Abstract
Although pineapple (Ananas comosus var. comosus) shoot tips have been cryopreserved but the possible effect of this process at the molecular level has not been studied. This communication describes the growth (plant fresh and dry weights; stem height; leaf length, width and area; and stem base diameter) and the Inter Simple Sequence Repeat (ISSR) analysis of pineapple plantlets of A. comosus MD-2; Red Spanish Florencia; and Hybrid 54 (Smooth Cayenne/Red Spanish) after 45 d of acclimatization. From each of these varieties, the acclimatized plants were obtained from: (1) conventional micropropagation (control 1); (2) from shoot tips submitted to pre-cryostorage conditioning treatments but not exposed to liquid nitrogen (LN) (treatment 2); and (3) from shoot tips exposed to cryostorage including use of LN (treatment 3). The ISSR-PCR method was used to study the genetic stability. There were no statistically significant differences between treatments for the phenotype indicators evaluated. On average, 45 day-old pineapple plants had 0.5 g fresh weight; 1.85 g dry weight; 12.2 cm stem height; 9.1 cm leaf length; 1.6 cm leaf width; 7.1 cm2 leaf area; and 1.4 cm stem base diameter. Also, the potential effects of cryopreservation at the DNA level were not revealed with the eight ISSR markers used, as no polymorphic bands were recorded, which represents 100% genetic stability. As far as we know, this is the first publication on ISSR analysis of pineapple plantlets after cryopreservation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pineapple is an important tropical fruit (Chen et al. 2019). Pineapple production in 2019 worldwide was around 1,250,00 ha with gross production value around US$ 12 b (FAOSTAT, 2021), indicating that this is a profitable crop. Pineapple is vegetatively propagated and in order to support future production, existing high performance varieties need to be stored for long periods. Therefore, long-term conservation of pineapple genetic resources using cryopreservation storage in liquid nitrogen (LN) should be encouraged (Villalobos-Olivera et al. 2019). As pineapple production increases and gets more intensive, it is threatened by several factors, including microbial pathogens. To support the continued success of pineapples, new varieties with increased microbial pathogen resistance and abiotic stress tolerance should be developed (Yabor et al. 2020). Conservation of pineapple genetic resources can therefore also aid future breeding programs.
Cryopreservation using shoot tips is carried out for many plant species (Engelmann and Ramanatha, 2012), including pineapple (Martínez-Montero et al. 2012; Souza et al. 2015, 2018; Villalobos-Olivera et al. 2019). However, there is no literature on the genetic stability of pineapple-regenerated plantlets following cryopreservation using LN. LN may affect subsequent plant growth and should be studied for its effect on DNA stability before large-scale usage in constructing a cryobank.
The inter simple sequence repeat (ISSR)-PCR method was used to study DNA stability. ISSR is an inter-microsatellite sequence that generates highly polymorphic multilocus markers. The advantage of this method is that it is simple, quick and does not require prior knowledge of the genome, thus combining most of the advantages of other possible methods like microsatellites (SSRs), amplified fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) (Kaya 2016). ISSR markers produce many bands (polymorphic) useful in studies on genetic diversity, phylogeny, gene tagging, genome mapping, and evolutionary biology and can be used to identify mutations of in vitro grown material (Reddy et al. 2002).
ISSR has been recently used to study DNA of different plant species, such as sugarcane (Shingote et al. 2019), Nilgirianthus ciliatus (Rameshkumar et al. 2019) and Polianthes tuberosa L. (Nalousi et al. 2019). These markers have also been employed to study genetic stability of pineapple not exposed to LN (Carlier et al. 2004; Tapia et al. 2002; Vanijajiva 2012) and cryopreserved materials of other plant species (Atmakuri et al. 2009; Espasandin et al. 2019; Lambardi et al. 2004; Liu et al. 2008; Rao et al. 2007; Yamuna et al. 2007). In these studies, genetic differences caused by cryopreservation were not reported.
This communication describes the growth variables and the inter simple sequence repeat (ISSR) analysis of pineapple plantlets of three cultivars after 45 days of acclimatization. From each of these cultivars, the acclimatized plants were obtained from: (1) conventional micropropagation (control 1); (2) from shoot tips submitted to pre-cryostorage conditioning treatments but not exposed to LN (treatment 2); and (3) from shoot tips exposed to cryostorage including use of LN (treatment 3).
