Abstract
Gastric diseases are a common medical issue; they can be detected using endoscopy equipment. Computer-aided diagnosis (CADx) systems can help internists identify gastric diseases more accurately. In this paper, we present a CADx system that can detect and classify gastric diseases such as gastric polyps, gastric ulcers, gastritis, and cancer. The system uses a deep learning model as a GoogLeNet based on an Inception module. The fast and robust fuzzy C-means (FRFCM) and simple linear iterative clustering (SLIC) superpixel algorithms are applied for image segmentation during preprocessing. The FRFCM algorithm, which is based on morphological reconstruction and membership filtering, is much faster and more robust than fuzzy C-means. In addition, the SLIC superpixel algorithm adapts the k-means clustering method to efficiently generate superpixels. These two approaches produce a feasible method of classifying normal and abnormal gastric lesions. The areas under the receiver operating characteristic curves were 0.85 and 0.87 for normal and abnormal lesions, respectively. The proposed CADx system also performs reliably.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The National Cancer Registration Statistics that were released by the Ministry of Health and Welfare in 2018 [1] show an increase in the incidence of gastric cancer based on the cancer age standardized incidence rate (ASR) up to 2016. According to these statistics, it is now the first and fourth most frequently occurring gastric cancer in men and women, respectively. The ASR of gastric cancer around the world is shown in Fig. 1, based on data from the International Agency for Research on Cancer (IARC), which is affiliated with the World Health Organization (WHO) [2]. East Asian countries occupy the top positions and South Korea has the highest incidence. Early diagnosis of gastric lesions is very important because gastric cancer does not have any significant symptoms until it has progressed to a later stage. In order to prevent gastric cancer, it is necessary to diagnose the gastric lesions that cause the cancer. Most of these diagnoses are done via gastric endoscopy [3].
Recently, computer-aided diagnosis (CADx) systems, which assist doctors in the characterization of lesions, have been actively studied and applied. The number of available endoscopy images has increased as endoscopy equipment has been improved, but the fatigue of doctors observing the lesions and the diagnostic time have also increased. At present, the characterization of endoscopy images is highly dependent on the experience of doctors. An effective CADx system could increase the early diagnosis of gastric lesions, which would improve the quality of life of patients by preventing gastric cancer [4].
A CADx system study is currently underway that uses endoscopy images to classify Helicobacter pylori infections based on a deep learning model [5] and other studies have used convolutional neural networks to classify tumors and adenomas [6]. In addition, there studies have compared the results of multiple convolutional neural networks for various lesion classes [7]. Another study has used a support vector machine (SVM) to classify local binary pattern texture features that were extracted by a wavelet transformation of wireless capsule endoscopy images [8]. Furthermore, other studies have extracted various color and texture features from segmented images in order to identify rare features among them and classify them using an SVM [9]. We have designed a CADx system that uses a new algorithm to classify abnormal and normal results based on endoscopy images. To achieve this, we used the deep learning model, which is based on the Inception module [10]. The gastric lesions are somewhat irregular, and show different characteristics and sizes. We expect that a segmented image identified as normal or abnormal by an internist can provide more detailed information about the characteristics of lesions. Segmentation was conducted as a pre-processing technique using the simple linear iterative clustering (SLIC) superpixel algorithm [11] as well as the fast and robust fuzzy C-means (FRFCM) algorithm [12]. The results of this method were compared to those from a previous study [13], in which segmentation was not applied.
2 Methods
2.1 Dataset
With institutional review board approval, a dataset was collected from the files of patients who had undergone gastric endoscope imaging in the Department of Internal Medicine at Gyeongsang National University, South Korea. For this study, 940 normal and 465 abnormal endoscopy images from 90 patients were collected. All of the images were chosen and classified by internists. We randomly divided the patient cases into two subsets, the training set and the test set. Table 1 shows the frequency of each disease seen in the abnormal images.
