Abstract
Rice (Oryza sativa L.) plant is vulnerable to a number of pest and diseases. Among them sheath blight disease caused by Rhizoctonia solani, insect pest and brown plant hopper (BPH) (Nilapavata lugens) are the most devastating agents and major challenge to rice cultivation. Plant growth-promoting rhizobacteria (PGPR) are associated with plant roots which augment plant productivity and immunity. Protein analysis was carried out to study the molecular mechanisms underlying PGPR mediated pest and disease resistance and growth promotion. Plants were treated with and without Pseudomonas fluorescens strain Pf1 and challenged with pest and pathogen at 0, 6, 24, 48, 72, and 96 h after inoculation. The comparative analysis of relative abundances of protein bands between inoculated and non-inoculated samples was carried out. Five proteins were upregulated and 15 were differentially regulated in PGPR-primed plants challenged with BPH. In PGPR-primed plants challenged with pathogen, 27 proteins were upregulated. The differential protein bands were sequenced by Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS). Protein sequencing results showed high-percent homology with chloroplastic aldolase, fructose-bisphosphate aldolase, peroxidase, 2-cys peroxiredoxin bas1, chloroplastic-like, small subunit of ribulose-1,5-bisphosphate carboxylase, Os12g0291400 and hypothetical protein OsI_38046. Western blotting detected the presence of PR protein chitinase. The analysis confirmed the presence of chitinase of molecular weight 17, 20, and 35 kDa in PGPR primed plant challenged with R. solani. These results showed that the differentially expressed proteins possibly play role in biotic stress defense in plants challenged with biotic stress. Expression proteins remarkably influenced by Pf1 colonization, which might be a key element for induced systemic tolerance by PGPR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is one of the major food crops for about 65% of the world’s population and is the staple food for an expansive part of the world, particularly in Asia (Chatterjee et al. 2021). Biotic stress including pest and diseases is the major concern that pulls down the productivity of rice. Among the pest, brown plant hopper (Nilaparvata lugens) is one of the most destructive pests of rice throughout Asia. In addition to reducing yields through the direct sucking of plant sap, it acts as a vector for viral diseases like rice grassy stunt and ragged stunt. Brown planthopper (BPH) is re-emerging as a key pest threatening rice production in tropical Asia (Otuka 2013). Furthermore, the soilborne Basidiomycete fungus Rhizoctonia solani (teleomorph: Thanatephorus cucumeris) is a large species complex containing related but genetically distinct groups of pathogens affecting many plants species worldwide. It causes sheath blight in rice, which is considered to be one of the most devastating diseases on a global scale (Li et al. 2021). A predisposing infection with a pathogen causing a hypersensitive reaction is the classic way to induce induced resistance. This elevated resistance response is known as systemic acquired resistance (SAR), which results from challenge inoculation of distant plant parts. SAR was first characterized in tobacco plants that expressed increased resistance systemically after infection by tobacco mosaic virus (Ross 1961). Pathogen-induced SAR is associated with an early increase in endogenously synthesized salicylic acid (SA) (Tripathi et al. 2019). Accumulation of SA is critical in the signaling pathway that controls SAR, since plants that do not accumulate SA are incapable of expressing induced resistance (Gruner et al. 2018). Furthermore, SAR is characterized by the activation of so-called SAR genes (Delaney et al. 1994), including genes that encode pathogenesis-related (PR) proteins (Ali et al. 2018), which are often used as markers for the state of induced resistance. Both PR genes and induced resistance are expressed in plants treated with SA (Molinari and Leonetti 2019). Plant growth-promoting rhizobacteria (PGPR) suppress a variety of root and vascular diseases caused by soilborne pathogens (Bhattacharyya and Jha 2012). PGPR suppress disease by antagonizing pathogens via different mechanisms such as antibiosis, competition for iron or car-bon, and production of lytic enzymes. PGPR also activate plant defense resulting in systemic protection against different fungal, bacterial, and viral pathogens (Pieterse et al. 1996), a phenomenon termed induced systemic resistance (ISR). An alternative approach to inducing systemic resistance was reported by Alstrom (1991), Peer et al. (1991), and Wei et al. (1991). These authors independently demonstrated that selected strains of nonpathogenic plant growth-promoting rhizobacteria colonize the rhizosphere of the plant and are able to elevate plant resistance. Moreover, these bacteria, mainly fluorescent Pseudomonas spp., had been studied for their ability to control soilborne pathogens through competition for nutrients, siderophore-mediated competition for iron, or antibiosis (Ran et al. 2005). It appeared that these bacteria could also provide protection against disease by elevating systemic resistance within the plant. Induced systemic resistance (ISR) mediated by nonpathogenic rhizobacteria has been demonstrated in several plant species (Choudhary et al. 2007) and shown to be effective against bacterial, viral, and fungal diseases. Maurhofer et al. (1994) showed that ISR induced by Pseudomonas fluorescens strain CHA0 in tobacco is associated with PR protein accumulation, suggesting that nonpathogen-induced ISR and pathogen-induced SAR share similar mechanisms. Recent development of high-throughput techniques, omics data are providing opportunities for research into the molecular mechanisms of biological phenotypes (Kumar et al. 2015; Stephen et al. 2022). The present study was undertaken to analyze proteome with special reference to biotic stress tolerance in a popular high-yielding rice variety Jyothi through application of P. fluorescens. Protein analysis was carried out to study the molecular mechanisms underlying the PGPR mediated pest and disease resistance and growth promotion.
