Abstract
The effects of the short-term application of Ascophyllum nodosum-fermented seaweed fertilizer on the bacterial community, soil nitrogen contents, and plant growth in maize rhizosphere soil were evaluated. The changes in the bacterial community composition and nitrogen contents including those of total nitrogen (TN), nitrate nitrogen (NO3−-N) and ammonium nitrogen (NH4+-N) in rhizosphere soils in response to treatment with seaweed fertilizer were determined. Furthermore, soil enzymatic activity and crop biomass were analyzed. The relative abundance of the dominant phyla varied regularly with fertilization, and bacterial α-diversity was apparently influenced by seaweed fertilizer amendment. The TN contents of all soil samples decreased gradually, and the NO3−-N and NH4+-N contents of the soils treated with seaweed fertilizer were much higher than those of the control soils. Similarly, the enzymatic activities of dehydrogenase, nitrite reductase, urease, and cellulase in the soil were significantly increased on day 3, day 8, and day 13 after the application of seaweed fertilizer to the maize rhizosphere soil. However, there was no difference in the activity of soil sucrase between the treatment group and the control group. In this study, the growth of maize seedlings was confirmed to be greatly promoted by the utilization of seaweed fertilizer. These results deepen our understanding of plant-microbe interactions in agroecosystems and should benefit the wide use of seaweed fertilizer in sustainable agricultural production.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The application of chemical fertilizers has greatly increased the grain production per unit area of farmland, resulting in the feeding of more people worldwide (Zhu and Chen 2002). However, the long-term excessive use of chemical fertilizers has also led to the destruction of soil structure and the pollution of water bodies (Atafar et al. 2010). When a large amount of nitrogen fertilizer is applied to crops, nitrate nitrogen is easily lost under rain or leaching, eventually resulting in water pollution (Dinnes et al. 2002). Additionally, the ammonia and nitrogen oxides produced by denitrification will be released into the atmosphere (Fischer et al. 2010; Guo et al. 2010; Velthof et al. 2009). Considering the increasingly serious levels of energy consumption and environmental pollution, it is necessary to find alternatives to chemical fertilizers.
Seaweed is an environmentally friendly natural resource, and the potential to utilize seaweed extract as organic fertilizer has been confirmed by resultant increases in chlorophyll b levels and a better nitrogen use efficiency in crops (Latique et al. 2013; Sangeetha and Thevanathan 2010). Seaweed is extremely rich in nutrients, including mineral elements, trace elements, auxin, polyamines, and other active substances (Lötze and Hoffman 2016; Khan et al. 2009). It can regulate the balance of plant endogenous hormones and greatly promote the growth of plants. In addition, some growth regulators contained in seaweed fertilizer can improve soil microecology and enhance fertilizer efficiency, and the contribution of green-engineered seaweed production systems to the mitigation of the ongoing and historical anthropogenic disturbances of nutrient flows has been highlighted (Zhu et al. 2019; Zhang and Thomsen 2019). Seaweed extract has multiple positive effects on crop yield, product quality and the resistance of plants to diseases and abiotic stresses (Rathore et al. 2009; Jayaraj et al. 2008; Munshaw et al. 2006). Because of its high fertilizer efficiency and richness in nutritional elements, seaweed fertilizer presents a good performance-to-price ratio in terms of sustainable agricultural production. Therefore, to protect soil structure and avoid the pollution of water bodies caused by the excessive application of chemical fertilizers, such as eutrophication, it is feasible to decrease the consumption of chemical fertilizer via the utilization of seaweed fertilizer and seaweed extract.
As a component of soil biodiversity, soil enzyme activities are the direct expression of the soil community to metabolic requirements and available nutrients (Caldwell 2005). Soil enzymes regulate ecosystem functioning and play a key role in nutrient cycling in particular (Makoi and Ndakidemi 2008). Soil enzymes vary in their composition and quantities due to differences in plant communities, animal activities, and ecological environments (Kotroczó et al. 2014; Shackle et al. 2000; An and Kim 2009). Therefore, the soil enzyme system represents a specific biological community. The metabolism of soil microorganisms requires the participation of soil enzymes, and biochemical processes, redox reactions, material transformation, and energy metabolism depend on soil enzymatic activities. Soil enzymes are among the most active organic components of soil aggregates (Marx et al. 2001) and are also a parameter for the assessment of soil fertility and the change rate of the chemical composition. Soil enzymatic activity reflects the microbial contents, and there is growing interest in the application of soil enzymes as early indicators of soil quality change under contrasting agricultural management practices (García-Ruiz et al. 2008).
Although seaweed fertilizer has been widely proven to be effective for crop growth, it is not very clear how seaweed fertilizer affects bacterial diversity and community structure in plant rhizosphere soils and how to modulate soil nutrients, especially nitrogen nutrients to promote plant growth. To investigate these problems in greater detail, we conducted deep 16S rRNA sequencing, and the dynamics of nitrogen content and enzymatic activity in soil were also monitored.