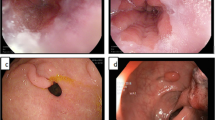
The training set included a total of 738 images, 493 normal and 245 abnormal. The test set included a total of 667 images, 447 normal and 220 abnormal. The types of lesions included in the dataset are shown in Fig. 2. The abnormal images included different gastric diseases such as gastric cancer, gastric ulcers, gastric cancer, and gastric bleeding. All of the lesions were cancerous gastric lesions [14].
2.2 Image Segmentation
The training data were applied to the deep learning model after image segmentation. Image segmentation is a process in which similar features within an image are grouped together; in this work, the SLIC superpixel and FRFCM algorithms were used for the segmentation. Deep learning was then performed using the segmented images. It is important to optimize image segmentation because there is a risk of under- or over-segmentation. In the case of over-segmentation, it is difficult to extract the image features because the area is too small. On the other hand, under-segmentation does not provide enough information about the desired area. In Fig. 3, the areas containing the desired characteristics are shown in circles, Fig. 3a shows the optimal-segmentation. When information is extracted from a circle containing a desired feature, over-segmentation results in information also being extracted from other areas (Fig. 3b). On the other hand, under-segmentation does not extract all of the information about the desired feature as the segment is smaller than the feature (Fig. 3c). Therefore, it is important to find the optimum segmentation value through several repetitions in order to extract the information effectively. Figure 4 provides the flowchart summarizing the proposed model. First, the segmentation parameter was set to 9 in the proposed CADx system. Given that the optimal segmentation parameters vary from one algorithm to another, it is possible to assess how two algorithms perform for the same parameter.
Figure 5 shows the configuration of the segmentation process and convolutional neural network (CNN), which retains a Google Inception V3 model.
2.3 SLIC Superpixel Segmentation
The SLIC superpixel algorithm is commonly used for image segmentation. This method segments the original image into groups of pixels with similar characteristics and then splits them into similar uniform areas. Each uniform area is treated as a superpixel. The shape of the superpixel is controlled by various features such as compactness, boundary precision, boundary recall, minimization of under-segmentation, and uniformity. Superpixel algorithms can be categorized as graph-based or gradient-ascent-based methods. The SLIC superpixel algorithm can be categorized as the latter.
The SLIC superpixel algorithm reduces the number of calculations required by limiting the range used in the calculations. It is beneficial to adjust the size and compactness of the superpixels by applying different weights to the difference between color information and local information. As shown in Fig. 6, this method is quick because it is calculated by limiting the area in a different way to the standard k-means method. First, the RGB (red–green–blue) input image is converted to CIELAB (International Commission on Illumination) color space. Then, the superpixels generated will have a similar size and cluster center value \(C_{i}\). Equation (1) performs clustering based on the lightness L, green to red a, and blue to yellow b values of the CIELAB color space and the x and y values of the pixel’s coordinates:
The center point of C is calculated for the same interval as Eq. (2). Where N is the number of image pixels and K is the number of superpixels to be segmented. In Eq. (2), S is the spacing between the centers of the clusters. As shown in Fig. 6, k-means clustering is performed across the entire image area; in contrast, the SLIC algorithm performs clustering based on the limited area 2S × 2S:
The distance is calculated for \(L_{i}\), \(a_{i}\), and \(b_{i}\), then the distances \(x_{i}\) and \(y_{i}\) are calculated to obtain the center of the clustered superpixel, \(C_{i}\) [11]. The SLIC superpixel algorithm was applied to the endoscopy images and the segmentation results are shown in Fig. 7. The segmented image was compared with the ground truth specified by the internist; hence the abnormal superpixels were classified and training was performed.
