Abstract
Overuse of chemical fertilizers and pesticides has led to severe ecological and food safety problems. The application of seaweed fertilizer meets the needs of green and sustainable agriculture. In the present study, we aimed to evaluate whether seaweed fertilizer was suitable for long-term application as a green fertilizer. Seaweed fertilizer derived from brown algae residues was applied to the okra planting field for 2 years, and the soil’s physical and chemical properties, enzyme activity indicators, and bacterial flora structure were tested. We found that seaweed fertilizer could effectively improve the soil pH, water content, organic carbon, alkali nitrogen, and the activities of various enzymes, such as catalase, sucrase, and cellulase. In addition, compared with the short-term use of seaweed fertilizer previously studied, our study showed that long-term fertilization could more significantly improve soil bacterial diversity. The abundance of some bacteria involved in polysaccharide degradation and nitrogen fixation was significantly increased, such as Acidobacteria and Cyanobacteria at the phylum level, and Methylobacterium, norank Acidobacteriales, and Micromonospora at the genus level. This study showed that the long-term application of seaweed fertilizer was one of the effective ways to develop sustainable green agriculture.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Under global climate change, crops face various challenges, such as diseases, insect pests, drought, and high- or low-temperature stress. In traditional agriculture, chemical pesticides and fertilizers play a vital role in ensuring crop yield, with the characteristics of fast efficiency and low cost. However, people have gradually realized that the overuse of chemical fertilizers and pesticides has caused serious environmental, ecological, and food safety problems, such as soil structure damage, fertility decline, soil microbial diversity reduction, groundwater pollution, greenhouse gas emissions, increasing resistance of pathogens, and the increase of pathogenic varieties (Kahrl et al. 2010; Kaur et al. 2008; Nakasone et al. 2000; Sharma and Singhvi 2017). Given the ongoing progress in pollution-free and environmentally sustainable agriculture, there is an urgent demand for organic and biological agricultural fertilizers.
Seaweed fertilizer is produced and processed with marine algae as raw materials, in which a certain amount of nitrogen (N), phosphorus (P), potassium (K), and medium and trace elements are added. Its active ingredient is processed into small-molecule compounds, which can be quickly absorbed and utilized by crops in a few hours. Due to its outstanding environmental protective effect and high efficiency, seaweed is widely used as fertilizer and soil conditioner in land-based farming (Hong et al. 2007). Seaweed and its extract are rich in seaweed polysaccharides, plant hormones, alginic acid, and other plant growth-promoting factors, and their beneficial effects on plants and soil have been fully proved. They can not only promote plant growth (Kumari et al. 2011; Sridhar and Rengasamy 2010), reduce the damage caused by adversity to plants (Ibrahim et al. 2014), but also improve the structure of soil and microbial communities, facilitating the self-healing function of soil in response to pollutants (Sudharshan et al. 2013).
Soil microbial communities play a crucial role in soil chemical and physical properties and are a significant driver of soil health and quality (Six et al. 2004). The biological process of the soil microbial community is key to maintaining the function of the soil ecosystem (Xue et al. 2013). Studies have shown that soil microorganisms are closely related to plants (Esitken et al. 2010; Franche et al. 2009; Madsen 2005). The nutrients and energy produced in the microbial cycle can maintain the nutritional needs of other residents of the biosphere (Franche et al. 2009). Some bacteria can provide nutrients and growth-promoting substances for plants (Esitken et al. 2010). In addition, the resistance and resilience of soil microbial communities directly affect the anti-interference ability of soil, and soil health and ecosystem functional diversity are closely related to plant diversity (Griffiths and Philippot 2013; Wagg et al. 2014). Therefore, it is necessary to study the effects of seaweed fertilizer application on the composition and diversity of soil microbial communities. At present, some scholars have assessed the effect of the short-term application of seaweed fertilizer on the soil microbial community and found that seaweed fertilizer can improve the diversity of the bacterial community (Chen et al. 2020; Wang et al. 2018). However, the effect of the long-term application of seaweed fertilizer on soil remains largely unexplored.
In order to assess the viability of seaweed fertilizer as a continuous and effective biological fertilizer in agricultural production, it is crucial to conduct further research on the long-term effects of seaweed fertilizer on soil microbial communities. This investigation would provide valuable insights into the potential benefits or drawbacks associated with the prolonged utilization of seaweed fertilizer and help determine its sustainability in agricultural practices. In a previous study, we find that seaweed fertilizer can improve the yield and quality of okra Abelmoschus moschatus (Li et al. 2019). In the present study, seaweed fertilizer was applied to the experimental field of okra for 2 consecutive years, and the changes in soil physicochemical properties and enzyme activity (PPEA), such as urease, catalase, sucrase, and cellulase activity, were detected. In addition, the soil bacterial diversity and community composition were analyzed by 16S rRNA sequencing. We aimed to (1) investigate the impact of the prolonged application of seaweed fertilizer on soil bacterial diversity and community composition, (2) explore the correlation between bacterial community composition and soil physical and chemical properties following the long-term use of seaweed fertilizer, and (3) assess the potential improvements in soil structure and bacterial composition resulting from long-term seaweed fertilization and to determine whether these effects surpass those observed with short-term fertilization.