Materials and methods
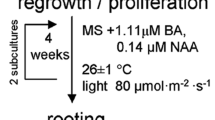
This research was based on the pineapple micropropagation protocol established by Daquinta and Benegas (1997) as described in Gomez et al. (2017), the acclimatization according to Pino et al. (2014), and the droplet-vitrification technique developed by Souza et al. (2015) and Villalobos-Olivera et al. (2019). Pineapple buds (cvs. MD-2, Red Spanish Florencia, Hybrid 54 (Smooth Cayenne/Red Spanish) were initiated as described in Gomez et al. (2017). In summary, the fruit and leaves were removed from pineapple crowns, which were then sterilized with 1% (w:v) Ca(ClO)2 for 10 min and the buds excised (Daquinta and Benegas 1997). These bud explants were placed in 300 ml glass jars with 5 ml liquid culture medium (one explant per jar). MS (Murashige and Skoog 1962) salts, 100 mg l−1 myo-inositol, 0.1 mg l−1 thiamine-HCl, 30 g l−1 sucrose, 4.4 µM 6-benzyladenine (BA), and 5.3 µM naphthaleneacetic acid (NAA) were included in the initiation medium (MS1). After 45 days on liquid initiation medium, shoots were transferred to multiplication medium (as described above except it had 9.3 µM BA and 1.6 µM NAA and 2 g/l gelrite (MS2). At 45-days intervals, shoots were subcultured for 6 months (referred to as the control—C1). Nodal propagules were placed on rooting medium, MS medium without growth regulators (MS3) for 4 weeks and then were hardened.
Three treatments were compared: (1) conventional micropropagation (control as summarized above); (2) plants from shoot tips that were submitted to pre-cryostorage conditioning treatments but not LN (treatment 2); and (3) plants from shoot tips exposed to treatment 2 and LN (treatment 3). Treatments 2 and 3 are similar to Villalobos-Olivera et al. (2019) and are summarized below. Rooting medium, MS medium without growth regulators (MS3) was the same for all three methods and was used for 45 d (Daquinta and Benegas 1997).
Treatment 2: The droplet-vitrification technique was used (Villalobos-Olivera et al. 2019). In it, the shoot tips (1 mm long) excised from in vitro pineapple plantlets after incubation for 24 h in MS with 2.0 M glycerol and 0.4 M sucrose (MS4) were transferred to poly-propylene cryovials (volume: 2 ml; shoot tips/vial: 10) containing 1 ml MS4; these were incubated for 20 min at 25 ± 2 °C. Shoot tips were transferred to pieces of aluminum foil (5 shoot tips/piece) containing micro-drops (0.1 ml) of PVS3 solution (pre-cryostorage treatment) for 60 min. The aluminum foils containing the shoot tips were placed in a tin containing ice cubes for 1 h. Shoot tips were placed in the regeneration medium, MS1 (Daquinta and Benegas 1997). Plantlets were hardened for 45 days.
Treatment 3: The pre-cryostorage conditioning treatment was carried out as for treatment 2. The aluminum foils containing the shoot tips on ice for 1 h were then transferred to 2-ml cryovials and immersed in LN for 24 h. Shoot tips were recovered at room temperature by discarding the PVS3 solution and replacing it with MS5 medium (MS + 1 M sucrose, 1 ml, 25 ± 2 °C, 20 min). They were then transferred to MS1 medium (same as initiation medium) to recover plantlets from shoot tips.
Plantlets from MS1 (taken from the three treatments compared) were rooted on MS medium (MS3) transferred for the hardening stage (Yanes-Paz et al. 2000). Rooted plantlets of at least 5 cm height, 5–8 leaves, 4 roots and 4.5 g were removed from MS3 medium. The acclimatization trial, following a completely randomized design, included four reps (15 plants each) per treatment (3) and genotype (3). Plastic trays with 4 holes of 0.5 cm Ø for drainage contained 82 cm3 red ferric soil and filter cake (1:1) per plant. Microject sprayed automated irrigation for 25 s every 30 min was applied (45 days). The photosynthetic photon flux density was 458 μmol m−2 s−1 for 45 days. Chemical fertilizers were not used. The experiment was repeated twice. At 45 days after transfer from culture, the following indicators were recorded: fresh and dry weights per plantlet; stem height; D leaf (middle-aged leaf) (Ebel et al.2016) length, width and area; and diameter of stem base. SPSS (Version 8.0 for Windows, SPSS Inc., New York, NY) was used to perform ANOVA (p = 0.05).