Materials and methods
Seaweed fertilizer and pot experimental design
Commercial Ascophyllum nodosum-fermented seaweed fertilizer (Lannengliang®, Qingdao Brightmoon Seaweed Group Corporation) was used for short-term fertilization treatment of maize seedlings in this study. The nutritional composition of this powder product was as follows: 14% N, 2% P2O5, 20% K2O, and 3.0% humic acid. The seeds of maize (Zea mays L.) were soaked in sterile water for approximately 4 h and then subjected to surface disinfection for 2 min in a 75% ethanol solution. Subsequently, these seeds were thoroughly rinsed with sterile water and soaked in a 5% sodium hypochlorite solution for approximately 5 min. Finally, the seeds were fully washed with sterile water and placed on double layers of wet filter paper for germination in a sterile 9-cm (diameter) Petri dish. After germination, the maize seedlings were planted in plastic flowerpots (20 cm in diameter and 20 cm in height, four seedlings per pot).
Preparation of maize rhizosphere soil
A total of 32 maize seedlings were divided into two groups: a treatment group and a control group. The seedlings (approximately 10 cm tall) of the treatment group were subjected to fertilization. A 25-mL volume of the seaweed fertilizer (0.3 g/100 mL) was carefully added to the maize rhizosphere zone using a sterile syringe. The control group was treated with an equal volume of aseptic water. All seedlings were treated once a week, and the maize rhizosphere soil was collected after five treatments. Soil samples at depths between 3 and 5 cm within a 0.5-cm range around the maize roots were gently collected using a stainless steel spoon (referred to as rhizosphere soil samples hereafter). The rhizosphere soils were collected at three time points, day 3, day 8, and day 13 (the last treatment time as day 0). For the comparison of the two groups at each sampling time point, approximately 60 g of rhizosphere soil was harvested and subjected to DNA extraction.
To determine the nitrogen content and enzymatic activity in the maize rhizosphere soils, air-dried soil samples at room temperature (more than 20 g) were first ground using an electric grinder and then passed through a 100-mesh nylon sieve. The prepared soil samples were kept at 4 °C until they were used.
Soil DNA extraction and PCR amplification
To explore the bacterial diversity and community structure of the maize rhizosphere soils and characterize the soil microbial properties in response to the short-term application of seaweed fertilizer, deep 16S rRNA sequencing was performed. High-purity genomic DNA was isolated from the maize rhizosphere soil using the E.Z.N.A.® Soil DNA Kit (Omega Bio-TEK, USA) according to the manufacturer’s instructions. The final DNA concentration and purity were determined with a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3–V4 hypervariable regions of the bacterial 16S rRNA gene were amplified with the primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by using a thermocycler PCR system (GeneAmp® 9700, ABI, USA). The amplification reactions were conducted using the following program: 3 min of denaturation at 95 °C, 27 cycles of 30 s at 95 °C, 30 s of annealing at 55 °C, and 45 s of elongation at 72 °C, and a final extension at 72 °C for 10 min. The reactions were performed in triplicate in a 20-μL volume containing 4 μL of 5× FastPfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu polymerase, and 10 ng of template DNA. The resulting PCR products were subjected to electrophoresis on a 2% agarose gel and then extracted and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using the QuantiFluor™-ST system (Promega, USA) according to the product manual.
Illumina MiSeq sequencing and the deposition of raw reads
Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to standard protocols by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). All raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP215409).
Processing of sequencing data
Raw FASTQ files were quality filtered with the Trimmomatic software and merged with FLASH software. Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version 7.1, http://drive5.com/uparse/) with a novel algorithm that performs chimera filtering and OTU clustering simultaneously. The taxonomy of each 16S rRNA gene sequence was analyzed with the RDP classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database using a confidence threshold of 70%.
Measurement of soil enzymatic activity
A total of five soil enzymes were used to measure soil fertility and microbial activity in the maize rhizosphere soil. Soil enzymatic activities, including those of cellulase (S-CL), nitrite reductase (S-NIR), sucrase (S-SC), and urase (S-UE) were determined using commercial testing kits from Suzhou Comin Biotechnology Cooperation (Suzhou, China) with catalogue numbers S-CL-2-Y, S-NIR-2-G, S-SC-2-Y, and S-UE-2-Y, respectively. All assays were carried out in triplicate and evaluated by spectrophotometry according to the product manual.
Soil dehydrogenase activity (S-INT) was evaluated by the method of Casida et al. (1964). Colorless soluble 2,3,5-triphenyltetrazolium chloride (TTC) can be reduced to form insoluble 1,3,5-triphenylformazan (TPF) with a red color by dehydrogenase. The generated TPF presents a specific absorption peak at a wavelength of 492 nm. Two grams of rhizosphere soil was fully mixed with 2 mL of 0.1 mol/L of glucose, 2 mL of 1% TTC, and 2 mL of Tris-HCl buffer (pH 7.4) in a 50-mL centrifuge tube, followed by incubation at 37 °C for 24 h in darkness. 0.5 mL of formaldehyde solution was added to terminate the reaction, and the mixture was subjected to extraction for 30 min with 5 mL of toluene on a rotating shaker (150 rpm) followed by centrifugation at 4000×g for 5 min. Finally, the produced TPF was determined at 492 nm using a Cary 60 UV-Vis spectrophotometer from Agilent Technologies (Agilent, Australia). The enzymatic activity of soil dehydrogenase was expressed in μg/g/day. The measurements were duplicated three times.