2 Materials and Methods
2.1 Sampling Design and Treatments
The study area was located at Fan’s farm, Fenghua District, Ningbo City, Zhejiang Province, China (29°41′34″N, 121°24′17″E), which is an open-air farm. The soil type was cohesive soil, and the climate profile is shown in Table 1. There were three treatments in the experiment, namely, the control treatment without algal fertilizer (FH_CK), treatment with algal fertilizer for 1 year (FH_1), and treatment with algal fertilizer for 2 years (FH_2). The experimental year ranged from 2017 to 2019, and the FH_CK, FH_1, and FH_2 specimens were collected in 2017, 2018, and 2019, respectively. Okra was harvested from July to October every year, and the yield was counted. After 3 months of abandoned cultivation, soil samples were collected in December every year, and then seaweed fertilizer was applied.
In December of each year, the compound fertilizer was applied to furrow at the dosage of 15 kg·667 m−2. In 2017 and 2018, seaweed fertilizer was added to the treatment groups (FH_1 and FH_2) at 300 kg·667 m−2. Compound fertilizer was a high-concentration potassium sulfate produced by Guangzhou Kaimirui Chemical Fertilizer Co., Ltd., China. Seaweed fertilizer was processed from brown algae residues and provided by Shandong Jiejing Group Co., Ltd., China. Seaweed fertilizer was made of wild Kelp and Ascophyllum as the major materials and fermented after adding probiotics. The characteristics of seaweed fertilizer were pH 7–8, beneficial bacteria of 2 × 108 colony-forming units (CFU)·g−1, seaweed protein ≥ 20%, seaweed polysaccharide ≥ 3%, alginic acid ≥ 2%, auxin ≥ 0.08%, mannitol ≥ 0.03%, sodium alginate ≥ 0.05%, and phenolic polymer ≥ 0.02%. Each treatment had three plots with an area of 223 m2, and about 600 okra plants were planted. The interval between treatment groups, plots, and plants was 1.2 m, 0.5 m, and 0.3 m, respectively. The field management was the same as the routine.
2.2 Sample Collection
After 3 months of abandoned cultivation, the samples were collected according to the “S”-shaped line. Random multi-point location sampling was conducted in each test field area, and the sampling depth was the surface soil (0 ~ 20 cm).
Briefly, 500 g of well-mixed soil was taken, impurities were removed, and the specimens were placed into a sealed sample bag and transported to the laboratory in an ice box. Six parallels were set for each sampling. Each sample was divided into three parts: (1) dried through a 2-mm sieve for physicochemical property testing; (2) stored at 4 °C, and the soil enzyme activity was measured within 7 days; and (3) stored at − 80 °C for high-throughput sequencing of soil microbes.
2.3 Soil Physical and Chemical Analysis
The pH value was determined by the potentiometer method, and the water content was measured by the weighing method. In addition, the contents of soil organic carbon (OC), total nitrogen (TN), alkali nitrogen (AN), total phosphorus (TP), available phosphorus (AP), total potassium (TK), and available potassium (AK) were determined according to Bao 2000.
2.4 Detection and Analysis of Soil Enzyme Activity
Briefly, 1 g of fresh soil was mixed with 9 mL of PBS (pH 7.2 ~ 7.4, 0.01 moL·L−1). The sample was homogenized (Bertin Precellys 24, Bertin Technologies Co., Ltd., France) in an ice bath. Subsequently, the sample was centrifuged at 3000 r·min−1 for 15 min, and the supernatant was collected for inspection. The contents of soil sucrase, catalase, urease, and cellulase were detected by respective ELISA kits (Jiangsu Meibiao Biotechnology Co., Ltd., China). The kit utilized an enzyme-linked immunosorbent assay (ELISA) and comprised microwells coated with specific antibodies for the corresponding enzyme. Horseradish peroxidase (HRP)-labeled detection antibodies were subsequently added to the coated microwells, followed by an incubation period and thorough washing. Color development was achieved using the substrate trimethylbenzene (TMB), which was catalyzed by peroxidase and converted to a blue color, ultimately turning yellow when exposed to acid. The intensity of the color was directly proportional to the enzyme activity present in the sample. The absorbance (OD value) of the sample was measured at a wavelength of 450 nm using a microplate reader (Thermo Fisher Varioskan Flash, Thermo Fisher Scientific Co., Ltd., USA).
2.5 Amplification and Sequencing of Soil DNA
The total DNA was extracted by the CTAB method (Doyle and Doyle 1987). First, the DNA concentration was diluted to 1 ng·μL−1 with sterile water, followed by PCR amplification. The primers for V3-V4 amplification of 16S rRNA were as follows: 338F: 5′-ACTCCTACGGGAGGCAGCAG-3′ and 806R: 5′-GGACTACHVGGGTWTCTAAT-3′ (Du et al. 2021). All PCR reactions were conducted in a 30-μL reaction system consisting of 15 μL of Phusion High-Fidelity PCR Master Mix (New England Biolabs Co., Ltd., USA), 0.2 μM of forward and reverse primers, and about 10 ng template DNA. Briefly, after an initial denaturation step at 95 °C for 3 min, the amplifications were carried out with 25 cycles at a melting temperature of 95 °C for 30 s, an annealing temperature of 55 °C for 30 s, and an extension temperature of 72 °C for 30 s, followed by preservation at 16 °C for 2 min.