DNA isolation was carried out following Kobayashi et al. (1998) with several modifications established by Yanes-Paz et al. (2012). Leaf samples (250 mg) taken from the 45 days old hardened plants were macerated in 200 ml LN. The powder was re-suspended in 650 μl extraction buffer (50 mmol l−1 Tris–Cl, pH 7.5; 20 mmol l−1 EDTA, pH 8.0; 0.3 mmol l−1 NaCl; 2% (v/v) sarcosil; 0.5% (v/v) sodium dodecyl sulphate; and 4.8 mol l−1 urea). Then, an equal volume (650 μl) of phenol–chloroform-isoamilic alcohol (25:24:1, v:v:v) was added and mixed by inverting the tube several times. The mixture was centrifuged at 28,241×g for 15 min at room temperature in a tabletop centrifuge (Tyfon II PRO R from the Republic of Argentina). DNA in the liquid phase was precipitated by the addition of 0.8% (v/v) isopropanol at room temperature, centrifuged for 10 min at 11,854g (RCF), then the pellet was washed with 70% ethanol (v/v) and the DNA re-suspended in 50 μl DNase-free water, containing 10 μg ml−1 RNAse A. The quality and integrity of the DNA were checked by electrophoresis on a 0.8% (w/v) agarose gel. In addition, both parameters were checked by spectrophotometric analysis.
The amplifications were developed in an Applied Biosystems® Veriti® 96-Well Fast Thermal Cycler under the following conditions: 3 min at 94 °C, followed by 39 cycles 94 °C for 45 s, 48 °C for 45 s then 72 °C for 1 min. The final extension was developed at 72 °C and 14 °C for 7 min each. The amplification products were first separated in a gel with 2% (w/v) of ultra-pure agarose to check for PCR products. Fluorescence-based ISSR analyzes of capillary electrophoresis were conducted on an ABI PRISM® 3130 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The primers in the sequence direction were labeled with FAM (blue) or HEX (green), and the Gen Scan TM 500 ROX TM standard (red). To estimate the size of the variant, Peak Scanner software (Applied Biosystems, version 1.0) was used. In the Table 1 show the eight ISSR primers were tested hybridization temperature.
Results and discussion
Statistically significant differences (ANOVA, p > 0.05) for the phenotypic indicators evaluated (Table 2, Fig. 1). In general, 45 day-old pineapple plants averaged 10.5 ± 1.1 (SE) g fresh weight; 1.85 ± 0.9 g dry weight; 12.2 ± 1.2 cm stem height; 9.1 ± 1.0 cm leaf length; 1.6 ± 0.8 cm leaf width; 7.1 ± 0.5 cm2 leaf area; and stem base diameter of 1.4 ± 0.4 cm.
Typical phenotype of plantlets at 45 days of acclimatization. Statistically significant differences (ANOVA, p > 0.05, data not shown). Average information: 10.5 g fresh weight per plantlet; 1.85 g dry weight per plantlet; 12 cm stem height; 9.1 cm D leaf length; 1.6 cm D leaf width; 7.1 cm2 D leaf area; and 1.4 cm diameter of steam base. (ABC) cv. MD-2, (DEF) Red Spanish Florencia, (GHI) Hybrid 54 (Smooth Cayenne/Red Spanish) (ADG), conventional micropropagation—derived plants (C1: control) (BEH) Shoot tips never exposed to LN (−NL) (C2: control), (CFI) Shoot tips exposed to LN (LN+). The line is equivalent to 1 cm
The plants of the three genotypes show an expanded foliar and root system, which leads to adaptation of ex vitro acclimatization condition. The phenotypic characteristics obtained by pineapple plants from cryopreserved shoot tips at 45 days of acclimatization correspond to the results of Aragón et al. (2013), Villalobos et al. 2012 and Pino et al. (2014). This result demonstrates stability in the morphological characters of the plants of the three cultivars in acclimatization (Fig. 1).