2.4 FRFCM Clustering Segmentation
The FRFCM algorithm is more advanced than other fuzzy C-means (FCM) algorithms. The FCM algorithm does not assign a pixel to a specific cluster; instead, it uses an algorithm that provides information about clustering by calculating how much belongs to each defined cluster. The value data to be clustered using the FCM algorithm can be expressed as A = \(\left\{ { a_{1} ,a_{2} , \ldots ,a_{N} } \right\} \subseteq R^{p}\), where R is the vector space, p is the feature dimension, and N represents the total number of data. Each pixel of a color image is represented by a feature vector such as \(\varvec{x}_{\varvec{k}} = \left( {x_{k1} ,x_{k2} , \ldots x_{kp} } \right)\). The cluster center can be expressed as C = \((c_{1} , \ldots c_{m} )\) where M is the number of clusters, equal to the number of clustering areas in the image.
The FCM algorithm is a method of obtaining the matrix U when the function \(F_{FCM} (\varvec{U},\varvec{C} | A)\) in Eq. (3) is minimized by applying iterative optimization algorithms with dataset A and cluster center C. In Eq. 2, n is a constant that indicates the degree of fuzzification and \(\left| {\left| {{\text{a}}_{\text{k}} - {\text{c}}_{\text{i}} } \right|} \right|^{2}\) is a measure of the distance between a and c. Euclidean distance is used in the FCM algorithm. The membership value satisfies the condition for Eq. (4) and the sum of the affiliation values should be one. The values from Eq. (3) are repeated to satisfy the condition for Eq. (4) while the values of C and U are updated, and clustering is carried out with the optimal values. The cluster center value c is equal to Eq. (5) and m is the weight. When the center point c does not change, the operation ends:
The fast and robust FCM, or FRFCM, algorithm used in this paper was created by linking spatial information with the FCM algorithm in order to reduce the noise in the existing FCM algorithm. The FRFCM algorithm is robust to noise from morphology reconstruction, and it is effective for fast and efficient noise image segmentation using local membership filtering. It preserves image detail through morphological reconstruction before clustering and it does not calculate the distance between the local spatial constraint and the cluster center point. It also uses membership filtering, which only depends on the local spatial constraint. The proposed FRFCM algorithm is much simpler and faster because it does not calculate between the local spatial constraint and the cluster center point. That is, it divides the
image based on the clustering method that minimizes the objective function [12]. Since the FRFCM algorithm suppresses noise, we believe that clustering the gastric endoscopy images will improve the segmentation of the lesion area. The resulting segmentation is shown in Fig. 8. The image is segmented into clusters that are different to those generated by the SLIC superpixel algorithm. Figures 7 and 8 show that SLIC superpixel partitions of a uniform size can be identified by finding the cluster center point, which can be achieved by limiting the clustering space. On the other hand, the FRFCM algorithm, which relies on local spatial constraints, does not have space limitations. Thus, the splitting results are different to those generated using the SLIC superpixel algorithm.
2.5 Training Process
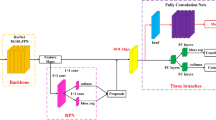
In this study, we used convolutional neural networks, one of the deep learning models, to classify endoscopy images. Their performance improves as they become deeper. However, as the network becomes deeper it also becomes more complicated and various problems such as overfitting or gradient vanishing can occur. To combat this, we used GoogLeNet, a convolutional neural network that is a network of Inception modules. As shown in Fig. 9, the Inception modules consist of multiple 1 × 1 convolution layers, 3 × 3 and 5 × 5 convolution layers, and 3 × 3 max pooling layers.
These modules play an important role in efficiently extracting image features. The Inception module limits the problems described above by simplifying the network. As the network becomes deeper, complex computations can be effectively reduced through the 1 × 1 convolution layer and other layers of the Inception module. The module maintains performance while reducing the amount of computation required. The Inception-v3 model was selected as this model improves performance by adding batch normalization to version v2 [10]. Figure 10 shows a flowchart of the training process. The SLIC superpixel and FRFCM algorithms were applied to the pre-processing step. The segmented images were then inputted in the deep learning model based on the ground truth set by the internist. Only the abnormal areas are used for training, hence not all of the segmented areas are used.
