Abstract
The Iguaçu National Park is the largest remnant of the Atlantic Forest in southern Brazil, providing habitats for a wide variety of species, including bedbugs of suborder Heteroptera. These insects have an great capacity for adapting and spreading over widely different habitats, resulting in high species diversity. However, little is known about the genetic structure of the group. In this paper, we analyzed the cytochrome c oxidase subunit I (COI) mitochondrial gene of two species, Loxa viridis (Palisot de Beauvois 1805) and Loxa virescens Amyot & Serville 1843, collected in Iguaçu National Park. The 32 COI sequences analyzed in this study were grouped into six haplotypes, that were exclusive to each collection site. The analysis of molecular variance showed tree polymorphisms for each species and variations among populations was 100%. Maximum Likehood test analysis showed two large groups, with L. viridis and L. virescens from the same collection points tend to be closer together. The results obtained contributed to the identification of the species and populations of each collection site are suffering local selection pressure in the Iguaçu National Park. The conservation actions carried out by the Iguaçu National Park are being important for the maintenance of biodiversity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The world’s biodiversity is increasingly threatened by environments devastation, especially in the neotropical region, were the natural environmental fragmentation has been intensified by antropogenic activities (Kruess and Tscharntke 1994). Natural habitats are shrinking, affecting radically the composition and abundance of species. In some cases, there are drastic effects on the natural processes of communities, sometimes leading to total extinction (Young et al. 1996, Fahrig 2003).
The Atlantic Forest is a prime example of this phenomenon, where only 11 to 16% of the original forest remains (Ribeiro et al. 2009). The Iguaçu National park (INP) is one of the few remnants of Atlantic Forest that is still conserved, being considered a World Heritage, with an exemplary integration among economic and environmental sustainability (D'Oliveira et al. 2002). The INP is known for its great diversity of species, especially insects, which are considered particularly important for assessing environmental impact (Thomazini and Thomazini 2000).
Several processes threaten insects, and any change in their habitat has the potential to affect sensitive or specialized species (New 1995; Freitas et al. 2006). Information on the ecology of most invertebrate groups is scarce. Thus, the only practical option for conserving diversity involves protecting as many environments as possible (New 1995, 1997). Insects respond to almost any kind of environmental change, regardless of its intensity, therefore they are considered the best indicators of their own conservation status (Freitas et al. 2006).
The order Hemiptera is currently classified in four suborders: Auchenorrhyncha, Coleorrhyncha, Sternorrhyncha and Heteroptera (Carver et al. 1991; Forero 2008). Regarding the last suborder, the family Pentatomidae is the fourth biggest and most diverse, being composed by over 4.700 species distributed in around 900 genera (Schuh & Slater, 1995; Grazia and Fernandes 2012; Rider 2011).
The Family Pentatomidae can be considered, among Hemiptera, one of the most appropriate insect groups for monitoring environmental changes in Brazilian Atlantic Forest. They represent an efficient system for observing changes, due to sensitivity to humidity, availability of nutrients and specific plant growth cycles and their chemical compounds (Brown Jr 1997).
Loxa Amyot & Serville is a genus of insects within the family Pentatomidae that occurs mainly in Central America and Mexico, but is also found in Texas, Florida, and South America. Some of them are pests of small cultures, as Loxa deducta Walker, 1867. However, most species are phytophagous which do not cause significant economic and ecological vegetal damages, thus not being considered pests (Panizzi et al. 2000). A few examples are Loxa viridis (Palisot de Beauvois, 1805) and Loxa virescens Amyot and Serville, 1843.
Partial mitochondrial DNA sequences, such as cytochrome c oxidase subunit I (COI), have been used to identify new species. Several studies show that a 648 bp fragment of COI gene might be used as a “barcode” in the identification process (Hebert et al. 2004a; Ward et al. 2005). COI 5′ extremity works as a molecular barcode, in which animal species might be distinguished without morphological identification keys (Park et al. 2011). Using a small-standardized DNA fragment, Barcode DNA might be considered a promising tool to diagnose diversity, demonstrating nucleotide diversity and even highlighting interspecific geographical barriers.
Despite the ecological importance and adaptability of Pentatomidae, advanced molecular studies can elucidate its biodiversity and population dynamics. Besides, they can provide valuable information for conservation planning for several other organisms, especially for species that live in conservation areas. Thus, this paper aimed to characterize the population structure of two species of the suborder Heteroptera: L. viridis and L. virescens collected at INP and highlight the importance of monitoring biodiversity in conservation units.