The phenotypic characteristics of the plants show de efficient recovery after applying the cryopreservation technique. The cryopreservation induces stress to the pineapple shoot tips, with repercussion on the morphological development of regenerated plants during in vitro and ex vitro conditions (Martínez-Montero et al. 2012). The correct pre-conditioning of donor in vitro plants generate an efficient regeneration of cryopreserved shoot tips during conditions in vitro and ex vitro (Villalobos-Olivera et al. 2019). Process guarantees the phenotypic stability by the plants of the three genotypes after 45 days of ex vitro acclimatization.
Importantly, the possible effects of cryopreservation were also not evident at the DNA level as there was no sign of polymorphism using the ISSR markers (Fig. 2). There was no difference in band pattern among the pre-treated or cryo-preserved shoot tips and control plants as shown by the absence of polymorphic bands with all the markers tested (Fig. 2).
Inter simple sequence repeats (ISSR) observed in plantlets at 45 days of acclimatization. ISSR markers: 31 ISSR TriCAC5′CR, 32 r ISSR TriCAC5′CY, 34 ISSR TriCAG3′RC, 37 ISSR TriCAG5′CY, 47 ISSR TriTGT5′CY, 57 ISSR TriACC3′RC 57, 72 ISSR TriTCC3′RC, and 92 ISSR TriGAC3′RC. (A) cv. MD-2, (B) Red Spanish Florencia, (C) Hybrid 54 (Smooth Cayenne/Red Spanish) (1), Conventional micropropagation—derived plants (C1: control) (2) Shoot tips never exposed to LN (−NL) (C2: control), (3) Shoot tips exposed to LN (LN+). Polymorphic bands were not recorded which represented 100% integrity. Potential effects of cryopreservation at DNA level were not revealed with the ISSR markers used
Cryopreservation seems to be genetically stable. Cryopreserved shoot tips of plantain (Agrawal et al. 2014), potato (Wang et al. 2014a, b) and sugarcane (Kaya and Souza 2017) tested by use of SSR and ISSR markers, respectively, showed no differences compared to controls.
Cryopreservation is used to break the dormancy of recalcitrant species, such as Teramnus labialis (Acosta et al. 2019) and Neonotonia wightii (Acosta et al. 2020), without showing genetic variations (Matsumoto et al. 2015). LN has also been employed in cryotherapy to clean plant materials (banana sucker meristems) of pathogens, and has not affected their genetic stability (SSR markers) (Wang et al. 2014a, b). Also, ISSR markers have been used to confirm genetic stability after exposure to LN in citrus (Lambardi et al. 2004), Morus species (Rao et al. 2007), ginger (Yamuna et al. 2007), apple (Liu et al. 2008) and mulberry (Espasandin et al. 2019).
Contrasting to our results (Fig. 1), some articles claim that cryopreservation can introduce variations in the genome of plant material. These are Channuntapipat et al. (2003) in Prunus dulcis (Mill), DeVerno et al. (1999) in Picea glauca (Moench) Voss, Kaity et al. (2008) in Carica papaya L, and Johnston et al. (2009) in Ribes rubrum L. These authors cryopreserved unorganized tissues (callus and cell suspensions) and measured genetic variation with microsatellites. These changes are probably due to the use of poorly organized tissues such as callus or cell suspensions.
The genetic stability expressed by the plants is of great importance for the cryopreservation an micopropagation of the pineapple crop. The result for the first time that the process from pre-conditioning donor plants in vitro to the exposure in LN does not induce polymorphic variations plants. It also corresponds to what is established by (Kaya and Souza 2017), the cryopreservation does not induce genetic variations in the regeneration and adaptation of vegetable material.
The primers used in the research are of stable response and do not allow the loss of genetic information of the analyzed DNA fragment (da Silva et al., 2016). The characteristics of representing stability or polymorphic variation in DNA, allows the reliability of the results (Souza et al. 2017). In addition, its characteristics are not affected by the development stages of the plants (Silva et al. 2019).
Our results indicate that the cryopreservation procedure, especially the shoot tip exposure to LN did not alter the phenotype or genotype of three important pineapple cultivars, 45 days after transfer from tissue culture. These results support cryopreservation as an important tool for conservation of pineapple germplasm.