Equal volumes of 1X loading buffer (containing SYB green) and PCR products were mixed and subjected to electrophoresis on 2% agarose gels. Samples of 460 bp in size (V3-V4) were chosen for further experiments. PCR products were purified with Thermo Scientific™ GeneJET Gel Extraction Kit (Thermo Fisher Scientific Co., Ltd., USA). Sequencing libraries were generated using the NEB Next Ultra DNA Library Prep Kit for Illumina (New England Biolabs Co., Ltd., USA) following the manufacturer’s recommendations, and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Life Technologies, CA, USA) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina MiSeq platform, and 250-bp/300-bp paired-end reads were generated.
2.6 Data Analysis
The sequencing data were spliced using Flash v1.2.11 for pair-end double-ended sequences, and Qiime v1.9.1 was used to generate taxonomic level abundance tables and β diversity distance calculation. Uparse v7.1 was adopted to cluster the sequences with the operational taxonomic unit (OTU) at a similarity level of 97%. The classification analysis of OTU representative sequences was carried out using Ribosomal Database Project (RDP) Classifier v2.11, with a confidence threshold of 0.7. SILVA (release138) database was used for species annotation. Origin 2018 was adopted to make charts of α diversity. Majorbio cloud platform (Majorbio Co., Ltd., China) was employed to make relevant charts of β diversity data. The correlation between PPEA and bacteria was calculated using the “psych” package of the R language, and the co-relation network was made using Cytoscape v3.5.1. Single-factor analysis of variance (ANOVA) of SPSS (20.0) software was used to compare the significant differences between samples in α diversity and PPEA indicators. The statistical method was Duncan (d) and Dunnett’s T3 (3) test, and P < 0.05 was considered statistically significant.
3 Results
3.1 Okra Yield, Soil Physicochemical Characteristics, and Enzyme Activity (PPEA)
Table 2 shows that the yield of okra was increased year by year after seaweed fertilizer was applied, indicating that seaweed fertilizer had a specific promoting effect on crop yield. Seaweed fertilizer treatment had a significant impact on most PPEA indicators of soil. The soil pH, water content, catalase activity (SOD), and sucrase activity (sucrase) were significantly increased with the extension of the treatment time (P < 0.05). In addition, the contents of OC, AN, and cellulase activity (cellulase) were significantly increased after 1 year of treatment (P < 0.05), which tended to be stable in the second year (P > 0.05). Besides, seaweed fertilizer treatment had no significant effect on soil phosphorus and potassium contents.
3.2 Analysis of Soil Microbial Flora Characteristics
After 16S rRNA amplification and sequencing of soil bacteria, reads of 1,048,902 bp were obtained, with an average of 58,272 ± 7,037-bp reads per sample. The average length of reads was about 436 bp. Supplementary Fig. S1 illustrates the Coverage and Shannon rarefaction curves, indicating adequate sequencing depth. Figure 1 shows that the application of seaweed fertilizer could significantly improve the soil microbial flora in the α diversity. Shannon (Fig. 1a), ACE (Fig. 1c), and Chao (Fig. 1d) indexes were significantly increased with the years of fertilization (P < 0.05). Simpson index (Fig. 1b) was decreased slightly, although there was no significant difference (P > 0.05).
Effects of seaweed fertilizer treatment in different years on soil bacterial diversity. a Shannon diversity, b Simpson diversity, c Ace diversity, d Chao diversity. “FH_CK” represents the soil sample without algal fertilizer. “FH_1” and “FH_2” represent the soil samples applied with seaweed fertilizer for 1 and 2 years, respectively
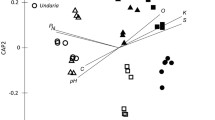
According to principal coordinate analysis (PCoA) and analysis of similarities (ANOSIM) (Fig. 2b, c), the application of seaweed fertilizer had a significant impact on the structure of soil microbial flora. The R-value of ANOSIM was 0.7142, and the P value was 0.001. Moreover, the results of PCoA showed that the structure of soil flora exhibited a progressive trend with the increase in fertilization time. The distance between FH_ CK and FH_2 was greater than that between FH_ CK and FH_ 1.
a Systematic clustering diagram of alkali nitrogen (AN), sucrase, and cellulase indicators. After data homogenization, systematic clustering was adopted. The clustering method was an inter-group connection, and the measurement standard was square Euclidean distance. b Principal coordinate analysis (PCoA) of bacterial community structure at the operational taxonomic unit (OTU) level based on Bray–Curtis. c Analysis of similarities (ANOSIM) at the OTU level based on Bray–Curtis. “Between” represents the distance between all groups. d Redundancy analysis (RDA) based on the OTU level. See Table 2 for the full name of PPEA indicators. For example, “FH_CK” represents the soil sample without algal fertilizer. “FH_1” and “FH_2” represent the soil samples applied with seaweed fertilizer for 1 and 2 years, respectively
3.3 Correlation Analysis Between PPEA and Bacterial Community Structure
All soil PPEA factors were screened through variance inflation factor (VIF), and the factors with small collinearity (IF value < 10) were selected. Redundancy analysis (RDA) of OTU level was carried out with the above factors. The results showed that most indicators were positively correlated, among which AN, sucrase, cellulase, and urease had the highest correlation with sample distribution (Fig. 2d). A mental test was used to further screen the above four factors (two-sided, 999 displacements) most significantly related to the difference in flora structure, that is mantel R statistic > 0.6, P value < 0.05. Three indicators of AN, sucrase, and cellulase were screened (Table 3). Besides, the combined effect of these three indicators was also significantly correlated with the structure of microbial flora (mantel R statistic = 0.873, P value = 0.001). After clustering AN, sucrase, and cellulase, we found that sample FH_CK was quite different from the other two samples, and the clustering of the three factors had a good projection relationship with the PCoA chart (Fig. 2a, b). The above results indicated that seaweed fertilizer significantly changed β diversity and soil PPEA indicators of the soil bacterial community, and three indicators, such as AN, sucrase, and cellulose, had a significant correlation with the structure of soil flora.
