–12.
Sung M, Jeong M, Choi Y, Kim D, Lee J, Kang J. BERN2: an advanced neural biomedical named entity recognition and normalization tool. Bioinformatics. 2022;38(20):4837–9.
Wen Y, Fan C, Chen G, Chen X, Chen M. A survey on named entity recognition. In: Communications, signal processing, and systems: proceedings of the 8th international conference on communications, signal processing, and systems. Springer Singapore; 2020. p. 1803–1810.
Giorgi JM, Bader GD. Towards reliable named entity recognition in the biomedical domain. Bioinformatics. 2020;36(1):280–6.
Song B, Li F, Liu Y, Zeng X. Deep learning methods for biomedical named entity recognition a survey and qualitative comparison. Brief Bioinf. 2021;22(6):bbab282.
Naseem U, Musial K, Eklund P, Prasad M. Biomedical named-entity recognition by hierarchically fusing biobert representations and deep contextual-level word-embedding. In: 2020 International joint conference on neural networks (IJCNN). IEEE; 2020, July. p. 1–8.
Eftimov T, Koroušić Seljak B, Korošec P. A rule-based named-entity recognition method for knowledge extraction of evidence-based dietary recommendations. PLoS One. 2017;12(6): e0179488.
Asghari M, Sierra-Sosa D, Elmaghraby AS. BINER: A low-cost biomedical named entity recognition. Inf Sci. 2022;602:184–200.
Cho M, Ha J, Park C, Park S. Combinatorial feature embedding based on CNN and LSTM for biomedical named entity recognition. J Biomed Inform. 2020;103: 103381.
Naseem U, Khushi M, Reddy V, Rajendran S, Razzak I, Kim J. Bioalbert: a simple and effective pre-trained language model for biomedical named entity recognition. In: 2021 International joint conference on neural networks (IJCNN). IEEE; 2021, July. p. 1–7.
Hong SK, Lee JG. DTranNER: biomedical named entity recognition with deep learning-based label-label transition model. BMC Bioinform. 2020;21:1–11.
Ning G, Bai Y. Biomedical named entity recognition based on Glove-BLSTM-CRF model. J Comput Methods Sci Eng. 2021;21(1):125–33.
Wei H, Gao M, Zhou A, Chen F, Qu W, Wang C, Lu M. Named entity recognition from biomedical texts using a fusion attention-based BiLSTM-CRF. IEEE Access. 2019;7:73627–36.
Çelikmasat G, Aktürk ME, Ertunç YE, Issifu AM, Ganiz MC. Biomedical named entity recognition using transformers with biLSTM+ CRF and graph convolutional neural networks. In: 2022 International conference on innovations in intelligent systems and applications (INISTA). IEEE; 2022, August. p. 1–6.
Zhang Z, Chen AL. Biomedical named entity recognition with the combined feature attention and fully-shared multi-task learning. BMC Bioinform. 2022;23(1):1–21.
Khan MR, Ziyadi M, Abdel Hady M (2020) Mt-bioner: multi-task learning for biomedical named entity recognition using deep bidirectional transformers. arXiv preprint arXiv:2001.08904.
Harnoune A, Rhanoui M, Mikram M, Yousfi S, Elkaimbillah Z, El Asri B. BERT based clinical knowledge extraction for biomedical knowledge graph construction and analysis. Comput Methods Programs Biomed Update. 2021;1: 100042.
Acknowledgements
The authors acknowledged the JSS Academy of Technical Education, Bengaluru, affiliated to VTU Belagavi, India for supporting the research work by providing the facilities.
Funding
No funding received for this research.
Author information
Authors and Affiliations
Contributions
The dedicated efforts and valuable contributions of all authors involved enabled this collaborative work, significantly enriching the study's outcome through their collective input.
Corresponding author
Ethics declarations
Conflict of Interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Advances in Computational Approaches for Image Processing, Wireless Networks, Cloud Applications and Network Security” guest edited by P. Raviraj, Maode Ma and Roopashree H R.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pooja, H., Jagadeesh, M.P.P. A Deep Learning Based Approach for Biomedical Named Entity Recognition Using Multitasking Transfer Learning with BiLSTM, BERT and CRF. SN COMPUT. SCI. 5, 482 (2024). https://doi.org/10.1007/s42979-024-02835-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02835-z