References
Acosta Y, Hernández L, Mazorra C, Quintana N, Zevallos BE, Cejas I, Fontes D (2019) Seed cryostorage enhances subsequent plant productivity in the forage species Teramnus labialis (L.F.) Spreng. CryoLetters 40:36–44
Acosta Y, Santiago F, Escalante D, Mazorra C, Cejas I, Martínez-Montero ME, Fontes D (2020) Cryo-exposure of Neonotonia wightii Wigth & Am seeds enhances field performance of plants. Acta Physiol Plant. https://doi.org/10.1007/s11738-019-3010-y
Agrawal A, Sanayaima R, Singh R, Tandon R, Verma S, Tyagi R (2014) Phenotypic and molecular studies for genetic stability assessment of cryopreserved banana meristems derived from field and in vitro explant sources. In Vitro Cell Dev Biol Biol Plant 50:345–356. https://doi.org/10.1007/s11627-014-9606-4
Aragón C, Pascual P, González J, Escalona M, Carvalho L, Amancio S (2013) The physiology of ex vitro pineapple (Ananas comosus L. Merr. var MD-2̕′) as CAM or C3 is regulated by the environmental conditions: proteomic and transcriptomic profiles. Plant Cell Rep 32:1807–1818. https://doi.org/10.1007/s00299-013-1493-3
Atmakuri AR, Chaudhury R, Malik S, Kumar S, Ramachandran R, Qadri S (2009) Mulberry biodiversity conservation through cryopreservation. In Vitro Cell Dev Biol 45:639. https://doi.org/10.1007/s11627-008-9185-3
Carlier JD, Reis A, Duval MF, D’Eeckenbrugge CG, Leitão JM (2004) Genetic maps of RAPD, AFLP and ISSR markers in Ananas bracteatus and A. comosus using the pseudo-testcross strategy. Plant Breed 123:186–192. https://doi.org/10.1046/J.1439-0523.2003.00924.X
Channuntapipat C, Sedgley M, Collins G (2003) Changes in methylation and structure of DNA from almond tissues during in vitro culture and cryopreservation. J Am Soc Hortic Sci 128:890–897. https://doi.org/10.21273/JASHS.128.6.0890
Chen H, Hu B, Zhao L, Shi D, She Z, Huang X, Qin Y (2019) Differential expression analysis of reference genes in pineapple (Ananas comosus L.) during reproductive development and response to abiotic stress. Trop Plant Biol 12:1–11. https://doi.org/10.1007/s12042-019-09218-2
da Silva RL, Ferreira CF, da Silva Ledo CA, de Souza EH, da Silva PH, de Carvalho Costa MAP, Souza FVD (2016) Viability and genetic stability of pineapple germplasm after 10 years of in vitro conservation. Plant Cell Tissue Organ Cult 127:123–133
Daquinta M, Benegas R (1997) Brief review of tissue culture of pineapple. Pineap News 3:7–9
DeVerno L, Park Y, Bonga J, Barrett J, Simpson C (1999) Somaclonal variation in cryopreserved embryogenic clones of white spruce [Picea glauca (Moench) Voss.]. Plant Cell Report 18:948–953. https://doi.org/10.1007/s002990050689
Ebel AI, Giménez L, González AM, Alayón Luaces P (2016) Evaluación morfoanatómica de hojas" D" de piña (Ananas comosus (L.) Merr.) en respuesta a la implantación de dos sistemas de cultivo en Corrientes. Acta Agron 65:390–397. https://doi.org/10.15446/ACAG.V65N4.50560
Engelmann F, Ramanatha R (2012) Major research challenges and directions for future research. In: Normah MN, Chin HF, Reed BM (eds) Conservation of tropical plant species. Springer, Berlin, pp 511–526. https://doi.org/10.1007/978-1-4614-3776-5_20
Espasandin FD, Brugnoli EA, Ayala PG, Ayala LP, Ruiz OA, Sansberro PA (2019) Long-term preservation of Lotus tenuis adventitious buds. Plant Cell Tissue Organ Cult 136:373–382. https://doi.org/10.1007/s11240-018-1522-6
FAOSTAT. (2021) FAO Statistics Division [online]. Disponible. http://faostat.fao.org/site/567/. Accessed 9 Oct 2021
Gómez D, Hernández L, Valle B, Martínez J, Cid M, Escalona M, Lorenzo JC (2017) Temporary immersion bioreactors (TIB) provide a versatile, cost-effective and reproducible in vitro analysis of the response of pineapple shoots to salinity and drought. Acta Physiol Plant 39:277