Materials and methods
Study materials
The high-yielding rice variety of Kerala, Jyothi (PTB-39), susceptible to blight and BPH, was used in this study. A commercial P. fluorescens strain, Pf1, developed by Kerala Agricultural University (Thrissur, India) and found to exhibit plant growth promotional activity in rice under both laboratory and field conditions, was used.
Biopriming with Pseudomonas fluorescens Pf1 strain
Thirty-five-day-old rice plants were treated with Pf1, and untreated plants served as the control. The experiment was conducted in a completely randomized block design with five pots per treatment and three plants per pot. Plants were treated with Pf1 by two different modes, first as seed treatment and second as foliar spray at 35 days after sowing. For the seed treatment, rice seeds were soaked in the sterile distilled water containing talc-based formulation (10 g/kg of seed). After 24 h, the suspension was drained off, and the seeds were dried under shade for 30 min and sown. For the foliar spray, the talc-based formulation was dissolved in water (20 g/L; 2%) and sprayed.
Isolation and culturing of Rhizoctonia solani
Rice sheath blight pathogen Rhizoctonia solani was isolated from infected rice plants collected from the fields of Agriculture Research Station, Mannuthy (Kerala, Thrissur). The portion of the leaf sheath showing typical lesions was cut into small pieces, surface sterilized with 0.1% sodium hypochlorite for 1 min, and washed thrice before placing on Potato Dextrose Agar (PDA) medium. The axenic culture of the pathogen was obtained by single hyphal tip technique and maintained in PDA slant at 30˚C under laboratory conditions for further use.
Challenge inoculation with Rhizoctonia solani
Forty-five-day-old plants were inoculated with immature white sclerotia obtained from the 4-day-old R. solani culture. The immature sclerotia were placed between the leaf sheath and stem without any injury and wrapped with moist cotton; the inoculated plants were covered with polythene to maintain humidity. Both Pf-treated and non-treated plants were inoculated with R. solani culture. The experiment was conducted in a completely randomized block design with five pots per treatment and three plants per pot. Sheath blight intensity 7 days after inoculation was calculated based on a 0–9 grading scale of Standard Evaluation System for rice, IRRI (1980). The percent disease incidence was calculated using the following formula Wheeler (1969).
Mass rearing of brown plant hopper
Mass rearing of BPH was done following the method described. Rice seedlings were raised in earthen pots, and the pots were kept partially immersed in a plastic tray containing water. These trays were kept in insect rearing cages (3 m × 2.5 m × 2.75 m) with netting. BPH adults were collected from the rice fields of the Regional Agricultural Research Station (Pattambi, Kerala) and released onto individual plants kept inside the rearing cages for oviposition; plants damaged by BPH were periodically replaced with fresh plants. The plants were always maintained free of spiders, mired bugs, and ants for effective BPH rearing. Nymphs emerged a week after oviposition.
Challenge inoculation with BPH
The efficiency of Pf1 treatment on rice plants in protecting against BPH was assessed using five pots per control and Pf treatment. The fourth and fifth instar nymphs of BPH were carefully collected from the cage using an aspirator, starved for 5 h, and released (10 nymphs/pot) on to sheath of Pseudomonas treated rice plants covered with Mylar film and allowed to feed for 7 days. Percent mortality and damage caused by BPH was scored from undamaged (level 0) to complete hopper burn (level 9) based on Inger scale (Inger et al. 1996). Damage rating was done as per Standard Evaluation System for rice (IRRI 2002). The shoot length, root length, number of tillers, fresh weight, and dry weight of the control and treated plants were also measured 30 days after sowing. Statistical analysis was carried out using t-test.
Challenge inoculation with insect and pathogen
Forty-five-day-old rice plants were inoculated with the pathogen R. solani and the insect BPH. After 24 h of inoculation, pest/disease development was assessed based on symptoms. Leaf sheaths from different treatments, including PGPR treatment and challenged with R. solani (T1), PGPR treatment and challenged with BPH (T2), inoculation with R. solani alone (T3), inoculation with BPH alone (T4), PGPR alone (T5), and absolute control (T6), were collected in liquid nitrogen.
Assay of defense-related enzymes
Samples collected from rice plants under different treatments were used to study the induction of defense enzymes in response to pest and pathogen attack. Leaf and sheath tissues from each treatment were collected at 0, 6, 24, 48, 72, and 96 h intervals and immediately homogenized with liquid nitrogen. One gram of the powdered sample was extracted with 2 mL of 0.1 M sodium citrate buffer (pH 5.0) at 4 ºC, and the homogenate was centrifuged for 20 min at 10,000 rpm to collect the supernatant. Protein content in the supernatant was determined by the Bradford method using bovine serum albumin (Thermo Scientific) as the standard.
Assay of peroxidase enzyme
Leaf- sheath sample (1 g) was homogenized in 2 mL of 0.1 M phosphate buffer (pH 7.0) at 4 °C. The homogenate was centrifuged at 16,000 g at 4 °C for 15 min, and the supernatant was used as the enzyme source. The reaction mixture consisting of 1.5 mL of 0.05 M pyrogallol, 0.5 mL of enzyme extract, and 0.5 mL of 1% H2O2 was incubated at room temperature (28 ± 2 °C). The change in absorbance at 420 nm was recorded every 30 s for 3 min. The enzyme activity was expressed as the change in the absorbance min−1 mg−1protein.