Determination of soil nitrogen contents
To examine whether the application of seaweed fertilizer greatly altered nitrogen cycling in the maize rhizosphere soil, soil nitrogen contents including total nitrogen (TN), ammonium nitrogen (NH4+-N), and nitrate nitrogen (NO3−-N) were assayed at three sampling time points. A spectrophotometric method for the assessment of ammonium nitrogen and nitrate nitrogen contents in soil was adopted based on extraction with a potassium chloride solution according to the procedures of the National Environmental Protection Standards of the People’s Republic of China (HJ 634-2012). These standards were slightly modified in accordance with the processing approaches of ISO/TS14256-1:2003(E).
Ammonium nitrogen was extracted from the soil with a potassium chloride solution. The ammonia ion can react with phenol to form a blue-indigo dye in the presence of hypochlorite ions under alkaline conditions, which exhibits maximum absorption at a wavelength of 630 nm. The absorbance value is in accordance with Lambert–Beer’s law for a certain range of ammonium concentrations. To determine the contents of nitrate nitrogen in the soil, the samples were first extracted with a potassium chloride solution, after which the nitrate nitrogen in solution was reduced to form nitrite nitrogen through a reduction column. Finally, all the nitrite nitrogen in the extraction solution reacts with sulfonamide to form a diazo salt, and the resulting product couples with N-(1-naphthyl) ethylenediamine hydrochloric acid to form a red dye under acidic conditions, which exhibits maximum absorption at 543 nm. The actual nitrate nitrogen content of soil was determined from the total nitrite content in the final step minus the original nitrite nitrogen content.
Determination of maize seedling biomass
To evaluate the effect of seaweed fertilizer application on the growth of maize seedlings, the mean biomass values of ten seedlings were measured. The seedlings were carefully removed from pots on day 40 after fertilization (the treatment group) and then washed thoroughly in double distilled water to remove any soil on the root surface, air-dried at room temperature, and eventually subjected to the determination of plant biomass, including the measurement of seedling height, above-ground fresh weight, and root fresh weight.
Statistical analysis
The 16S rRNA sequencing data were analyzed at the free online Majorbio I-Sanger Cloud Platform (www.i-sanger.com), developed by the Shanghai Majorbio Bio-Pharm Technology Cooperation (Ltd). ADONIS based on 999 permutations was applied by using the Bray–Curtis (Bray and Curtis 1957) algorithm to quantify community differences between groups.
Pearson’s correlation coefficient analysis was conducted to assess the correlation between rhizospheric nitrogen content and soil enzymatic activity by using the SPSS 24.0 software.
One-way analysis of variance (ANOVA) was applied to evaluate the differences in the biomass values of seedlings between different treatments by using the SPSS 22.0 software, and the statistical significance level was set at P < 0.05.
Results
Dynamics of nitrogen contents in the rhizosphere soils of maize
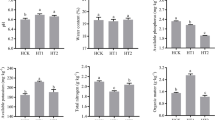
The results showed that the contents of NO3−-N (Fig. 1a), NH4+-N (Fig. 1b), and TN (Fig. 1c) in the rhizosphere soil varied obviously during the test period. The TN content in the treatment group was significantly higher than that in the control group (treated with sterile water) on day 3 (P < 0.05). However, on day 8 and day 13, although the TN content of the treatment group was higher than that of the control group, the difference was not statistically significant. Regarding the content of NO3−-N, the difference between the two groups was extremely significant at two time points, day 3 and day 8 (P < 0.01), and on day 13, the difference remained significant (P < 0.05). In the comparison of NH4+-N contents between the two groups, the difference was highly significant on day 3 (P < 0.01) and remained significant on day 8 and day 13 (P < 0.05). These results suggested that the nitrogen contents of the maize rhizosphere soil were increased significantly after seaweed fertilizer application.
Dynamics of nitrogen contents in the maize rhizosphere soil at different time points after seaweed fertilizer application. The contents of nitrate nitrogen, ammonium nitrogen and total nitrogen in the soil are shown in a, b, and c, respectively. BE, before the experiment; T, soils of the treatment groups (treated with seaweed fertilizer) sampled on day 3, day 8, and day 13, respectively, after seaweed fertilizer application; CK, soils of the control groups (treated with sterile water) sampled on day 3, day 8, and day 13, respectively, after seaweed fertilizer application
Effect of seaweed fertilizer application on soil enzymatic activity
With the growth of plants, all the tested enzymatic activities in the soil samples increased (Fig. 2). The enzymatic activities of dehydrogenase (S-INT) (Fig. 2b) and nitrite reductase (S-NIR) (Fig. 2c) in the soil were increased greatly at three sampling time points, and the difference in enzymatic activity between the two groups reached a statistically significant level (P < 0.01). Soil urease (S-UE) activities in the treatment group were all significantly increased compared with those in the control group (P < 0.05) (Fig. 2e). The soil cellulase (S-CL) activity of the treatment group was increased significantly on day 3 (P < 0.01), after which the difference gradually decreased (Fig. 2a). On day 13 after fertilization, no significant difference in soil cellulase activity was found between the two groups. It should be noted that there was no significant difference in soil sucrase (S-SC) activity between the two groups (Fig. 2d).