Johnston JW, Benson EE, Harding K (2009) Cryopreservation induces temporal DNA methylation epigenetic changes and differential transcriptional activity in Ribes germplasm. Plant Physiol Biochem 47:123–131. https://doi.org/10.1016/j.plaphy.2008.10.008
Kaity A, Ashmore S, Drew R, Dulloo M (2008) Assessment of genetic and epigenetic changes following cryopreservation in papaya. Plant Cell Report 27:1529–1539. https://doi.org/10.1007/s00299-008-0558-1
Kaya E (2016) ISSR analysis for determination of genetic diversity and relationship in some Turkish olive (Olea europaea L) cultivars. Not Bot Hortic Agrobot Lujapoca 43:96–99. https://doi.org/10.15835/nbha4319818
Kaya E, Souza FVD (2017) Comparison of two PVS2-based procedures for cryopreservation of commercial sugarcane (Saccharum spp.) germplasm and confirmation of genetic stability after cryopreservation using ISSR markers. In Vitro Cell Dev Biol Plant 53:410–417. https://doi.org/10.1007/s11627-017-9837-2
Kobayashi T, Heck DJ, Nomura M, Horiuchi T (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12:3821–3830. https://doi.org/10.1101/gad.12.24.3821
Lambardi M, De Carlo A, Biricolti S, Puglia A, Lombardo G, Siragusa M, De Pasquale F (2004) Zygotic and nucellar embryo survival following dehydration/cryopreservation of Citrus intact seeds. CryoLetters 25:81–90
Liu Y-G, Liu L-X, Wang L, Gao A-Y (2008) Determination of genetic stability in surviving apple shoots following cryopreservation by vitrification. CryoLetters 29:7–14
Martínez-Montero M, Engelmann F, González-Arnao M (2012) Cryopreservation of tropical plant germplasm with vegetative propagation-review of sugarcane (Saccharum spp.) and pineapple (Ananas comusus (L.) Merrill) cases. In: Katkov I (ed) Current frontiers in cryopreservation. London England, pp 260–396. https://doi.org/10.5772/32047
Matsumoto T, Yamamoto S, Fukui K, Rafique T, Engelmann F, Niino T (2015) Cryopreservation of persimmon shoot tips from dormant buds using the D cryo-plate technique. Hortic J 84:106–110. https://doi.org/10.2503/HORTJ.MI-043
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 5:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Nalousi AM, Hatamzadeh A, Azadi P, Mohsenpour M, Lahiji HS (2019) A procedure for indirect shoot organogenesis of Polianthes tuberosa L. and analysis of genetic stability using ISSR markers in regenerated plants. Sci Hortic 244:315–321. https://doi.org/10.1016/J.SCIENTA.2018.09.066
Pino Y, Concepción O, Santos R, González J, Rodríguez R (2014) Effect of Previcur® energy fungicide on MD-2 pineapple (Ananas comosus var. comosus) plantlets during the acclimatization phase. Pineapple News 1:1–24
Rameshkumar R, Pandian S, Rathinapriya P, Selvi CT, Satish L, Gowrishankar S, Ramesh M (2019) Genetic diversity and phylogenetic relationship of Nilgirianthus ciliatus populations using ISSR and RAPD markers: implications for conservation of an endemic and vulnerable medicinal plant. Biocatal Agric Biotechnol 18:101072. https://doi.org/10.1016/J.BCAB.2019.101072
Rao AA, Chaudhury R, Kumar S, Velu D, Saraswat R, Kamble C (2007) Cryopreservation of mulberry germplasm core collection and assessment of genetic stability through ISSR markers. Int J Ind Entomol 15:23–33
Reddy MP, Sarla N, Siddiq E (2002) Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128:9–17. https://doi.org/10.1023/A%3A1020691618797
Shingote PR, Mithra SA, Sharma P, Devanna NB, Arora K, Holkar SK, Sharma T (2019) LTR retrotransposons and highly informative ISSRs in combination are potential markers for genetic fidelity testing of tissue culture-raised plants in sugarcane. Mol Breed 39:25. https://doi.org/10.1007/s11032-019-0931-5