In order to further analyze the relationship between the three indicators and soil bacterial diversity, we conducted a linear regression analysis of AN (Fig. 3a), sucrase (Fig. 3b), cellulase (Fig. 3c), and Shannon index of bacterial communities. The results indicated that these three indicators had a significant linear relationship with soil bacterial diversity (R2 > 0.5, P < 0.01).
3.4 Analysis of Bacterial Community Composition at the Phylum and Genus Levels
Figure 4a illustrates the dominant phylum of bacteria in different samples. Proteobacteria, which occupied the largest niche, accounted for 29.18–31.09% in different samples. Other bacteria with high abundance were Actinobacteria (15.43–22.68%), Chloroflex (13.82–14.55%), and Acidobacteria (11.25–16.62%). After application of seaweed fertilizer for 1 year, the abundance of Actinobacteria (CK: 22.68%; FH_1: 15.43%) and Firmicutes (CK: 5.40%; FH_1: 3.77%) was significantly decreased (corrected P value < 0.05). The abundance of Acidobacteria (CK: 11.25%; FH_1: 16.62%), Cyanobacteria (CK: 0.42%; FH_1: 1.76%; FH_2: 1.76%), Rokubacteria (CK: 0.5766%; FH_1: 1.00%), Dadabacteria (CK: 0.33%; FH_1: 0.43%), and Latescibacteria (CK: 0.19%; FH_1: 0.44%; FH_2: 0.39%) was increased significantly after fertilization (corrected P value < 0.05). Notably, the abundance of Cyanobacteria was increased by more than four folds.
a Bacterial components and significant differences analysis at the phylum level in different samples. Single asterisk indicates that there is a significant difference between this sample and the FH_CK sample (corrected P value < 0.05). Wilcoxon rank-sum test was used to calculate the significant difference between groups, and the P value was corrected by Benjamin Hochberg (BH) method. “FH_CK” represents the soil sample without algal fertilizer. “FH_1” and “FH_2” represent the soil samples applied with seaweed fertilizer for 1 and 2 years, respectively. b Correlation analysis between bacteria (at the phylum level) and soil physicochemical properties and enzyme activity (PPEA) indicators using Spearman correlation coefficient. Single asterisk indicates significant correlation (|R| value > 0.6, P value < 0.05)
The results of correlation analysis between dominant bacteria and soil PPEA indicators showed that Cyanobacteria and Latescibacteria were significantly positively correlated with AN, sucrase, and cellulase (Fig. 4b, P < 0.05). Acidobacteria was significantly positively correlated with cellulase (P < 0.05). Actinobacteria was significantly negatively correlated with sucrase and cellulase (P < 0.05).
Figure 5 exhibits that norank Xanthobacteriae was the genus with the highest abundance (with an average abundance ranging from 7.14 to 8.80% in different groups), followed by norank Acidobacteriales under Acidobacteria and Mycobacterium and norank Gaillales under Acidobacteria. The abundance of Mycobacterium was significantly decreased after fertilization from 4.90% in the CK group to 2.99% in the FH_1 group and 2.71% in the FH_ 2 group (corrected P value < 0.05). The clustering results of samples were similar to those of PCoA. The composition of soil flora after fertilization was significantly different from that of unfertilized samples.
Bacterial components and significant difference analysis at the genus level in different samples. Single asterisk indicates that there is a significant difference between this sample and the FH_CK sample (corrected P value < 0.05). Welch’s T-test was used to calculate the significant difference between groups, and the P value was corrected by Benjamin Hochberg (BH) method. “FH_CK” represents the soil sample without algal fertilizer. “FH_1” and “FH_2” represent the soil samples applied with seaweed fertilizer for 1 and 2 years, respectively
Figure 6a shows that many bacterial genera were significantly positively correlated with sucrase, including Methylobacterium, unclassified Acidobacteriales, and Dactylosporangium. Rubellimicrobium and other genera were significantly positively correlated with AN, and Micromonospora and other genera were positively correlated with sucrase. The abundance of the above genera was increased after fertilization, and the abundance of unclassified Acidobacteriales and Rubellimicrobium was significantly increased (Fig. 6b, corrected P value < 0.05). Mycobacterium, Methylocystis, and other bacteria were negatively correlated with AN, while Defluvicoccus was negatively correlated with sucrase. Norank Burkholderiaceae and norank Gaiellales were significantly negatively correlated with cellulase and sucrase, and the abundance of norank Burkholderiaceae was significantly decreased in the 2nd year of fertilization (Fig. 6b, corrected P value < 0.05).