Enzymatic activities in the maize rhizosphere soil after seaweed fertilizer application. The changes in the enzymatic activities of soil cellulase (S-CL), dehydrogenase (S-INT), nitrite reductase (S-NIR), sucrase (S-SC), and urase (S-UE) are displayed in a, b, c, d, and e, respectively. BE, before the experiment; T, soils of the treatment groups (treated with seaweed fertilizer) sampled on day 3, day 8, and day 13, respectively, after seaweed fertilizer application; CK, soils of the control groups (treated with sterile water) sampled on day 3, day 8, and day 13, respectively, after seaweed fertilizer application
Analyses of operational taxonomic unit and α-diversity
The bacterial communities in the maize rhizosphere soil were detected by deep sequencing of bacterial 16S rRNA gene amplicons. A total of 957,943 sequences with a mean length of 416 bp were obtained and rarefied for diversity analysis. A total of 5988 OTUs of the soil bacterial communities were identified; these OTUs belonged to 41 phyla, 1015 classes, 224 orders, 441 families, 871 genera, and 1814 species.
The relative abundance of the dominant phyla changed after fertilization. All the soil samples were dominated by Proteobacteria (27.59–37.83%), Actinobacteria (27.10–32.96%), Chloroflexi (9.54–15.19%), Acidobacteria (7.47–14.99%), and Bacteroidetes (3.64–4.02%), accounting for 88.1–89.7% of the bacterial sequences (Fig. 3). The performance of two dominant phyla, Chloroflexi and Acidobacteria was completely different between the treatment group (MT1, MT2, and MT3) and the control group (MCK1, MCK2, and MCK3). The relative abundance of Chloroflexi (9.54–15.19%) and Acidobacteria (8.87–12.24%) increased remarkably after fertilization; however, in the control group, their relative abundance decreased greatly. In particular, the relative abundance of three other phyla, Proteobacteria (28.28–33.96%), Actinobacteria (27.10–32.96%), and Bacteroidetes (3.64–3.97%), increased gradually in the control group during the test period.
Relative abundance of the dominant phyla in rhizosphere soils of maize sampled on day 3, day 8, and day 13 after seaweed fertilizer application. MT1, MT2, and MT3 are soil sampling on day 3, day 8 and day 13, respectively, after seaweed fertilizer application. MCK1, MCK2, and MCK3 are soils of the control groups (treated with sterile water) sampled on day 3, day 8, and day 13, respectively, after seaweed fertilizer application
The results of 16S rRNA gene sequencing revealed an obvious change in the composition of the maize rhizobacteria. The α-diversity analysis showed that the bacterial community richness indices (Sobs, Chao1, and Ace) and community diversity index (Shannon) varied markedly between the two groups (Table 1). Seaweed fertilizer application increased the bacterial Sobs (P = 0.02) and Shannon indices (P = 0.01) on day 13 after fertilization (Table 1). It can be concluded that the bacterial α-diversity was significantly influenced by the fertilization treatment.
Structure of the maize rhizobacterial community
The non-metric multi-dimensional scaling (NMDS) analysis at the OTU level indicated notable variation in the structure of the maize rhizobacterial community after short-term fertilization treatment. In the treatment group, the bacterial community composition in the soils on day 3 after fertilization (MT1) was distinctly different from that on day 8 (MT2) and day 13 (MT3) (Fig. 4). ADONIS analysis based on Bray–Curtis distance showed a significant difference in the bacterial community composition between the treatment and control groups after fertilization (P = 0.1, Table 2).
To evaluate the effect of seaweed fertilizer on the microbial species composition and community structure in maize rhizosphere soils, we evaluated 18 soil samples and compared the relative abundance of bacterial species between them. The results showed that Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, and Bacteroidetes were the predominant phyla in all soil samples. There were significant differences in species richness between all tested groups, from MT1 to MT3 and MCK1 to MCK3. Significant differences in the relative abundance of phylum Proteobacteria and four genera, Sphingomonas, Anaerolineaceae, Mycobacterium, and Lysobacter, existed between the soil groups at the 0.05 level (Fig. 5a, b). Additionally, the differences in the relative abundance of microbial species in the treatment group and the control group were analyzed at the phylum level by using Student’s two-sided t test. On day 3, the MT1 and MCK1 samples showed a statistically significant difference in the relative abundance of the phyla Proteobacteria (P < 0.01), Acidobacteria (P < 0.05), Chloroflexi (P < 0.05), Bacteroidetes (P < 0.05), Planctomycetes (P < 0.05), and Latescibacteria (P < 0.05) (Fig. 6a). There was no significant difference in species richness between the MT2 and MCK2 groups (Fig. 6b). Finally, significant differences in the relative abundance of microbial species in two phyla, Proteobacteria (P < 0.05) and Acidobacteria were detected (P < 0.05) (Fig. 6c).