Silva JM, Lima PR, Souza FV, Ledo CA, Souza EH, Pestana KN, Ferreira CF (2019) Genetic diversity and nonparametric statistics to identify possible ISSR marker association with fiber quality of pineapple. An Acad Bras Ciênc 91:144–154
Souza F, Kaya E, de Jesus VL, de Souza E, de Oliveira AV, Skogerboe D, Jenderek M (2015) Droplet-vitrification and morphohistological studies of cryopreserved shoot tips of cultivated and wild pineapple genotypes. Plant Cell Tissue Organ Cult 124:351–360. https://doi.org/10.1007/s11240-015-0899-8
Souza CPF, Ferreira CF, de Souza EH, Neto ARS, Marconcini JM, da Silva Ledo CA, Souza FVD (2017) Genetic diversity and ISSR marker association with the quality of pineapple fiber for use in industry. Ind Crops Prod 104:263–268
Souza FVD, de Souza EH, Kaya E, de Jesus VL, da Silva RL (2018) Cryopreservation of pineapple shoot tips by the droplet vitrification technique. Plant Cell Cult Protocols 124:269–277. https://doi.org/10.1007/s11240-015-0899-8
Tapia C, Gutiérrez G, Warbourton L, Uriza A, Rebolledo M (2002) Characterization of pineapple germplasm (Ananas spp.) by mean AFLPs. In: IV international pineapple symposium 666, pp 109–114
Vanijajiva O (2012) Assessment of genetic diversity and relationships in pineapple cultivars from Thailand using ISSR marker. J Agric Technol 8:1829–1838
Villalobos A, González J, Santos R, Rodríguez R (2012) Morpho-physiological changes in pineapple plantlets (Ananas comosus (L.) Merr.) during acclimatization. Ciência e Agrotecnol 36:624–630. https://doi.org/10.1590/S1413-70542012000600004
Villalobos-Olivera A, Martínez J, Quintana N, Zevallos BE, Cejas I, Lorenzo JC, Montero MEM (2019) Field performance of micropropagated and cryopreserved shoot tips-derived pineapple plants grown in the field for 14 months. Acta Physiol Plant 41:34. https://doi.org/10.1007/s11738-019-2825-x
Wang B, Li JW, Zhang ZB, Wang RR, Ma YL, Blystad DR, Wang QC (2014a) Three vitrification-based cryopreservation procedures cause different cryo-injuries to potato shoot tips while all maintain genetic integrity in regenerants. J Biotechnol 184:47–55. https://doi.org/10.1016/j.jbiotec.2014.04.021
Wang B, Wang RR, Cui ZH, Bi WL, Li JW, Li BQ, Wang QC (2014b) Potential applications of cryogenic technologies to plant genetic improvement and pathogen eradication. Biotechnol Adv 32:583–595. https://doi.org/10.1016/j.biotechadv.2014.03.003
Yabor L, Pérez L, Gómez D, Villalobos-Olivera A, Mendoza JR, Martínez J, Lorenzo JC (2020) Histological evaluation of pineapple transgenic plants following eight years of field growth. Euphytica 216:1–8. https://doi.org/10.1007/s10681-020-2555-6
Yamuna G, Sumathi V, Geetha S, Praveen K, Swapna N, Nirmal Babu K (2007) Cryopreservation of in vitro grown shoots of ginger (Zingiber officinale Rosc.). CryoLetters 28:241–252
Yanes-Paz E, González J, Sánchez R (2000) A technology of acclimatization of pineapple vitroplants. Pineap News 7:24
Yanes-Paz E, Gil K, Rebolledo L, Rebolledo A, Uriza D, Martínez O, Simpson J (2012) Genetic diversity of Cuban pineapple germplasm assessed by AFLP markers. Crop Breed Appl Biotechnol 12:104–110. https://doi.org/10.1590/S1984-70332012000200002
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Villalobos-Olivera, A., Ferreira, C.F., Yanes-Paz, E. et al. Inter simple sequence repeat (ISSR) markers reveal DNA stability in pineapple plantlets after shoot tip cryopreservation. Vegetos 35, 360–366 (2022). https://doi.org/10.1007/s42535-021-00327-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42535-021-00327-6