a Correlation network between soil physicochemical properties and enzyme activity (PPEA) indicators and bacteria (at the genus level). The solid line represents a significant positive correlation, and the dotted line represents a significant negative correlation (|R| value > 0.8, corrected P value < 0.05). The spearman correlation coefficient was used to calculate the correlation between PPEA and bacteria, and the Benjamin Hochberg (BH) method was used to correct the P value. The genera with |R| value > 0.8 and corrected P value < 0.05 were screened out and use Cytoscape to make a network map. b Heat map of the relative abundances of genera in Fig. 6a. Single asterisk indicates that there is a significant difference between this group and the FH_CK group. Welch’s t-test was used to calculate significant differences. P value was corrected by Benjamin Hochberg (BH) method
4 Discussion
Many studies have demonstrated that the application of seaweed fertilizer or seaweed extract can improve soil properties and nutrients (Hamouda et al. 2022; Kholssi et al. 2022). Acid rain caused by atmospheric pollution and disorderly application of inorganic fertilizers accelerate the process of soil acidification (Liu et al. 2021; Bai et al. 2020). Seaweed fertilizer is usually neutral or alkaline. Our research showed that long-term application of seaweed fertilizer could significantly increase soil pH and regulate acidic soil to a certain extent. Furthermore, seaweed fertilizer significantly improved the water content of the soil. It might be attributed to the fact that seaweed contains sugars, such as seaweed gum and seaweed polysaccharide, which can chelate metal ions to form polymers (Ghosh et al. 2021). These polymers can further absorb water and expand to maintain soil water content and improve the physical structure of soil.
Studies have found that some growth regulators in seaweed fertilizers can improve soil microecology and enhance fertilizer utilization (Zhang and Thomsen 2019; Zhu et al. 2019). We also found that the application of seaweed fertilizer could significantly improve some nutrients in the soil, such as organic nitrogen and OC. Furthermore, in the current study, the prolonged utilization of seaweed fertilizer demonstrated a substantial enhancement in the activities of various soil enzymes, including sucrase, cellulase, and SOD. Sucrase plays a crucial role in the soil’s carbon cycle by increasing soluble nutrients (He et al. 2019). It is closely associated with soil organic matter content, nitrogen and phosphorus levels, microbial population, and soil respiration intensity. Sucrase serves as an indicator for assessing soil maturity and fertility levels. Ge et al. (2009) have proposed that soil cellulase is a key determinant of soil fertility. Considering the outcomes of the PPEA analysis, we could infer that the long-term application of seaweed fertilizer significantly improved soil fertility, which could be a contributing factor to the increased production of okra.
As one of the most abundant and diverse communities on earth, soil bacterial communities are responsible for essential ecosystem processes, including decomposition, nutrient cycling, and detoxification. Our study revealed that the extended use of seaweed fertilizer had a notable and enduring influence on the composition of the soil bacterial community. This finding suggested that the altered bacterial community structure could be a significant internal driving force behind the enhancement of soil fertility.
For instance, we observed a substantial increase in the abundance of Acidobacteria following the application of seaweed fertilizer. Acidobacteria is a highly diverse bacterial phylum found worldwide, particularly thriving in acidic soils. Many genera within this phylum possess the capability to encode a range of carbohydrate-active enzymes, which are involved in the breakdown, utilization, and synthesis of various carbohydrates (Dedysh and Sinninghe Damsté, 2018). Moreover, the abundance of Cyanobacteria also exhibited a significant increase after fertilization, exhibiting a strong positive correlation with alkali nitrogen (AN) levels. Cyanobacteria are known as primary contributors to biological nitrogen fixation in both aquatic and terrestrial ecosystems, acting as key nitrogen-fixing microorganisms in the topsoil (Fujita and Uesaka 2022).
Furthermore, by analyzing the correlation network between PPEA and bacteria at the genus level, we discovered numerous genera that exhibited functions related to polysaccharide degradation and nitrogen fixation, and their abundance was increased following the application of seaweed fertilizer. For instance, Methylobacterium showed a positive relationship with sucrase and AN, and its nitrogen fixation capability can stimulate plant biomass and seed production (Madhaiyan et al. 2015). It possesses both methylotrophic and methanotrophic properties while also being able to utilize glucose as a carbon source (Madhaiyan et al. 2015). Acidobacteriales have the capacity to utilize various sugars and polysaccharides due to their hydrolytic capabilities (Dedysh and Sinninghe Damsté, 2018). Micromonospora plays a crucial role in biological control, plant growth promotion, maintenance of the root ecosystem, and decomposition of plant cell wall materials (Hirsch and Valdés, 2010). Additionally, hydrolases enable Micromonospora to actively participate in the degradation of organic matter in its natural habitat (Hirsch and Valdés, 2010). The application of seaweed fertilizer can partially enhance the abundance of beneficial bacteria involved in polysaccharide degradation and nitrogen fixation in the soil. This could be attributed to the rich protein, amino acids, and polysaccharide content of seaweed, such as sodium alginate, brown algal polysaccharide, and kelp protein (Mahmoud et al. 2019).