The analysis of significant differences between groups at the phylum (a) and genus (b) levels. An asterisk (*) indicates a significant difference in the relative abundance of microbial species (P < 0.05). The multiple testing correction method for P values was the false discovery rate (FDR). The Kruskal–Wallis H test was used for significance testing between groups
The analysis of significant differences between two groups at the phylum level. a MT1 and MCK1. b MT2 and MCK2. c MT3 and MCK3. The two-tailed Student’s t test was used for significance testing between two groups. An asterisk (*) indicates a significant difference in relative abundance at a statistical level of 0.05
Effect of seaweed fertilizer application on the growth of maize seedlings
When treated with seaweed fertilizer, the maize seedlings exhibited greater biomass values compared with the control seedlings treated with equal volumes of sterile water. The differences in seedling biomass values, including seedling height (P < 0.01), above-ground fresh weight (P < 0.01), and root fresh weight (P < 0.05), between the two groups were statistically significant (Fig. 7). The application of seaweed fertilizer had a significant promotion effect on the growth of maize seedlings under the experimental conditions.
The maize seedling biomass on day 40 after seaweed fertilizer application. FW and RFW represent the above-ground seedling fresh weight and root fresh weight, respectively. T, seedlings of the treatment groups (treated with seaweed fertilizer) sampled on day 40 after seaweed fertilizer application; CK, seedlings of the control groups (treated with sterile water) sampled on day 40 after seaweed fertilizer application
Discussion
The application of seaweed-fermented fertilizer or a seaweed extract has proven beneficial for the growth and yield of crops in agricultural production (Kumar et al. 2012; Zodape et al. 2011). This study evaluated the effects of the root-zone application of an Ascophyllum nodosum-fermented seaweed fertilizer on the rhizobacterium communities and the growth of maize seedlings in a pot trial experiment. The results showed that the biomass of maize seedlings increased significantly after fertilization. Compared to the control group, the root fresh weight (RFW) in the treatment group was increased greatly (P < 0.05), and seedling height and above-ground seedling fresh mass were increased significantly (P < 0.01). Seaweed fertilizer had an obvious promotion effect on the growth of maize in this study. Seaweeds contain abundant nitrogen nutrients and other major and trace elements, and their nutrients and organic matter contents are very beneficial for improving the nature of soil and enhancing its moisture-retaining capacity (Simpson and Hayes, 1958). Seaweeds also contain many growth-promoting hormones such as cytokinin and auxins (Tay et al. 1985 ). Seaweed fertilizer has been widely used in agricultural production due to these properties. Considering that the contents of nitrate nitrogen and ammonium nitrogen in the soils treated with the seaweed fertilizer were apparently increased compared to those in the control, we assume that the promotion effect of the seaweed fertilizer on the growth of maize seedlings may be partially attributable to the improvement of rhizosphere soil nutrition.
Deep 16S rRNA sequencing is a key tool for the investigation of bacterial communities and can provide useful insights into the thousands of uncultivated microbial communities for which only marker gene surveys are currently available. High-throughput sequencing has facilitated major advances in our understanding of microbial ecology and is now widespread in biotechnological applications (Langille et al. 2013). Although seaweed and its derivatives have been used as natural, harmless resources in sustainable agricultural production for many years, the impact of seaweed fertilizer application on the rhizobacterial community structure of plants and the plant-microbe interaction in rhizosphere zones remains unclear. Recently, a field trial in tomato was carried out to explore the effects of seaweed fertilizer application on soil microbial communities (Wang et al. 2018). The results showed that bacterial α-diversity was strongly influenced by Sargassum horneri-fermented seaweed fertilizer amendment after 60 days. Similar phenomena were observed in our study; the bacterial community compositions and relative abundance at the phylum level in the rhizosphere soil were significantly altered by the application of Ascophyllum nodosum-fermented seaweed fertilizer.
Many factors could cause the changes in rhizospheric enzymatic activities in soils such as antibiotics, pesticides, heavy metals, field management, and organic wastes (Liu et al. 2009; Sardar et al. 2007; Ros et al. 2003; Bankdick and Dick 1999). Soil enzyme activities are often used as indicators of microbial growth in soil. A previous study showed that dehydrogenase activity in soil was significantly related to microbial respiration (P < 0.01) and that urease activity was positively correlated with total nitrogen (TN) and negatively correlated with organic carbon content (P < 0.05) (Frankenberger Jr and Dick 1983). In this work, the highest TN content was observed on day 3 after seaweed fertilizer application. Although the TN content generally remained stable between different treatments, the contents of nitrate and ammonium nitrogen in the seaweed fertilizer-treated soils were increased significantly compared to the control. Considering the marked increases in the biomass values of the seedlings treated with seaweed fertilizer, we deduce that more nitrogen was absorbed and utilized by plants and that the TN content was therefore not significantly increased in the seaweed fertilizer-treated group. Dehydrogenase activities in the treatment group were significantly increased (P < 0.01), and urease activity in this group notably increased (P < 0.05) during the test period. Seaweed fertilizer clearly increases the content of nutrient elements including organic