Protein profiling by SDS-PAGE
Fresh leaf tissue ground using pestle and mortar in liquid nitrogen reconstituted with 0.2 M phosphate buffer (pH 7.3) and extracted total proteins from both treated and untreated plants. The protein extracted from different treatment samples was normalized to 1000 µg/mL, and the proteome in these samples was analyzed using SDS-PAGE. The protein profile was visualized using a white light transilluminator (Biorad) and analyzed using Quantity 1 software.
Development of polyclonal antibody for chitinase enzyme
The polyclonal antibodies was developed in 1-year-old New Zealand rabbits using crude protein extracted from Trichoderma harzianum following the method described by Acharya (2016). The pre-immune and post-immune serum collected from the animal was purified and used to detect the antibody titer by Outcherlony double immunodiffusion assay. The antibody was further used for Western blot analysis.
Western blot analysis
Approximately 15 µg of total protein from each sample was run on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and electro blotted onto nitrocellulose membrane (Millipore, India) in western transfer buffer (25 mM Tris, 150 mm glycine, 20% methanol and 0.04% SDS) for 3 h at a constant voltage of 50 V. A commercially available NAP (non-animal protein) blocker (G-Biosciences) was used to accomplish the blocking. The membrane was hybridized with a 1:3000 dilution of specific primary antibody and then with 1:6000 dilutions of horseradish peroxide-conjugated secondary antibody. The membrane was washed four times (10 min each time) with 1× PBST after blocking with the secondary antibody. After incubation with secondary antibody, the substrate TMB (3,30,5,50-Tetramethylbenzidine) was added and incubated in dark for about 30 min to form blue colored bands.
Characterization of differentially expressed proteins
The upregulated and the differentially expressed protein bands were sequence analyzed by MALDI-TOF/MS at the Sandor Proteomics Pvt. Ltd. (Hyderabad, India). The obtained peaks were analyzed with the bioinformatics tool, MASCOT/MS peptide search engine (https://www.matrixscience.com/search_form_select.html) was used to characterize proteins.
Results
Pathogenicity test for R. solani
Under favorable conditions, R. solani inoculation in rice resulted in typical sheath blight symptoms within 7 days. Initially, minute specks and small lesions were found, which gradually enlarged to form lesions with a grayish white center with brown margin. As the disease progressed, several lesions coalesced to form blights. Infection started near the water line and slowly spread to the upper leaf sheath and leaf blades. Severely affected plants dried up completely after 96 h (Fig. 1). A comparison of the control and Pf1-primed plants showed that the PDI of the control plants (47.33%) was remarkably higher than the plants treated with Pfl (14.19%; Tables 1 and 2). The significantly low PDI of the Pfl-primed plants indicated that Pfl induces resistance to R. solani in rice.
Percent mortality and damage due to BPH
The application of Pfl highly influenced the BPH feeding preference and extent of damage on rice plants. The damage assessment 7 days after pest release revealed 28% BPH mortality in Pfl-treated plants, whereas no BPH mortality was noticed in the control plants (Tables 2 and 3). BPH actively fed the control plants, resulting in damage, whereas the pest caused no noticeable damage to plants treated with Pfl. The Pfl-treated plants exhibited a convincingly high level of resistance to BPH attack, unlike the control plants, which were moderately susceptible.
Measurement of biometric parameters
The growth parameters such as shoot length, root length, number of tillers, fresh weight, and dry weight of treated and control rice plants were measured 30 days after sowing. The observed average shoot length, root length, fresh weight, and dry weight were significantly higher in Pfl-treated plants than the control plants; 4.08, 28.01, 13.14, 14.83, and 7.06% increase in shoot length, root length, number of tillers, and dry weight of Pfl-primed plants was observed, while the control plants failed to attain any gain in these parameters. Thus, the indisputable role of PGPR in plant growth promotion was upheld in rice plants (Table 3 and Fig. 2).
Analysis of Pf1-induced protein expression profiles
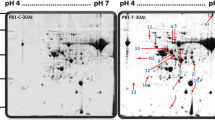
The protein bands of rice plants treated with Pseudomonas bacteria strain Pf1were analyzed after challenge inoculation with BPH. The banding pattern of Pf1-treated plants subsequently challenged with BPH was more predominant than the uninoculated control plants. Analysis of the differences in proteome profile between the Pf1 treated and non-treated rice leaves challenged with BPH revealed that 100 proteins were upregulated while 5 were down regulated in BPH challenged plants, and 15 differential proteins were noticed in PGPR-primed plants challenged with BPH compared to unprimed plants challenged with BPH (Figs. 3 and 4).