It is well-known that soil microorganisms are the primary source of most soil enzymes, although plant roots and soil animals also contribute to enzyme secretion in the soil. In our present study, we observed significant correlations between soil enzymatic activities (such as AN, cellulase, and sucrase) and bacterial diversity, as well as the composition and structure of the bacterial community. Sucrase and cellulase enzymatic products, namely glucose, serve as nutrients for plants, and the increase in nitrogen-fixing bacteria could enhance soil fertility. Based on these findings, we hypothesized that the long-term application of seaweed fertilizer could enhance the diversity of soil flora and enrich the presence of bacteria with functions related to polysaccharide degradation and nitrogen fixation. Consequently, this process could improve the content of organic matter, including carbon and nitrogen elements, in the soil, ultimately enhancing soil fertility.
In line with the findings regarding between-habitat diversity (β diversity), our study also demonstrated a significant improvement in within-habitat diversity (α diversity) of soil bacterial communities due to long-term fertilization. Previous studies have found that the change in soil microbial community diversity under long-term fertilization management is the critical factor driving crop yield and soil function (Fan et al. 2021). Therefore, it is of great practical significance to protect soil biodiversity, especially the diversity of vital microbial groups. Many studies have shown that long-term nitrogen application or enrichment can reduce bacterial and plant diversity (Song et al. 2023). Nitrogen accumulation may result in the decline of biodiversity by stimulating the expansion of nitrophilic species and the competitive exclusion of other species (Bobbink et al. 2010). In our current work, the long-term application of seaweed fertilizer significantly increased the content of soil available nitrogen. In contrast, the diversity of the soil bacterial community was increased year by year, avoiding the adverse consequences caused by the application of nitrogen fertilizer alone. Besides, compared with the short-term application of seaweed fertilizer within several tens of days, the increase of soil bacterial community diversity after continuous application of seaweed fertilizer for 1 to 2 years was more pronounced (Chen et al. 2020; Wang et al. 2018).
In addition, long-term use of seaweed fertilizer seems to reduce the abundance of some potential pathogens. In the present study, the abundance of Actinobacteria was significantly decreased after fertilization. Mycobacterium is the most abundant genus under Actinobacteria, including many well-known human pathogens or opportunistic pathogens, and soil is one of their living environments (Larsen et al. 2021; Shen et al. 2021; Verma et al. 2022). The abundance of Burkholderiaceae was significantly reduced in the 2nd year of fertilization, which contained many environmental saprophytes, plant pathogens, opportunistic pathogens, and major pathogens for humans and animals (Coenye 2014).
5 Conclusions
Our research showed that long-term use of seaweed fertilizer could not only significantly improve the yield of crop okra and soil properties but also ameliorate soil bacterial diversity and community structure (Fig. 7). Through the long-term application of seaweed fertilizer, the alkaline or neutral properties of seaweed could effectively counteract soil acidification. The unique sugar components present in seaweed could form polymers that help retain soil moisture while also enhancing the presence of probiotics that possess polysaccharide-degrading enzymes, like sucrase and cellulase. Furthermore, there was a notable increase in the abundance of bacteria with nitrogen-fixing capabilities. Both of these probiotics could effectively enhance the organic matter in the soil containing carbon (C) and nitrogen (N), thereby providing essential nutrients for plant growth. Additionally, these probiotics, along with those added during the production of seaweed fertilizer, might be key factors contributing to the significant increase in within-habitat diversity (α diversity) of soil flora. Moreover, it appeared that the long-term application of seaweed fertilizer led to a decrease in the abundance of certain potentially pathogenic bacteria in the soil. Therefore, in production practice, the continuous use of seaweed fertilizer seemed to have no side effects and was conducive to plant development and soil health. Collectively, it was an effective way to develop sustainable green agriculture.
Mechanism assumption of soil improvement with algal fertilizer. The red upward arrow represents the increased composition of the soil after applying seaweed fertilizer, while the blue downward arrow represents the reduced substance. α diversity represents the within-habitat diversity of soil bacterial communities
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. The raw data of 16S rRNA sequencing are available in the NCBI Sequence Read Archive database (SRA) under the BioProject accession number PRJNA865061.
References
Bai Y, Chang YY, Hussain M, Lu B, Pei D (2020) Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 8:694. https://doi.org/10.3390/microorganisms8050694
Bao S (2000) Soil and agricultural chemistry analysis (the, 3rd edn. China Agricultural Press, Beijing
Bobbink R, Hicks K, Galloway J, Spranger T, Alkemade R, Ashmore M, Bustamante M, Cinderby S, Davidson E, Dentener F, Emmett B, Erisman J-W, Fenn M, Gilliam F, Nordin A, Pardo L, De Vries W (2010) Global assessment of nitrogen deposition effects on terrestrial plant diversity: a synthesis. Ecol Appl 20:30–59. https://doi.org/10.1890/08-1140.1
Chen Y, Li J, Huang Z, Su G, Li X, Sun Z, Qin Y (2020) Impact of short-term application of seaweed fertilizer on bacterial diversity and community structure, soil nitrogen contents, and plant growth in maize rhizosphere soil. Folia Microbiol 65:591–603. https://doi.org/10.1007/s12223-019-00766-4
Coenye T (2014) The Family Burkholderiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: Alphaproteobacteria and Betaproteobacteria. Springer, Berlin Heidelberg, Berlin, Heidelberg
Dedysh SN, Sinninghe Damsté JS (2018) Acidobacteria. In: eLS John Wiley & Sons, Ltd: Chichester. https://doi.org/10.1002/9780470015902.a0027685.