Material and methods
Samples and collection sites
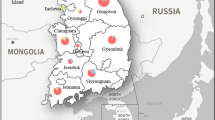
Samples were collected in the Iguaçu National Park (INP), located in the west of Paraná state, Brazil (25°27′37.6”S 53°49′04.2”W). The collection sites were: (1) tourist trails near to the Poço Preto research base (25°36′33.2”S 54°25′51.3”W), Foz do Iguaçu, considered a preserved region within the park; (2) tourist trails and closed forest region around the information and post control, Céu Azul (25°07′15.1”S 53°48′21.5”W), that although it is inserted in the conservation area, but it is close to a highway and near to the urban center of Céu Azul city; (3) dense forest region in Santa Tereza do Oeste (25°06′18.5”S 53°41′04.2”W) (Fig. 1). The specimens were collected manually and by light trap. Thirty individuals were collected from the Pentatomidae family: 15 individuals of L. viridis (5 in each collection site) and 17 of L. virescens (5 in collection site 1, 6 in both collection sites 2 and 3). To identify the specimens, Eger (1978) identification key was used.
DNA extraction, amplification, and electrophoresis
Total DNA extraction from the whole individual was performed based on the phenol-chloroform-alcohol-isoamylic method (Suzuki et al. 2010). The extraction buffer, with 1% Sodium dodecyl sulfate (SDS), 200 mM Tris–HCl, 250 mM Sodium chloride (NaCl), 25 mM Ethylenediaminetetra-acetic acid (EDTA) pH 8, and 0.5 μl of proteinase K (20 μg / μL), was added and the samples were incubated at 62 °C by 2 h for tissue digestion.
Cytochrome c oxidase subunit I (COI) gene sequences were obtained by Polymerase Chain Reaction (PCR) using the primers LCO1490 (5’-GGTCAACA AATCATAAAGATATTG-3′) and HCO2198 (5′- TAAACTTCAGGGTGACCAAAAAATCA-3′) described by Folmer et al. (1994). In the PCR, 12.5 μL of GoTaq® Green MasterMix 2x, 2.5 μL of each primer (10 μM), DNA extracted at a concentration of less than 250 ng/μL and ultrapure water were used to complete the reaction to 25 μL. Thermal conditions of the PCR cycles included 60s denaturation at 94 °C, 60s annealing at 48 °C, 60s extension at 72 °C (25 cycles) and 6 min final extension at 72 °C. PCR products were visualized in 1% agarose gel electrophoresis stained with SYBR® Safe DNA Gel Stain (Invitrogen).
To guarantee the quality of the sequences obtained, the PCR products were purified using 7.5 M ammonium acetate and absolute ethanol, followed by centrifugation, with subsequent washing with 70% ethanol. After the purified fragments were eluted in 30 μl of ultrapure water.
Sequencing, alignment, and analysis of COI sequences
The purified PCR products were sequenced with BigDye Terminator (Applied Biosystems) following the manufacturer’s recommendations in a MegaBace™ 1000 Molecular Dynamics type sequencer (Amershan, Pharmacia Biotech) by the method of Sanger et al. (1977). The obtained sequence alignments were made using BioEdit Software version 7.0.5 (Hall 2005), using the ClustaIW tool (Thompson et al., 1994). The acquired sequences were compared to the ones available in GenBank, using TBlastx tool to confirm the amplified fragment. The relation between the haplotypes was inferred by constructing a haplotype network, created with the help of the Network Software (Bandelt et al. 1999). The neutrality tests (Tajima’s D) were calculated with DnaSP Software version 6 (Rozas et al. 2017). The analysis of molecular variance (AMOVA) was performed using Arlequin version 3.5 software and pairwise difference was applied (Excoffier and Lischer 2010). The populational structure was determined using Wright’s fixation index (FST, Wright 1921) and the genetic flow (Nm) was also obtained using Arlequin 3.5 (Excoffier and Lischer 2010).
L. viridis already had sequencing data deposited in GenBank for the COI region, which were used for comparing the sequences obtained in this study and confirmation of the morphological classification of the barcode tool. The 32 sequences obtained here were deposited in Genbank under the access numbers: KY022462 to KY022476 for L. viridis and MF537316 to MF537332 for L. virenscens (Table 1).