Expression analysis of Pf1-treated rice plants challenged with R. solani
The protein extracted from the rice plants treated with Pf1 and inoculated with R. solani was analyzed. The protein banding pattern was more prevalent in Pseudomonas-treated plants challenged with R. solani than the uninoculated control plants. Detailed analysis revealed that 27 protein bands were upregulated in R. solani challenged samples, whereas the banding pattern remained the same in both Pfl-primed and nonprimed plants. However, no difference was observed in R. solani-treated Pfl plants compared with BPH challenged + Pf1-treated plants (Figs. 3 and 4). In BPH challenged plants, 100 proteins were upregulated while 5 were downregulated. Fifteen differential proteins were noticed in PGPR-primed plants challenged with BPH. Two proteins bands of 19 and 30 kDa, differentially expressed at 24 and 48 h, in Pfl-primed plants were selected for sequence analysis out of the 15 differentially expressed bands. In R. solani treated plants, 27 proteins were upregulated, out of which one protein band of 29 kDa upregulated after 4 h in Pf1-primed plant was selected. The sequences of the three selected proteins were annotated to protein databases by NCBI BLASTp. One among the identified proteins showed 81% similarity with peroxidase of Oryza sativa Indica group. Remaining two protein sequences showed homology with the rice protein of japonica cultivars. The identified proteins were fructose-bisphosphate aldolase, Os02g0537700-Oryza sativa (Japonica Cultivar-Group), and Rubisco complexed with 2-carboxyarabinitol-1,5- bisphosphate. Protein band of 19 kDa (denominated 1) was selected from the Pfl-primed plant challenged with BPH. The sequence alignment revealed 99, 94, and 81% homology with chloroplastic aldolase, fructose-bisphosphate aldolase, and peroxidase, respectively. Protein band 2, of 29 kDa obtained from the Pf1-treated plants challenged with the pathogen, showed 84 and 86% homology with 2-cys peroxiredoxin bas and 2-cys peroxiredoxin basI, chloroplastic-like, respectively. The protein band 3 selected from the Pfl-primed plant infected with BPH was 30 kDa and exhibited 99% to the small subunit of ribulose-1,5-bisphosphate carboxylase and 100% similarity to Os12g0291400 and a hypothetical protein Osl_38046. Details of MALDI-TOF search alignment are given in Table 4 and the MALDI-TOF search alignment of protein bands are provided in supplementary file 1.
Analysis of peroxidase activity
The enhanced activities of defense-related enzyme peroxidase (PO) in the Pf1-treated rice plants indicate the development of induced systemic resistance against sheath blight disease and BPH insect in rice. An increase in PO activity was detected between 6 to 96 h after challenge inoculation in Pfl-primed plants. Exactly 43.62% boost in PO activity was observed in pathogen-challenged plants over the control and 21.9% in BPH challenged Pfl-primed plants.
Western Blot
Western blot analysis confirmed that the 17, 20 and 35 kDa proteins were expressed in Pfl-treated plants challenged with R. solani inoculation after 96 h, and there was no detection in control plant protein sample (Fig. 5). These banding patterns indicate the presence of chitinase in PGPR-primed plants challenged with R. solani and the expressed chitinase possibly provides resistance against the pathogen.
Discussion
Broad conglomeration of PGPR frequently colonizes the rhizosphere of plant species and provides beneficial effects, such as enhanced plant growth and reduced disease susceptibility. In addition, plants respond to PGPR by enduring physiological or biochemical changes, commonly referred to as ISR. This PGPR-induced ISR has suppressed various plant diseases under both greenhouse and field conditions (Yi et al. 2013). In the present study, rice plants that had been treated with Pf1 were resistance to sheath blight than controls. Significantly lower PDI (14.19) was observed in Pf1-treated plants than control plants (47.33), indicating that Pf1 induced resistance to R. solani infection in rice through ISR. These observations are reliable with the earlier reports. Insect or pathogen attack triggers an endogenous defense mechanism in all plants. Various defense genes are induced or activated in response to the stimuli or signals. Researchers have proven that the self-defense mechanisms in plants can be instigated by applying a biological inducer, which is used as a novel and efficient plant protection strategy. The fluorescent Pseudomonas that has been efficiently controlled various soilborne pathogens. Pseudomonas fluorenscens successfully demonstrated to control the damping-off of cucumber (Rezzonico et al. 2005), red rot of sugarcane (Anita and Samiyappan 2012), and sheath blight in rice (Vidhyasekaran and Muthamilan 1999). The studies also reported that the type III protein secretion system present in P. Fluorenscens effectively counter the effectors in pathogens. In the present study, the P. fluorescens isolate Pfl reduced the intensity of sheath blight in rice. Jaleel et al. (2007) had indicated that P. fluorescens serves as a strong elicitor of plant defense reactions. Studies have suggested that the pre-application of fluorescent Pseudomonas toughens host cell wall structure, thereby restricting pathogen invasion into plant tissues (Preston 2004). For instance, T3SS gene hrcV a type III protein secretion system present in P. fluorenscens reduced polygalacturonase activity in Pythium ultimum thus provided tolerance to cucumber from pathogen mediated damping-off (Couillerot et al. 2009). Our previous transcriptome studies demonstrated the effect of Pfl in enhancing abiotic stress tolerance (Saakre et al. 2017). In contrast, the present study was performed to validate the effect of Pfl against biotic stress at the protein level. The study clearly depicts that Pfl induces the activity of enzymes associated with phenylpropanoid metabolism and PR protein accumulation in rice in response to R. solani challenge inoculation. Seed treatment with PGPRs has triggered the plant to synthesize small amounts of defense-related compounds, which occurred as an immune response to pathogen inoculation. The production of defense-related proteins in Pf1-primed plants before challenge inoculation and greater accumulation of defense compounds in Pf1-treated plants after pathogen challenge indicates that biopriming can trigger ISR at time of infection. Thus, the present study proposes the application of fluorescent Pf1 in rice as a promising approach to induce resistance against sheath blight.