carbon in the rhizospheric soils of maize, thus improving dehydrogenase activity. The Pearson correlation coefficient (PCC) is commonly used for assessing the possible two-way linear association between two variables (Hauke and Kossowski 2011). Our study revealed that nitrate nitrogen content was strongly correlated with urease activity (i.e., showed a high positive correlation). Similarly, the ammonium nitrogen content was also highly correlated with nitrite reductase activity. The PCCs for these variables were 0.822 and 0.872 (P < 0.05), respectively (Table 3). The result of the PCC analysis suggested that nitrogen nutrition had a marked influence on S-CL, S-INT, S-NIR, and S-UE enzymatic activities. Soil enzymatic activities are influenced by different nitrogen forms and the ratio of inorganic to organic nitrogen (Nannipieri et al. 2012), and long-term nitrogen fertilization significantly increases the activities of β-glucosidase and acid phosphatase but decreases urease activity (Ajwa et al. 1999). However, in this study, only sucrase activities showed no significant difference between the groups, but the activities of other soil enzymes, including urease (P < 0.05), cellulase (P < 0.05), dehydrogenase, and nitrate reductase, were greatly increased (P < 0.01) after short-term seaweed fertilizer application (Table 3). We speculate that the difference in soil urease activity compared to that determined in the previous report may have resulted from the differences in the amount of fertilizer applied and the length of treatment time. Soil cellulase and urease activity have been considered the main indicators of soil fertility as evaluated by principal component analysis (Ge et al. 2009). In general, soil enzymatic activities (S-INT, S-NIR, S-UE, and S-CL) were increased significantly under seaweed fertilizer treatment in this study. However, there was no difference in soil sucrase activities between the treatment group and the control group. Unusual performance of soil sucrase activity under organic fertilizer treatment has also been found by other researchers. It has been reported that the soil sucrase activities in paddy soil are not consistently increased by treatment with organic manure alone. These scientists indicated that soil temperature, moisture, and pH as well as soil texture should be taken into account in evaluating the effects of long-term fertilization on soil biological activity, and they speculated that sucrase activity in soil may be strongly induced in response to treatments involving lower soil TN and ammonium nitrogen levels and a higher pH (Ge et al. 2009). Considering that the TN contents of the maize rhizosphere soil between treatments remained stable and the contents of ammonium nitrogen in the treatment group were always higher than those in the control group during the test period, it seems that soil texture is an important factor that affects the activity of soil sucrase, and the specific mechanisms involved need to be further studied.
Functional diversity is an aspect of the overall microbial diversity in soil and encompasses a range of activities. The relationship between microbial diversity and function in soil is largely unknown, but biodiversity has been assumed to influence ecosystem stability, productivity, and resilience towards stress and disturbance (Torsvik and Øvreås 2002). Most microbes in nature have not been studied because traditional methods for culturing microorganisms limit analysis to those that grow under laboratory conditions. A bacterial artificial chromosome (BAC) vector was previously used for constructing libraries of genomic DNA isolated directly from soil, and metagenomic libraries are a powerful tool for exploring soil microbial diversity, providing access to genetic information for uncultured soil microorganisms (Rondon et al. 2000). Additionally, comparative 16S rRNA gene sequencing analysis and structural genes such as amoA, as a functional marker of natural ammonia-oxidizing populations, have been utilized for the prediction of the functional profile of microbial communities (Lu and Domingo 2008; Rotthauwe et al. 1997). High-throughput sequencing has facilitated major advances in our understanding of microbial ecology and is now widespread in biological applications. PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) has been widely applied to predict the functional composition of a microbial community’s metagenome from its 16S profile (Langille et al. 2013). To explore the possible difference in the microbial community’s functional composition resulting from seaweed fertilizer application, deep 16S rRNA sequencing data were subjected to a potential functional diversity analysis of microbes using PICRUSt in the present study, and we found that the overall COG functional classification pattern was similar between the different soil samples. The predominant COG functions were energy production and conversion (accounting for 7.34 to 7.43% of the identified COG categories), transport and metabolism of carbohydrates (6.24 to 6.56%), amino acids (8.57 to 8.66%), inorganic ions (5.57 to 5.72%), and signal transduction mechanisms (6.42 to 6.62%) (Fig. S1). In particular, it should be noted that the relative abundance of functional proteins involved in carbohydrate transport and metabolism in the treatment groups was increased compared with that in the control treatments, suggesting more active growth of rhizobacterial cells. Thus, seedling growth may have been promoted due to the interaction between plants and rhizobacteria.
The predicted highly abundant functional enzymes were dehydrogenase reductase, histidine kinase, RNA polymerase, and acyl-CoA dehydrogenase (Fig. S2). In conclusion, the relative abundance of the microbiota varied significantly, and some functional components of the microbial communities in the soil were changed. The application of seaweed fertilizer was confirmed to be beneficial for the growth of maize seedlings and should provide a promising approach for the development of sustainable agriculture.