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15. https://doi.org/10.1371/journal.pone.0061185
Du J, Zhang P, Luo J, Shen L, Zhu L (2021) Dietary betaine prevents obesity through gut microbiota-drived microrna-378a family. Gut Microbes 13:1–19. https://doi.org/10.1080/19490976.2020.1862612
Esitken A, Yildiz HE, Ercisli S, FigenDonmez M, Turan M, Gunes A (2010) Effects of plant growth promoting bacteria (PGPB) on yield, growth and nutrient contents of organically grown strawberry. Sci Hortic 124:62–66. https://doi.org/10.1016/j.scienta.2009.12.012
Fan K, Delgado-Baquerizo M, Guo X, Wang D, Zhu Y-g, Chu H (2021) Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J 15:550–561. https://doi.org/10.1038/s41396-020-00796-8
Franche C, Lindström K, Elmerich C (2009) Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plantc Soil 321:35–59. https://doi.org/10.1007/s11104-008-9833-8
Fujita Y, Uesaka K (2022) Chapter 3 - Nitrogen fixation in Cyanobacteria. Cyanobacterial Physiology Academic Press, America: 29–45. https://doi.org/10.1016/B978-0-323-96106-6.00007-1.
Ge GF, Li ZJ, Zhang J, Wang LG, Xu MG, Zhang JB, Wang JK, Xie XL, Liang YC (2009) Geographical and climatic differences in long-term effect of organic and inorganic amendments on soil enzymatic activities and respiration in field experimental stations of China. Ecol Complex 6:421–431. https://doi.org/10.1016/j.ecocom.2009.02.001
Ghosh T, Singh R, Nesamma AA, Jutur PP (2021) Marine polysaccharides: properties and applications. Polysaccharides, Scrivener Publishing LLC, Canada: 37-60https://doi.org/10.1002/9781119711414.ch3
Griffiths BS, Philippot L (2013) Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev 37:112–129. https://doi.org/10.1111/j.1574-6976.2012.00343.x
Hamouda MM, Saad-Allah KM, Gad D (2022) Potential of seaweed extract on growth, physiological, cytological and biochemical parameters of wheat (Triticum aestivum L.) seedlings. J Soil Sc Plant Nut 22:1818–1831. https://doi.org/10.1007/s42729-022-00774-3
He J, Yu Z, Shi Y (2019) Effects of strip rotary tillage with subsoiling on soil enzyme activity, soil fertility, and wheat yield. Plant Soil Environ 65:1–7. https://doi.org/10.17221/240/2019-PSE
Hirsch AM, Valdés M (2010) Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol Biochem 42:536–542. https://doi.org/10.1016/j.soilbio.2009.11.023
Hong DD, Hien HM, Son PN (2007) Seaweeds from Vietnam used for functional food, medicine and biofertilizer. J Appl Phycol 19:817–826. https://doi.org/10.1007/s10811-007-9228-x
Ibrahim WM, Ali RM, Hemida KA, Sayed MA (2014) Role of Ulva lactuca extract in alleviation of salinity stress on wheat seedlings. Sci World J 2014:847290. https://doi.org/10.1155/2014/847290
Kahrl F, Li Y, Su Y, Tennigkeit T, Wilkes A, Xu J (2010) Greenhouse gas emissions from nitrogen fertilizer use in China. Environ Sci Policy 13:688–694. https://doi.org/10.1016/j.envsci.2010.07.006
Kaur T, Brar BS, Dhillon NS (2008) Soil organic matter dynamics as affected by long-term use of organic and inorganic fertilizers under maize–wheat cropping system. Nutr Cycl Agroecosys 81:59–69. https://doi.org/10.1007/s10705-007-9152-0
Kholssi R, Lougraimzi H, Grina F, Lorentz JF, Silva I, Castao-Sánchez OC, Marks EAN (2022) Green agriculture: a review of the application of micro- and macroalgae and their impact on crop production on soil quality. J Soil Sc Plant Nut 22(4):4627–4641. https://doi.org/10.1007/s42729-022-00944-3
Kumari R, Kaur I, Bhatnagar AK (2011) Effect of aqueous extract of Sargassum johnstonii Setchell & Gardner on growth, yield and quality of Lycopersicon esculentum Mill. J Appl Phycol 23:623–633. https://doi.org/10.1007/s10811-011-9651-x
Larsen SE, Reese VA, Pecor T, Berube BJ, Coler RN (2021) Subunit vaccine protects against a clinical isolate of Mycobacterium Avium in wild type and immunocompromised mouse models. Sci Rep 11:9040. https://doi.org/10.1038/s41598-021-88291-8
Li Q, Yang R, Chen H, Wu Y, Fan H, Chen J, Luo Q (2019) Effect of oligoagars on the growth and quality of Okar (Abelmoschus moschatus). J Nucl Agric Sci 33:1465–1471
Liu Z, Shan X, Wei H, Zhang J, Saleem M, Li D, Zhang Y, Ma R, He Y, Zhong J, Liu Y (2021) Idiosyncratic responses of microbial communities and carbon utilization to acid rain frequency in the agricultural and forest soil ecosystems. Glob Ecol Conserv: e01429. https://doi.org/10.1016/j.gecco.2020.e01429.