The application of Pf1 highly influenced the feeding preference and damage caused by BPH in the rice plants. The highest mortality of BPH occurred on plants treated with Pf1, whereas the BPH released on control plants remained alive. Consequently, the damage index was also significantly higher on control plants than the plants treated with Pf1. These observations suggest that the application of P. fluorescens bioformulation significantly reduced the pest incidence in rice. The ISR or SAR mechanisms or a combination of both may be involved in pest resistance of rice, consistent with numerous earlier reports. Management of insect pests by various Pseudomonas strains either as a bacterial suspension or other formulations have been reported (Senthilraja et al. 2013). It is evident that feeding pattern of various insect pests has altered due to the application of Pseudomonas strains on plants. Karthiba et al. (2010) reported 20% weevil mortality in chestnuts sprayed with P. fluorenscens suspension. Previous investigations have also proven that Pseudomonas-treated rice leaves altered the feeding behavior of leaf folder, reduced larval and pupal weight, and increased the larval mortality and incidence of malformed adults under in vitro conditions (Radjacommare et al. 2002). In addition, the PGPR possess the ability to produce plant growth hormones like auxins, cytokinins, gibberellins, and ethylene (Cassán et al. 2014; Nysanth et al. 2019). Rhizobacteria produced auxins promote plant growth. Ahirwar et al. (2015) reported an increase in root and shoot fresh weight of tomato, cucumber, lettuce, and potato after bacterization with Pseudomonas strains. Furthermore, direct growth stimulation occurred as a result of ACC deaminase enzyme secreted by PGPR that has the ability to lower plant ethylene level during biotic and abiotic stress (Glick 2014). In the present study, the plants treated with Pf1 showed an increase in all growth parameters, including plant height, active tillers, root length, wet weight, and dry weight, which confirms that the application of fluorescent Pseudomonas in rice promotes growth.
Further sequence analysis of the differentially expressed protein band 1 revealed significant homology with chloroplastic aldolase, fructose-bisphosphate aldolase, and peroxidase. The chloroplastic aldolase and fructose-bisphosphate aldolase belonging to class I adlolases possess a key lysine residue in the catalytic site required for the formation of a covalent intermediate in the form of a Schiff base with its substrate prior to the aldol cleavage (LowKam et al. 2010). They play vital roles in photosynthesis, especially in the Calvin cycle. In the present study, the high expression of the protein band 1 in the PGPR-treated rice leaf sheath tissues compared to control indicates that the photosynthetic activity might have facilitated attain greater growth and better defense.
The biochemical and molecular analysis of biomolecules associated with systemic resistance induced via Pf1 priming revealed an increase in the activity of the defense-related enzyme peroxidase (PO) against sheath blight disease and BPH insect in rice. Peroxidases are commonly associated with shielding plants from pathogens. They are responsible for the radical dehydrogenation of sinapyl alcohol and coniferyl alcohol during lignin synthesis. Peroxidase polymorphism could be used as a biochemical marker indicating different levels of field resistance (Lebeda et al. 2001). The enzyme participates in insolubilization of the extracellular matrix processes in the extracellular matrix (Singh et al. 2010). Their association with the cell wall has also been confirmed. Peroxidases remove the toxic hydrogen peroxide from tissues and participate in synthesis of phenolic compounds and in the building of intermolecular bonds during the organization of the cell wall at the sites of infection by pathogens (Kerchev et al. 2015). The peroxidase enzyme is also involved in the synthesis of ethylene, the concentration of which increases during pathogenesis (Prasannath 2017). The present study found increased peroxidase activity in Pf-treated plants challenged with the BPH. Anand et al. (2010) reported the higher PO activity in cucumber roots challenged with P. corrugate as compared with control plants. The present study also found that a PO was prominently expressed in P. fluorescens isolate Pf1 treated tissues against R. solani. This unique PO induced by Pf1 might have contributed to induced defense in rice sheath against BPH infection.