References
Ajwa HA, Dell CJ, Rice CW (1999) Changes in enzyme activities and microbial biomass of tallgrass prairie soil as related to burning and nitrogen fertilization. Soil Biol Biochem 31:769–777. https://doi.org/10.1016/S0038-0717(98)00177-1
An Y-J, Kim M (2009) Effect of antimony on the microbial growth and the activities of soil enzymes. Chemosphere 74:654–659. https://doi.org/10.1016/j.chemosphere.2008.10.023
Atafar Z, Mesdaghinia A, Nouri J, Homaee M, Yunesian M, Ahmadimoghaddam M, Mahvi AH (2010) Effect of fertilizer application on soil heavy metal concentration. Environ Monit Assess 160:83–89. https://doi.org/10.1007/s10661-008-0659-x
Bankdick AK, Dick RP (1999) Field management effects on soil enzyme activities. Soil Biol Biochem 31:1471–1479. https://doi.org/10.1016/S0038-0717(99)00051-6
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27(4):325–349
Caldwell BA (2005) Enzyme activities as a component of soil biodiversity: a review. Pedobiologia 49:637–644. https://doi.org/10.1016/j.pedobi.2005.06.003
Casida LCJR, Klein DA, Santoro T (1964) Soil dehydrogenase activity. Soil Sci 98:371–376
Dinnes DL, Karlen DL, Jaynes DB, Kaspar TC, Hatfield JL, Colvin TS, Cambardella CA (2002) Nitrogen management strategies to reduce nitrate leaching in tile-drained midwestern soils. Agron J 94:153–171. https://doi.org/10.2134/agronj2002.1530
Fischer G, Winiwarter W, Ermolieva T, Cao GY, Qui H, Klimont Z, Wiberg D, Wagner F (2010) Integrated modeling framework for assessment and mitigation of nitrogen pollution from agriculture: concept and case study for China. Agric Ecosyst Environ 136:116–124. https://doi.org/10.1016/j.agee.2009.12.004
Frankenberger WT Jr, Dick WA (1983) Relationships between enzyme activities and microbial growth and activity indices in soil. Soil Sci Soc Am J 47:945–951. https://doi.org/10.2136/sssaj1983.03615995004700050021x
García-Ruiz R, Ochoa V, Hinojosa MB, Hinojosa MB, Carreira JA (2008) Suitability of enzyme activities for the monitoring of soil quality improvement in organic agricultural systems. Soil Biol Biochem 40:2137–2145. https://doi.org/10.1016/j.soilbio.2008.03.023
Ge GF, Li ZJ, Zhang J, Wang LG, Xu MG, Zhang JB, Wang JK, Xie XL, Liang YC (2009) Geographical and climatic differences in long-term effect of organic and inorganic amendments on soil enzymatic activities and respiration in field experimental stations of China. Ecol Complex 6:421–431. https://doi.org/10.1016/j.ecocom.2009.02.001
Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, Christie P, Goulding KWT, Vitousek PM, Zhang FS (2010) Significant acidification in major Chinese croplands. Sci 327:1008–1010. https://doi.org/10.1126/science.1182570
Hauke J, Kossowski T (2011) Comparison of values of Pearson’s and Spearman’s correlation coefficients on the same sets of data. Quaest Geogr 30:87–93
Jayaraj J, Wan A, Rahman M, Punja ZK (2008) Seaweed extract reduces foliar fungal diseases on carrot. Crop Prot 27:1360–1366. https://doi.org/10.1016/j.cropro.2008.05.005
Khan W, Rayirath UP, Subramanian S, Jithesh MN, Rayorath P, Hodges DM, Critchley AT, Craigie JS, Norrie J, Prithiviraj B (2009) Seaweed extracts as biostimulants of plant growth and development. J Plant Growth Regul 28:386–399. https://doi.org/10.1007/s00344-009-9103-x
Kotroczó Z, Veres Z, Fekete I, Krakomperger Z, Tóth JA, Lajtha K, Tóthmérész B (2014) Soil enzyme activity in response to long-term organic matter manipulation. Soil Biol Biochem 70:237–243. https://doi.org/10.1016/j.soilbio.2013.12.028
Kumar NA, Vanlalzarzova B, Sridhar S, Baluswami M (2012) Effect of liquid seaweed fertilizer of Sargassum wightii Grev. on the growth and biochemical content of green gram (Vigna radiate (L.) R. Wilczek). Recent Res Sci Technol 4:40–45
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. https://doi.org/10.1038/nbt.2676
Latique S, Chernane H, Mansori M, Kaoua E (2013) Seaweed liquid fertilizer effect on physiological and biochemical parameters of bean plant (Phaesolus vul-garis var Paulista) under hydroponic system. Eur Sci J 9:174–193
Liu F, Ying GG, Tao R, Zhao JL, Yang JF, Zhao LF (2009) Effects of six selected antibiotics on plant growth and soil microbial and enzymatic activities. Environ Pollut 157:1636–1642. https://doi.org/10.1016/j.envpol.2008.12.021
Lötze E, Hoffman EW (2016) Nutrient composition and content of various biological active compounds of three South African-based commercial seaweed biostimulants. J Appl Phycol 28:1379–1386. https://doi.org/10.1007/s10811-015-0644-z
Lu JR, Domingo JS (2008) Turkey fecal microbial community structure and functional gene diversity revealed by 16S rRNA gene and metagenomic sequences. J Microbiol 46:469–477. https://doi.org/10.1007/s12275-008-0117-z
Makoi JHJR, Ndakidemi PA (2008) Selected soil enzymes: examples of their potential roles in the ecosystem. Afr J Biotechnol 7:181–191. https://doi.org/10.5897/AJB07.590
Marx MC, Wood M, Jarvis SC (2001) A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol Biochem 33:1633–1640. https://doi.org/10.1016/S0038-0717(01)00079-7