Madhaiyan M, Alex THH, Ngoh ST, Prithiviraj B, Ji L (2015) Leaf-residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas. Biotechnol Biofuels 8:222. https://doi.org/10.1186/s13068-015-0404-y
Madsen EL (2005) Identifying microorganisms responsible for ecologically significant biogeochemical processes. Na Rev Microbiol 3:439–446. https://doi.org/10.1038/nrmicro1151
Mahmoud SH, Salama DM, El-Tanahy AMM, Abd El-Samad EH (2019) Utilization of seaweed (Sargassum vulgare) extract to enhance growth, yield and nutritional quality of red radish plants. Ann Agric Sci 64:167–175. https://doi.org/10.1016/j.aoas.2019.11.002
Nakasone H, Yamasita I, Kuroda H, Kato T (2000) The effect of nitrogen fertilizer use at tea yards on water quality of an irrigation reservoir. J Jpn Soc Water Environ 23:374–377. https://doi.org/10.2965/jswe.23.374
Sharma N, Singhvi R (2017) Effects of chemical fertilizers and pesticides on human health and environment: a review. Int J Agric Environ Biotech 10:675–680. https://doi.org/10.5958/2230-732X.2017.00083.3
Shen K, Bai B, Liu Y, Lowary TL (2021) Synthesis of a tridecasaccharide lipooligosaccharide antigen from the opportunistic pathogen Mycobacterium kansasii. Angew Chem Int Ed Engl 60:24859–24863. https://doi.org/10.1002/anie.202111549
Six J, Bossuyt H, Degryze S, Denef K (2004) A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics. Soil Tille Res 79:7–31. https://doi.org/10.1016/j.still.2004.03.008
Song B, Li Y, Yang L, Shi H, Zhao Y (2023) Soil acidification under long-term n addition decreases the diversity of soil bacteria and fungi and changes their community composition in a semiarid grassland. Microb Ecol 85:221–231. https://doi.org/10.1007/s00248-021-01954-x
Sridhar S, Rengasamy R (2010) Significance of seaweed liquid fertilizers for minimizing chemical fertilizers and improving yield of Arachis hypogaea under field trial. Recent Res Sci Technol 2:73–80
Sudharshan S, Mallavarapu M, Bolan N, Naidu R (2013) Effect of seaweeds on degradation of DDT in soils. Water Air Soil Poll 224:1715. https://doi.org/10.1007/s11270-013-1715-x
Verma RN, Malik MZ, Singh GP, Subbarao N (2022) Identification of key proteins in host-pathogen interactions between Mycobacterium tuberculosis and Homo sapiens: a systematic network theoretical approach. Healthc Anal 2:100052. https://doi.org/10.1016/j.health.2022.100052
Wagg C, Bender SF, Widmer F, van der Heijden MGA (2014) Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci U S A 111:5266–5270. https://doi.org/10.1073/pnas.1320054111
Wang M, Chen L, Li Y, Chen L, Liu Z, Wang X, Yan P, Qin S (2018) Responses of soil microbial communities to a short-term application of seaweed fertilizer revealed by deep amplicon sequencing. Appl Soil Ecol 125:288–296. https://doi.org/10.1016/j.apsoil.2018.02.013
Xue K, Wu L, Deng Y, He Z, Van Nostrand J, Robertson PG, Schmidt TM, Zhou J (2013) Functional gene differences in soil microbial communities from conventional, low-input, and organic farmlands. Appl Environ Microbiol 79:1284–1292. https://doi.org/10.1128/aem.03393-12
Zhang X, Thomsen M (2019) Biomolecular composition and revenue explained by interactions between extrinsic factors and endogenous rhythms of Saccharina latissima. Mar Drugs 17:1–40. https://doi.org/10.3390/md17020107
Zhu Y, Ni H, Cao J, Li K, Yue Q, Zhao L (2019) A new PGA-seaweed water-retaining agent and its use in saline-alkali soils repairing. AIP Conf Proc 2110:020015. https://doi.org/10.1063/1.5110809
Funding
This work was supported by the Zhejiang Province Natural Science Foundation of China (LY22C190002), Scientific and Technological Project of Ningbo (2022S208), Major Scientific and Technological Project of Ningbo (2021Z004, 2021Z103), Major Scientific and Technological Project of Fenghua (20173502), China Agriculture Research System of MOF and MARA, Safe seaweed collation (TTSCSP), and K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Contributions
Conceptualization and funding acquisition: Yang Rui, Chen Haimin. Methodology: Sun Minxiu, Li Qian, Liu Qiqin. Writing—original draft preparation: Liu Qiqin. Writing—review and editing and supervision: Yang Rui. Resources: Zhou Huaguang, Fan Haijun.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhou Huaguang is co-first author.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qiqin, L., Huaguang, Z., Minxiu, S. et al. Improvement of Soil Structure and Bacterial Composition by Long-Term Application of Seaweed Fertilizer. J Soil Sci Plant Nutr 23, 5122–5132 (2023). https://doi.org/10.1007/s42729-023-01341-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42729-023-01341-0