The sequence analysis of protein band 2, which was differentially expressed in Pf1 treated plants challenged with R. solani, was 29 kDa and showed 84 and 86% homology to 2-cys peroxiredoxin bas and 2-cys peroxiredoxin bas1, chloroplastic-like, respectively. 2-Cys peroxiredoxins are a large family of peroxidases that reduce alkyl hydroperoxides and hydrogen peroxide. These enzymes were identified in plants as ubiquitously occurring group of enzymes (Tripathi et al. 2009). However, Bas1 expression is restricted to green tissues of the shoot (Baler and Dietz 1997) and bas1 gene expression is regulated by cellular redox state, which depends on the antioxidant function of the enzyme while oxidative stressors increase the expression only slightly. Researchers have correlated peroxiredoxins and signaling networks under various stress responses, including pathogen elicitors, insect feeding, wounding, high temperature, and ABA-associated stomatal closure (Larkindale and Knight 2002; Apel and Hirt 2004; Peng et al. 2004; Mateo et al. 2006; Lakshmi et al. 2023). Moreover, peroxiredoxins are generally expressed more under oxidative stress, which is one of the most rapid defence reactions to pathogen attack in plants (Apel and Hirt 2004). Therefore, the differential expression of protein band 2 in the PGPR-treated rice leaf sheath tissues challenged with pathogen indicates that 2-Cys peroxiredoxins was a part of the defense mechanism during the pathogen attack. The analysis of protein band 3 differentially expressed in Pf1-treated plants challenged with BPH (30 kDa) revealed high level of homology with the small subunit of ribulose-1,5-bisphosphate carboxylase (99%) and 100% similarity was observed to both Os12g0291400 and hypothetical protein OsI_38046; however, their roles in defense mechanism are not known. The level of RuBisCO proteins, one of the important enzymes in photosynthesis, is known to be reduced in infected plant cells under pest attack (Bilgin et al. 2010). RuBisCO proteins were also reported to be high in rice leaf sheath under wounding stress (Lee et al. 2006). Differential expression of band 3 proteins in the PGPR-treated rice leaf sheath tissues indicates increased photosynthetic activity that helped to attain greater growth and possibly related to plant defense. The chitinase enzyme was isolated from Trichoderma hazianum and purified by salt separation and dialysis. The purified chitinase was injected to rabbit to develop polyclonal antibody for chitinase. The presence of an antibody for chitinase was detected post-immune serum collected from rabbit by Ouchterlony double diffusion. The antibody developed was used to detect target protein in Western blot analysis. Results of the Western blot analysis also revealed that about 17, 20 and 35 kDa protein were developed in Pf-primed plants challenged with R. solani after 96 h of inoculation. These observations confirmed the upregulation of chitinase in PGPR-primed plants challenged with R. solani, indicating the role of chitinase in developing resistance against pathogen. Most chitinases have a molecular mass ranging between 15 and 43 kDa. Chitinase has been isolated from chickpea (Zarei et al. 2011), cucumber, barley (Kirubakaran and Sakthivel 2006), tobacco (Pu et al.1996), black turtle bean (Chu and Ng 2005), tomato (Wu and Bradford 2003), and grapes (Sluyter et al. 2005). Few studies also found expression of chitinase gene in response to cold stress (− 2 to − 5 ºC). The chitinases also have demonstrated significant antifungal activities against plant pathogenic fungi like Alternaria sp. of rice, Bipolaris oryzae of rice, Botrytis cinerea of tobacco, Curvularia lunata of clover, Fusarium oxysporum, F. udum, Mycosphaerella arachidicola, Pestalotia theae of tea, and R. solani of rice (Chu and Ng 2005; Saikia et al. 2005; Kirubakaran and Sakthivel 2006). The mode of action of PR-3 proteins is relatively simple and characterized. Chitinases cleave the cell wall chitin polymers in vivo, making the fungal cells osmotically sensitive (Prasannath 2017).
Conclusion
The present study found that the colonization of P. fluorescens influenced the differential expression of various proteins in rice plants; the differentially expressed chitinase, RuBisCO, and 2-cys peroxiredoxins provide resistance against the fungal pathogen, and chloroplastic aldolase, fructose-bisphosphate aldolase, and peroxidase provide resistance against BPH. Thus, P. fluorescens might be a key element for induced systemic resistance. The application of P. fluorescens will facilitate the rice plants to tolerate biotic stress via the expression of defense-related genes.
References
Acharya A (2016) Serological and molecular detection of foliar fungal pathogens of Persea bombycina Kost and activation of defense response using bioinoculants. Doctoral dissertation, University of North Bengal
Ahirwar NK, Gupta G, Singh V, Rawlley RK, Ramana S (2015) Influence on growth and fruit yield of tomato (Lycopersicon esculentum Mill.) plants by inoculation with Pseudomonas fluorescence (SS5): possible role of plant growth promotion. Int J Curr Microbiol Appl Sci 4(2):720–730
Ali S, Ganai BA, Kamili AN, Bhat AA, Mir ZA, Bhat JA, Tyagi A, Islam ST, Mushtaq M, Yadav P, Rawat S (2018) Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol Res 212:29–37
Alstrom S (1991) Induction of disease resistance in common bean susceptible to halo blight bacterial pathogen after seed bacterization with rhizosphere Pseudomonas. J Gen Appl Microbiol 37:495–501
Anand T, Chandrasekaran A, Kuttalam S, Senthilraja G, Samiyappan R (2010) Integrated control of fruit rot and powdery mildew of chilli using the biocontrol agent Pseudomonas fluorescens and a chemical fungicide. Biol Control 52(1):1–7
Anita B, Samiyappan R (2012) Induction of systemic resistance in rice by Pseudomonas fluorescens against rice root knot nematode Meloidogyne graminicola. J Biopesticides 5:53
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Baier M, Dietz KJ (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12(1):179–190
Bhattacharyya PN, Jha DK (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28(4):1327–1350
Bilgin DD, Zavala JA, Zhu JIN, Clough SJ, Ort DR, DeLUCIA EH (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33(10):1597–1613
Cassán F, Vanderleyden J, Spaepen S (2014) Physiological and agronomical aspects of phytohormone production by model plant-growth-promoting rhizobacteria (PGPR) belonging to the genus Azospirillum. J Plant Growth Regul 33(2):440–459
Chatterjee S, Stoy PC, Debnath M, Nayak AK, Swain CK, Tripathi R, Chatterjee D, Mahapatra SS, Talib A, Pathak H (2021) Actual evapotranspiration and crop coefficients for tropical lowland rice (Oryza sativa L.) in eastern India. Theor Appl Climatol 146(1): 155–171.