Munshaw GC, Ervin EH, Shang C, Askew SD, Zhang X, Lemus RW (2006) Influence of late-season iron, nitrogen, and seaweed extract on fall color retention and cold tolerance of four Bermudagrass cultivars. Crop Sci 46:273–283. https://doi.org/10.2135/cropsci2005.0078
Nannipieri P, Giagnoni L, Renella G, Puglisi E, Ceccanti B, Masciandaro G, Fornasier F, Moscatelli MC, Marinari S (2012) Soil enzymology: classical and molecular approaches. Biol Fertil Soils 48:743–762. https://doi.org/10.1007/s00374-012-0723-0
Rathore SS, Chaudhary DR, Boricha GN, Ghosh A, Bhatt BP, Zodape ST, Patolia JS (2009) Effect of seaweed extract on the growth, yield and nutrient uptake of soybean (Glycine max) under rainfed conditions. S Afr J Bot 75:351–355. https://doi.org/10.1016/j.sajb.2008.10.009
Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, Macneil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547. https://doi.org/10.1128/AEM.66.6.2541-2547.2000
Ros M, Hernandez MT, Garcia C (2003) Soil microbial activity after restoration of a semiarid soil by organic amendments. Soil Biol Biochem 35:463–469. https://doi.org/10.1016/S0038-0717(02)00298-5
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Sangeetha V, Thevanathan R (2010) Potential of traditional and Panchagavya amended with seaweed extract. J Am Sci 6:61–67
Sardar K, Qing C, El Latif HA, Yue X, Zheng HJ (2007) Soil enzymatic activities and microbial community structure with different application rates of Cd and Pb. J Environ Sci 19:834–840. https://doi.org/10.1016/S1001-0742(07)60139-9
Shackle VJ, Freeman C, Reynolds B (2000) Carbon supply and the regulation of enzyme activity in constructed wetlands. Soil Biol Biochem 32:1935–1940. https://doi.org/10.1016/S0038-0717(00)00169-3
Simpson K, Hayes SF (1958) The effect of soil conditioners on plant growth and soil structure. J Sci Food Agric 9(3):163–170
Tay SAB, Macleod JK, Palni LMS, Letham DS (1985) Detection of cytokinins in a seaweed extract. Phytochemistry 24(11):2611–2614
Torsvik V, Øvreås L (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5:240–245. https://doi.org/10.1016/S1369-5274(02)00324-7
Velthof GL, Oudendag D, Witzke HP, Asman WAH, Klimont Z, Oenema O (2009) Integrated assessment of nitrogen losses from agriculture in EU-27 using MITERRA-EUROPE. J Environ Qual 38:402–417. https://doi.org/10.2134/jeq2008.0108
Wang MP, Chen L, Li YT, Chen L, Liu ZY, Wang XJ, Yan PS, Qin S (2018) Responses of soil microbial communities to a short-term application of seaweed fertilizer revealed by deep amplicon sequencing. Appl Soil Ecol 125(4):288–296. https://doi.org/10.1016/j.apsoil.2018.02.013
Zhang XQ, Thomsen M (2019) Biomolecular composition and revenue explained by interactions between extrinsic factors and endogenous rhythms of Saccharina latissimi. Mar Drugs 17:107. https://doi.org/10.3390/md17020107
Zhu ZL, Chen DL (2002) Nitrogen fertilizer use in China-contributions to food production, impacts on the environment and best management strategies. Nutr Cycl Agroecosyst 63:117–127. https://doi.org/10.1023/A:1021107026067
Zhu YD, Ni HF, Cao J, Li KL, Yue QL, Zhao L (2019) A new PGA-seaweed water-retaining agent and its use in saline-alkali soils repairing. AIP Conf Proc 2110:020015. https://doi.org/10.1063/1.5110809
Zodape ST, Gupta A, Bhandari SC, Rawat US, Chaudhary DR, Eswaran K, Chikara J (2011) Foliar application of seaweed sap as biostimulants for enhancement of yield and quality of tomato (Lycopersicon esculentum Mill.). J Sci Ind Res 70:215–219
Acknowledgments
The authors appreciate the assistance of Baoyun Feng in the data processing and chart presentation.
Funding
This work was funded by the National Natural Science Foundation of China (No. 31870496) and the Open Foundation of the Ministry of Agriculture Key Laboratory of Seaweed Fertilizers (No. MAKLSF1810).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
The predicted COG function classification based on deep 16S rRNA sequencing data using PICRUSt. (PDF 19 kb)
Fig. S2
The predicted high-abundance functional enzymes based on deep 16S rRNA sequencing data using PICRUSt. COG1028, Dehydrogenase reductase; COG1309, Transcriptional regulator; COG2814, Major facilitator; COG0624, Histidine kinase; COG1595, RNA polymerase; COG0596, Alpha beta hydrolase; COG0745, Regulator; COG0438, Glycosyl transferase (Group 1); COG0583, Transcriptional regulator; COG2197, Two-component transcriptional regulator, luxR family; COG0515, Serine threonine protein kinase; COG2226, Methyltransferase required for the conversion of demethylmenaquinone to menaquinone; COG1960, acyl-CoA dehydrogenase; COG2204, Two-component sigma54-specific, transcriptional regulator, Fis family; COG1846, Transcriptional regulator. (PDF 15 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Li, J., Huang, Z. et al. Impact of short-term application of seaweed fertilizer on bacterial diversity and community structure, soil nitrogen contents, and plant growth in maize rhizosphere soil. Folia Microbiol 65, 591–603 (2020). https://doi.org/10.1007/s12223-019-00766-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-019-00766-4