2.6 Test Algorithm
The trained model was used to test the data from the test set. The test process is shown by the flowchart in Fig. 11. The test data were segmented using the SLIC superpixel and FRFCM algorithms. The segmented images were then inputted in the classification model where they were classified depending on whether the area was abnormal or normal. The classified results for the segmented areas were then used in the Abnormal Score. At this stage, if more than one-third of the segmented areas in a given image were classified as abnormal, then the image was classified as abnormal. The size of gastric lesions can vary significantly. The lesions can be a lesion that includes all of the segmented regions, and some that contain half of segmented regions. There are also lesions that contain one region. Therefore, classification was conducted using Eq. (6). The classification threshold value was set through experimentation:
3 Results and Discussion
Lesions of various sizes are discovered in endoscopy images. Therefore, a model without segmentation is not enough to classify abnormal images. Thus, we proposed using the SLIC superpixel and FRFCM algorithms to create a CADx system for gastric lesion diagnosis, as shown in Fig. 4. In the models with segmentation, the image is segmented using an algorithm, and training is conducted based on the internist’s ground truth for the segmented area. A comparison of the performance of the two segmentation algorithms and its counterpart without segmentation is shown in Figs. 11 and 12. All the segmentation parameters were set to 9 before segmentation was conducted. As shown in Fig. 12, the area under the curve (AUC) for the receiver operating characteristic (ROC) curve was 0.87 for the FRFCM algorithm, 0.85 for the SLIC superpixel algorithm, and 0.82 without segmentation. As shown by the ROC curves, the FRFCM algorithm performed the best. The results show that the models trained using segmentation based on the internist’s ground truth performed better than the model trained without segmentation. We note that the FRFCM algorithm, which carried out segmentation after clustering without any restrictions on area, performed better than the SLIC superpixel algorithm, which segmented the clusters after limiting the area.
Thus, we can see that abnormal images with lesions of various sizes are better categorized in models using segmentation algorithms than in the model without it. However, the two segmentation algorithms take slightly different features and proceed with the segmentation. As mentioned in Sect. 2.3, the SLIC superpixel is segmented into uniform sizes as clustering is conducted in a limited area (2S × 2S). In addition to the color space, when clustering segmentation is in progress, the values of the position coordinates affect the clustering. Thus, the partition is more likely to contain areas other than the lesions. By contrast, the FRFCM algorithm is not clustered in a limited area, and hence, clustering times are longer than those in the SLIC superpixel algorithm but are more likely to be classified as abnormal for lesions of varying sizes. Because gastric lesions of the same type have similar characteristics, lesions such as gastric cancer or gastritis spreading in a wide area, and early lesions spreading in a narrow area, are most likely to be included in the segmented region. For this reason, in order to classify abnormal images for various types of lesions, if one-third of the segmented regions in the Abnormal Score step are abnormal, then the images are classified as abnormal images.
4 Conclusion
We designed a deep learning model CADx system for gastric lesions using gastric endoscopy images. In order to provide better training for gastric lesions than previous studies, we applied a segmentation algorithm before the training data were input. The segmentation algorithms used were the simple linear iterative clustering (SLIC) superpixel algorithm and the fast and robust fuzzy C-means (FRFCM) algorithm. Comparisons were made with the model that did not use segmentation. The SLIC superpixel and FRFCM algorithms produced better results than previous models.