Results and discussion
In both studied species, fragments of approximately 660 bp were obtained, showing that the primers used, and the amplification methods were resolvable. The same conclusion was reached in the analysis of the mitochondrial COI gene of several species of butterfly, in which 650 bp of the same gene were also amplified (Hebert et al. 2004b). The diversity in the 650-bp nucleotide sequences was sufficient to discriminate closely related species of butterfly, demonstrating that analysis of COI gene can be effective in biodiversity and phylogeny studies.
The 32 COI sequences analyzed in this study were grouped into six haplotypes. It was possible to observe that the obtained haplotypes were exclusive to each collection point, and to each species (Fig. 2). For L. viridis populations, haplotypic diversity (Hd = 0.714) and nucleotide diversity (Pi = 0.002) results were almost like L. virescens populations (Hd = 0.706, Pi = 0.002). Tajima’s D neutrality test results (D = 1.631, L. viridis; D = 1.637, L. virescens) were not significant and were according to the model of neutral mutation (p > 0.10).
Haplotypes obtained in the evaluation of the 32 specimens of L. viridis and L. virescens. Six haplotypes can be observed; the red haplotypes are exclusive to collection point 1, while the blue ones are exclusive to collection point 2 and the yellow ones refer to exclusive haplotypes of collection point 3
Three segregating sites were observed in each species. For both species, the analysis of molecular variance (AMOVA, Fst = 1.000, p < 0.05) showed that variations among populations was 100%, while variations between populations were zero (Φst = 1.000; Φct = 0.000; Φsc = 0.000). This high Fst value evidences that there is a significant differentiation among populations. Gene flow was estimated and for both populations were Nm = 0.00. Garcia et al. (2003) reported that the differences in Triatoma infestans (Klug, 1834) were not necessarily related to gene flow in populations from different locations and could be the result of local selection pressure or genetic drift.
Maximum Likehood test analysis grouped the specimens by species into two large groups, showing that even though they are distinct species, L. viridis and L. virescens from the same collection points tend to be closer together, confirming the results of the haplotypes obtained (Fig. 3). We can observe that group 1 was formed only by L. virescens samples (MF537331 to MF537325); and group 2 was formed only by L. viridis samples (KY022470 to KY022476). There is a great difficulty in differentiating species of the genus Loxa, where they can often be classified as the same species.
Maximum Likelihood Tree for the COI-mtDNA of L. viridis and L. virescens. The sequences were identified by the GenBank accession number: KY022462 to KY022476 for L. viridis and MF537316 to MF537332 for L. virenscens. The colored squares represent the haplotypes showed in Fig. 2
Although the two groups formed were separated by species, within each group there was the formation of another 3 subgroups (Fig. 2), represented respectively by the same haplotypes observed in Fig. 3. The grouping of species in haplotypes coincide with the division of populations by each collection site. This shows that even separated into two large groups of different species (1 and 2), the collection sites location influences the characterization of haplotypes.
The same observation in respect of collection sites was made by Castanhole et al. (2013) for Rhagovelia zela (Drake, 1959), suggesting that the diversity may be due to the distance between collection sites, inducing differential selective pressure. However, additional studies are needed, as well as population analysis of L. viridis and L. virescens collected from other points, to determine whether the population is subjected to different selective pressures or whether diversification is a natural process.
We can conclude that the amplification of COI gene is an important tool in the identification of L. viridis and L. virescens, as well as showing how the populations of each collection point are suffering local selection pressure. For this reason, our results are important to suggest that conservation actions carried out in the Iguaçu National Park are contributing to the maintenance of biodiversity, due to the particularity of the populations of each collection site.