Choudhary DK, Prakash A, Johri BN (2007) Induced systemic resistance (ISR) in plants: mechanism of action. Indian J Microbiol 47(4):289–297
Chu KT, Ng TB (2005) Purification and characterization of a chitinase-like antifungal protein from black turtle bean with stimulatory effect on nitric oxide production by macrophages. Biol Chem 386:19–24
Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moënne-Loccoz Y (2009) Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol 48(5):505–512
Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gur-Rella M, Kessmann H, Ward E, Ryals J (1994) A central role of salicylic acid in plant disease resistance. Sci 266:1247–1250
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169(1):30–39
Gruner K, Zeier T, Aretz C, Zeier J (2018) A critical role for Arabidopsis MILDEW RESISTANCE LOCUS O2 in systemic acquired resistance. Plant J 94(6):1064–1082
Inger S, Ramsbotham W, Cliff RA, Rex DC (1996) Metamorphic evolution of the Sesia-Lanzo Zone, Western Alps: time constraints from multi-system geochronology. Contrib Mineral Petrol 126(1):152–168
IRRI (1980) Bacterial blight of rice. Proceedings of the International Workshop on Bacterial Blight of Rice 14–18 March 1988. International Rice Research Institute in collaboration with Administration Générale de la Coopération au Développement Ministère des Affaires Etrangères Belgium. 1–225.
IRRI I (2002) Standard evaluation system for rice. International Rice Research Institute, Philippine, pp 1–45.
Jaleel CA, Manivannan P, Sankar B, Kishorekumar A, Gopi R, Somasundaram R, Panneerselvam R (2007) Pseudomonas fluorescens enhances biomass yield and ajmalicine production in Catharanthus roseus under water deficit stress. Colloids Surf B Biointerfaces 60(1):7–11
Karthiba L, Saveetha K, Suresh S, Raguchander T, Saravanakumar D, Samiyappan R (2010) PGPR and entomopathogenic fungus bioformulation for the synchronous management of leaffolder pest and sheath blight disease of rice. Pest Manag Sci 66(5):555–564
Kerchev P, De Smet B, Waszczak C, Van Breusegem MJ (2015) Redox strategies for crop improvement. Antioxidants Redox Signal 23(14):1186–1205
Kirubakaran SI, Sakthivel N (2006) Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Expr Purif 52(1):159–166
Kumar A, Bimolata W, Kannan M, Kirti PBPPP, Qureshi IA, Ghazi IA (2015) Comparative proteomics reveals differential induction of both biotic and abiotic stress response associated proteins in rice during Xanthomonas oryzae pv. oryzae infection. Funct Integr Genomics 15(4):425–437.
Larkindale J, Knight M (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128:682–695
Lakshmi G, Beena R, Soni KB, Viji MM, Uday CJ (2023) Exogenously applied plant growth regulator protectsrice from heat induced damage by modulating plant defense mechanism. J Crop Sci Biotechnol. https://doi.org/10.1007/s12892-022-00162-4
Lebeda A, Luhová L, Sedlářová M, Jančová D (2001) The role of enzymes in plant-fungal pathogens interactions/Die Rolle der Enzyme in den Beziehungen zwischen Pflanzen und pilzlichen Erregern. Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz/J Plant Dis Protect, 89–111.
Lee J, Bricker TM, Lefevre M, Pinson SR, Oard JH (2006) Proteomic and genetic approaches to identifying defence-related proteins in rice challenged with the fungal pathogen Rhizoctonia solani. Mol Plant Pathol 7(5):405–416
Li D, Li S, Wei S, Sun W (2021) Strategies to manage rice sheath blight: lessons from interactions between rice and Rhizoctonia solani. Rice 14(1):1–15
LowKam C, Liotard B, Sygusch J (2010) Structure of a class I tagatose-1,6-bisphosphate aldolase: investigation into an apparent loss of stereospecificity. J Biol Chem 285(27):21143–21152
Mateo A, Funck D, Muhlenbock P, Kular B, Mullineaux PM, Karpinski S (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J Experimental Bot 57:1795–1807
Maurhofer M, Hase C, Meuwly P, Metraux JP, Defago G (1994) Induction of systemic resistance of tobacco to tobacco necrosis virus by the rootcolonizing Pseudomonas fluorescens strain CHA0: Influence of the gacA gene and of pyoverdine production. Phytopatholo 84:139–146
Molinari S, Leonetti P (2019) Bio-control agents activate plant immune response and prime susceptible tomato against root-knot nematodes. PLoS ONE 14(12):e0213230
Nysanth NS, Meenakumari KS, Syriac EK, Beena R (2019) Screening of pink pigmented facultative methylotrophs for growth enhancement in paddy. Biocatal Agric Biotechnol 18:101055
Otuka A (2013) Migration of rice planthoppers and their vectored re-emerging and novel rice viruses in East Asia. Front Microbiol 4(309):1–11
Peer VR, Niemann GJ, Schippers B (1991) Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Phytopathol 81:728–734
Peng JY, Deng XJ, Huang JH, Jia SH, Miao XX, Huang YP (2004) Role of salicylic acid in tomato defense against cotton bollworm, Helicoverpa armigera Hubner. Zeitschrift Fur Naturforschung C-a J Biosci 59:856–862
Pieterse CMJ, Wees SCM, Hoffland E, van Pelt JA, van Loon LC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogensis-related gene expression. Plant Cell 8:1225–1237