The SLIC superpixel is clustered in a limited area under the influence of the position coordinate value and is divided into a grid of a similar size. Therefore, there is a high probability of including areas other than lesions. On the other hand, the FRFCM algorithm does not conduct clustering in restricted areas and does not split them into similar shapes. In addition, because of morphological reconstruction, noise is suppressed, and clustering is performed so that the divided areas have similar color characteristics. The AUC values of the ROC curves for the model without segmentation, the SLIC superpixel algorithm, and the FRFCM algorithm were 0.82, 0.85, and 0.87, respectively. This value was 2% higher for the FRFCM algorithm than the SLIC superpixel algorithm. When we analyzed the classification results, we found that they were better classified by the segmentation models than by that without segmentation. This shows that models allow segmentation of gastric endoscopy images are superior to those that do not. Segmented images that are confirmed as either normal or abnormal by an internist facilitate focusing on the characteristics of the lesions to a greater extent in the training process. In addition, the FRFCM algorithm suppresses noise and unrestricted area clustering in a manner different from that of the SLIC superpixel algorithm, which results in better performance. In future studies, we will attempt to improve the computer-aided diagnosis system for a more accurate diagnosis of gastric lesions by factoring in different types of lesions. In order to increase the amount of data, we plan to collect additional data, apply simple image augmentation techniques (i.e., rotation), and use the Generative Adversarial Network (GAN) algorithm to augment the data in future studies.
References
Ministry of Health and Welfare, Korea National Cancer Center (2019) National cancer registration statistics 2016. [Online]. https://cancer.go.kr/lay1/S1T639C641/contents.do. Accessed 30 Apr 2019
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2019) Global cancer statistics 2018: GLOBOCAN Estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018. [Online]. https://www.wcrf.org/dietandcancer/cancer-trends/stomach-cancer-statistics. Accessed 30 Apr 2019
Choi IJ (2018) Helicobacter pylori eradication therapy and gastric cancer prevention. Korean J Gastroenterol 72:245–251
Lee TC, Lin YH, Uedo N, Wang HP, Chang HT, Hung CW (2013) Computer-aided diagnosis in endoscopy: a novel application toward automatic detection of abnormal lesions on magnifying narrow-band imaging endoscopy in the stomach. In: Engineering in medicine and biology society (EMBC), 2013 35th annual international conference of the IEEE, pp 4430–4433, 2013
Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T et al (2017) Application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine 25:106–111
Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T et al (2017) Computer-aided diagnosis based on convolutional neural network system for colorectal polyp classification: preliminary experience. Oncology 93:30–34
Pogorelov K, Randel KR, Griwodz C, Eskeland SL, de Lange T, Johansen D et al (2017) Kvasir: a multi-class image dataset for computer aided gastrointestinal disease detection. In: Proceedings of the 8th ACM on multimedia systems conference, 2017, pp 164–169
Yuan Y, Meng MQH (2017) Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys 44:1379–1389
Cong Y, Wang S, Liu J, Cao J, Yang Y, Luo J (2015) Deep sparse feature selection for computer aided endoscopy diagnosis. Pattern Recogn 48:907–917
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the Inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2818–2826, 2016
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34:2274–2282
Lei T, Jia X, Zhang Y, He L, Meng H, Nandi AK (2018) Significantly fast and robust fuzzy c-means clustering algorithm based on morphological reconstruction and membership filtering. IEEE Trans Fuzzy Syst 26:3027–3041
Kim DH, Cho CH (2018) Deep learning based computer-aided diagnosis system for gastric lesion using endoscope. Trans Korean Inst Electr Eng 67:928–933
Kim YL, Kim WH, Kim TJ, Choi KW (1992) Histotopographic characterization of chronic gastritis associated with early gastric carcinomas. Korean J Gastroenterol 24:216–223
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2017R1E1A1A03070297). This research was supported by the MSIT (Ministry of Science and ICT), Korea, under the ITRC (Information Technology Research Center) support program (IITP-2019-2018-0-01433) supervised by the IITP (Institute for Information & communications Technology Promotion).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, Dh., Cho, H. & Cho, Hc. Gastric Lesion Classification Using Deep Learning Based on Fast and Robust Fuzzy C-Means and Simple Linear Iterative Clustering Superpixel Algorithms. J. Electr. Eng. Technol. 14, 2549–2556 (2019). https://doi.org/10.1007/s42835-019-00259-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42835-019-00259-x