References
Bandelt H, Forster P, Rohl A (1999) Media-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16(1):37–48
Brown KS Jr (1997) Diversity, disturbance, and sustainable use of Neotropical forests: insects as indicators for conservation monitoring. J Insect Conserv 1:25–42
Carver M, Gross GF, Woodward TE (1991) Hemiptera. In: Naumann ID (ed) The insects of Australia: a textbook for students and research workers/ division of entomology, commonwealth scientific and industrial research organization, 2nd edn. Melbourne University Press & Cornell University Press, pp 429–509
Castanhole MMU, Marchesin SRC, Pereira LLV, Moreira FFF, Barbosa JF, Valério JR, Itoyama MM (2013) The first assess of the haplotypes from COI gene sequences in species of spittlebugs (Cicadomorpha: Hemiptera) and aquatic true bugs (Gerromorpha and Nepomorpha: Hemiptera) in Brazil. Genet Mol Res:5372–5381
D'Oliveira E, Bursztyn I, Badin L (2002) Parque Nacional do Iguaçu. Caderno Virtual de Turismo, Rio de Janeiro 2(4):1–10
Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour 10(3):564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34(1):487–515
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3(5):294–299
Forero D (2008) The systematics of the Hemiptera. Rev Colomb Entomol 34:1–21
Freitas AVL, Leal IR, Uehara-Prado M, Iannuzzi L (2006) Insetos como indicadores de conservação da paisagem. In: Rocha CFD, Bergallo HG, Van Sluys M, Alves MAS (eds) Biologia da conservação: essências. RiMa Editora, São Carlos, pp 357–384
Garcia BA, Manfredi C, Fichera L, Segura EL (2003) Short report: variation in mitochondrial 12S and 16S ribosomal DNA sequences in natural populations of Triatoma infestans (Hemiptera: Reduviidae). Am J Trop Med Hyg 68:692–694
Grazia J, Fernandes JAM (2012) Subordem Heteroptera. In: Rafael JA, Melo GAR, de Carvalho CJB, Casari SA, Constantino R. Insetos do Brasil: diversidade e taxonomia. Ribeirão Preto, Holos Editora, 810p
Hall T (2005) Bioedit v 7.0.5. Ibis therapeutics, a division of Isis Pharmaceuticals, Carlsbad. https://doi.org/10.12691/ajidm-4-3-3
Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004a) Identification of birds through DNA barcodes. PLoS Biol 2:e312
Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004b) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. PNAS USA 101(41):14812–14817
Kruess A, Tscharntke T (1994) Habitat fragmentation, species loss, and biological control. Science 264(5165):1581–1584
New TR (1995) An introduction to invertebrate conservation biology. Oxford University Press (no. 333.95516 N4)
New TR (1997) Are Lepidoptera an effective ‘umbrella group’ for biodiversity conservation? J Insect Conserv 1:5–12
Panizzi AR et al (2000) Stink Bugs (Pentatomidae). In: Schaefer CW, Panizzi AR (Org). Heteroptera of economic importance. Boca Raton: CRC Press, pp. 421–474
Park DS, Footit R, Maw E, Hebert PDN (2011) Barcoding bugs: DNA-based identification of the true bugs (Insecta: Hemiptera: Heteroptera). PLoS One 6(4):1–9
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM (2009) The Brazilian Atlantic Forest: how much is left, and how is the remaining forest distributed? Implications for conservation. Biol Conserv 142:1141–1153
Rider DA (2011) Pentatomoidea Home Page: number of genera & species of Pentatomidae. North Dakota: North Dakota State University. Accessed in July 20, 2020 em: https://www.ndsu.edu/pubweb/~rider/Pentatomoidea/Genus_Index/genus_index.htm
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J, Guirao-Rico S, Librado P, Ramos-Onsins S et al (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34(12):3299–3302. https://doi.org/10.1093/molbev/msx248
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci 74(12):5463–5467
Acknowledgments
The authors are grateful to the National Iguaçu Park staff members for their technical assistance and to Edson Mendes Francisco for his help with the sample collection. This work was supported by Universidade Estadual de Londrina (State University of Londrina).
Funding
This research was supported by State University of Londrina.
Author information
Authors and Affiliations
Contributions
Thayná Bisson Ferraz Lopes: Conceptualization, Formal analysis, Investigation, Writing - Original Draft, Visualization.
Felipe Cordeiro Dias: Investigation.
Joana Neres da Cruz Baldissera: Formal analysis, Investigation, Writing - Review & Editing.
Carlos Roberto Maximiano da Silva: Methodology, Formal analysis, Resources, Writing - Review & Editing.
José Antônio Marin Fernandes: Species identification.
Renata da Rosa: Conceptualization, Formal analysis, Methodology, Resources, Writing - Original Draft, Writing - Review & Editing, Supervision, Project administration.
Corresponding author
Ethics declarations
Conflicts of interest/competing interests
The authors declared that they have no conflict of interest.
Ethics approval
The researchers received permission (number 31946–4) from Instituto Chico Mendes de Conservação da Biodiversidade – ICMBio to collect insect specimens.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lopes, T.B.F., Dias, F.C., da Cruz Baldissera, J.N. et al. Genetic analysis in two species of Loxa Amyot & Serville 1843 (Pentatomidae) collected in Iguaçu National Park (Foz Do Iguaçu, Paraná, Brazil). Int J Trop Insect Sci 41, 759–764 (2021). https://doi.org/10.1007/s42690-020-00265-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42690-020-00265-x