Prasannath K (2017) Plant defense-related enzymes against pathogens: a review. Plant defence-related enzymes. https://doi.org/10.4038/agrieast.v11i1.33
Preston GM (2004) Plant perceptions of plant growth-promoting Pseudomonas. Philos Trans R Soc Lond 359(1446):907–918
Pu Z, Lu BY, Liu WY, Jin SW (1996) Characterization of the enzymatic mechanism of g-momorcharin, a novel ribosome-inactivating protein with lower molecular weight of 11,500 purified from the seeds of bitter gourd (Momordica charantia). Biochem Biophys Res Commun 229:287–294
Radjacommare R, Nandkumar R, Kandan A, Suresh S, Bharathi M, Raguchander T, Swamiyappan R (2002) Pseudomonads fluorencens based bioformulation for the management of sheath blight and leaffolder in rice. Crop Protect 21:251–256
Ran LX, Liu CY, Wu GJ, Van Loon LC, Bakker PAHM (2005) Suppression of bacterial wilt in Eucalyptus urophylla by fluorescent Pseudomonas spp. China Biological Control 32(1):111–120
Rezzonico F, Binder C, Défago G, Moënne-Loccoz Y (2005) The type III secretion system of biocontrol Pseudomonas fluorescens KD targets the phytopathogenic Chromista Pythium ultiumm and promotes cucumber protection. Mol Plant Microbe Interact 18(9):991–1001
Ross AF (1961) Systemic acquired resistance induced by localized virus infections in plants. Virol 14:340–358
Saakre M, Baburao TM, Salim AP, Ffancies RM, Achuthan VP, Thomas G, Sivarajan SR (2017) Identification and characterization of genes responsible for drought tolerance in rice mediated by Pseudomonas fluorescens. Rice Sci 24(5):291–298
Saikia R, Singh BP, Kumar R, Arora DK (2005) Detection of pathogenesis-related proteins—chitinase and α-1,3-glucanase in induced chickpea. Curr Sci 89(4):659–663
Senthilraja G, Anand T, Kennedy JS, Raguchander T, Samiyappan R (2013) Plant growth promoting rhizobacteria (PGPR) and entomopathogenic fungus bioformulation enhance the expression of defense enzymes and pathogenesis-related proteins in groundnut plants against leafminer insect and collar rot pathogen. Physiol Mol Plant Pathol 82:10–19
Singh P, Carraher C, Schwarzbauer JE (2010) Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol 26:397
Sluyter SV, Durako M J, Halkides CJ (2005). Comparison of Grape Chitinase Activities in Chardonnay and Cabernet Sauvignon with Vitis rotundifolia cv. fry. Am J Enol Vitic 56(1):81–85.
Stephen K, Beena R, Kiran AG, Shanija S, Saravanan R (2022) Changes in physiological traits and expression of key genes involved in sugar signaling pathway in rice under high temperature stress. 3 Biotech 12(2):183. https://doi.org/10.1007/s13205-022-03242-y
Tripathi S, Mehrotra GK, Dutta PK (2009) Physicochemical and bioactivity of cross-linked chitosan–PVA film for food packaging applications. Int J Biol Macromol 45(4):372–376
Tripathi D, Raikhy G, Kumar D (2019) Chemical elicitors of systemic acquired resistance-Salicylic acid and its functional analogs. Curr Plant Biol 17:48–59
Vidhyasekaran P, Muthamilan M (1999) Evaluation of a powder formulation of Pseudomonas fluorescens Pf1 for control of rice sheath blight. Biocontrol Sci Tech 9(1):67–74
Wei G, Kloepper JW, Tuzun S (1991) Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathol 81:1508–1512
Wu C, Bradford KJ (2003) Class I chitinase and α-1,3-glucanase are differentially regulated by wounding, methyl jasmonate, ethylene and gibberellin in tomato seeds and leaves. Plant Physiol 133:263–273
Yi HS, Yang JW, Ryu CM (2013) ISR meets SAR outside: additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spot in field-grown pepper. Front Plant Sci 4:122
Zarei M, Aminzadeh S, Zolgharnein H, Safahieh A, Daliri M, Noghabi KA, Ghoroghi A, Motallebi A (2011) Characterization of a chitinase with antifungal activity from a native Serratia marcescens B4A. Braz J Microbiol 42(3):1017–1029
Acknowledgements
We thank the Center for Plant Biotechnology and Molecular Biology, Bioinformatics Center, College of Horticulture, Kerala Agricultural University, Thrissur, India, for providing facilities for current work.
Author information
Authors and Affiliations
Contributions
Material preparation, data collection and analysis were performed by SS. The first draft of the manuscript was written by MS. The concept and project idea was conceived by APS and BR Pathological analysis was undertaken by HB. Insect bioassay was done by HB, and manuscript editing was done by PR and BR. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors do not have any conflict of interest in publishing this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Maria Rosa Simon.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shinde, S., Abida, P.S., Saakre, M. et al. Identification and comparative analysis of differential proteins expression in rice under biotic stress by protein sequencing. CEREAL RESEARCH COMMUNICATIONS (2023). https://doi.org/10.1007/s42976-023-00464-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42976-023-00464-5