Abstract
Cervical cancer, the fourth most common female cancer, is one of the major global health challenges. Because of the high prevalence rate, women are frequently screened for cervical cancer using Pap smears; however, the low sensitivity of Pap smear results in a late-stage diagnosis of cervical cancer. This study was designed to identify candidate biomarkers of cervical cancer, which can be explored for their utility as diagnostic markers and/or therapeutic targets. Cervical cancer and healthy tissue proteins were identified by tandem mass spectrometry (LC–MS/MS) and quantified by a label-free method. Candidate biomarkers were selected by applying stringent statistical methods. A total of 201 differentially expressed proteins were identified, which displayed statistical power to classify cervical cancer and control groups. The top proteins with higher abundance in cervical cancer included integrin beta-2, catenin alpha-1, stathmin, and tax1-binding protein 3; while the top proteins with higher abundance in healthy tissue samples included alpha-2-HS-glycoprotein, serotransferrin, afamin, hemopexin, plasminogen, apolipoprotein A-II, and immunoglobulin lambda variable 1–51. Stathmin and afamin expressions were also validated by western blot analysis. Upregulated proteins in cervical cancer were mainly associated with cell adhesion, protein folding, inflammatory disorders, and tumorigenesis. The KEGG pathways over-represented in cervical cancer were antigen processing and presentation, cell adhesion, spliceosome, adherens junction, and ferroptosis. Applying stringent statistical selection criteria, 46 DEPs were selected as candidate biomarkers of cervical cancer, which can be explored for their utility as diagnostic markers and/or therapeutic targets of cervical cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With more than 600 thousand new cases and 342 thousand deaths globally in 2020, cervical cancer is the 4th most common female malignancy and is considered as one of the major global health challenges (Sung et al. 2021). Of these, almost 90% of deaths occurred in low- and middle-income countries. Compared to developed regions of the world, mortality rate was 18 times higher in sub-Saharan Africa (Sung et al. 2021). In India, it is the second most common cancer among women and more than 450 million women aged 15 years and above are at risk of developing this cancer (Singh et al. 2022). Because of the high prevalence rate, women between the ages of 25 and 65 are frequently screened for cervical cancer using Pap smears; however, the low sensitivity of Pap smears can result in diagnosis of cervical cancer at a late stage and delayed diagnosis is responsible for the poor survival rate among cervical cancer patients (Einstein et al. 2009). Additionally, the non-availability of screening facilities in many regions is another challenge. Though significant advances are being made in the field, better diagnosis and prevention strategies are needed as cervical cancer can be cured if detected in the initial stages and treated promptly. About 95% of cervical cancer cases are associated with high-risk human papillomavirus (HPV) infections, an extremely common virus usually transmitted through sexual contact (Clifford et al. 2003). Among the high-risk strains, HPV types 16 and 18 are involved in 70% of the cases (Demarco et al. 2020). Therefore, risk factors of cervical cancer are primarily those associated with acquiring HPV infection, severely impaired immune response to HPV infection, or both (Grulich et al. 2007). An early age of sex debut, having multiple sex partners, giving many births, use of oral contraceptives for more than five years, immunosuppression, history of sexually transmitted infection or HPV-related vulvar or vaginal dysplasia, and smoking increase the risk of HPV infection. HPV infections initiate the development of high-grade cervical intraepithelial neoplasia (CIN), categorized as CIN I, II, and III based on cell abnormality levels, which may eventually lead to cervical cancer (Nguyen et al. 2005). Cancer treatment plans are highly focused and precisely aided by multiple diagnostic tests, such as CT scans, ultrasound, MRI, and biopsy along with physical examination used for detection and monitoring of the disease.
Despite significant progress in cancer screening technology, the cervical cancer morbidity rate remains high. The high morbidity and poor prognosis of cervical cancer transpire the need for molecular investigations to achieve a better understanding of the disease mechanism by identifying early screening molecular markers and novel and effective molecular targets for the treatment. In the last two decades, high-throughput proteomics technology has evolved significantly and applied to identify differentially expressed proteins (DEPs), involved in multifaceted biological functions associated with disease pathogenesis including tumorigenesis, as therapeutic targets and/or potential disease biomarkers. Identification of novel cancer biomarkers with high sensitivity and specificity for early detection can be useful to elucidate the underlying molecular events associated with the progression of cancer (Suh et al. 2010). In this direction, blood and tissue have been widely used for searching biomarkers of different cancers (Feng et al. 2006). For example, Annexin A2 (ANXA2) demonstrated as a factor linked to cell transformation and oncogenesis in cervical cancer was identified by two-dimensional gel electrophoresis (2-DE) and MALDI-TOF mass spectrometric analysis (Bae et al. 2005). An iTRAQ-based quantitative proteomics study identified 294 DEPs in cervical cancer tissue samples, including glucose-6-phosphate 1-dehydrogenase (G6PD), aldehyde dehydrogenase (ALDH3A1), signal transducer and activator of transcription 1-alpha/beta (STAT1), and heat shock protein beta-1 (HSPB1) (Ding et al. 2015). Another study by Ramirez-Torres et al. identified 622 DEPs in cervical cancer tissue samples and found that exocytosis-related proteins were the most over-represented (Ramirez-Torres et al. 2022). Based on their findings and downstream in vitro and in vivo analyses, they suggested reticulocalbin 3 (RAB14) and Ras-related protein Rab-14 (RCN3) proteins as potential biomarkers and therapeutic targets of cervical cancer. Recently, Aljawad et al. identified 336 DEPs in cervical cancer, including importin subunit alpha-1 (KPNA2), DNA replication licensing factor MCM2 (MCM2), collagen type I alpha-1 (COL1A1), and decorin (DCN) (Aljawad et al. 2022).
Despite a plethora of ongoing studies worldwide, cervical cancer prognosis and diagnosis remain a matter of concern for researchers globally and thus, entails an in-depth study of this disease at the molecular level. This study was envisioned to study cervical cancer tissue proteins for searching candidate biomarkers for cervical cancer. For this, cervical cancer and healthy tissue proteins were identified and quantified using tandem mass spectrometry (LC–MS/MS) and candidate biomarkers were selected applying stringent statistical methods. To understand the biological significance of DEPs, gene annotations and pathway analysis were also performed.
Methods
An overview of the experimental setup is presented in Fig. 1.
Subject recruitment and sample collection
This study was approved by the Institutional Ethics Committee, All India Institute of Medical Sciences (AIIMS), New Delhi (IECPG-119/24.02.2021, RT-09/24.03.2021). Cervical cancer patients and healthy controls were recruited from the Department of Obstetrics and Gynecology, AIIMS, New Delhi. Tissue samples of confirmed cervical cancer patients were collected at the time of hysterectomy, after the completion of the standard questionnaire and written informed consent (Supplementary Table 1). None of the recruited cervical cancer patients received chemotherapy, radiotherapy, or other medical treatments before surgery. Those patients who received preoperative radiotherapy and/or chemotherapy or had successive primary malignancies or concurrent were excluded. Healthy cervix tissue samples with no evidence of HPV infection and/or cervical cancer or any other type of cancer were collected from the patients attending Obstetrics and Gynecology OPD at AIIMS, New Delhi for the treatment of cervix benign diseases.
Extraction of tissue proteins
Surgically removed tissue samples were washed three times with phosphate buffer saline (PBS, pH 7.4). After washing, the pellet was resuspended in RIPA buffer (with added protease inhibitors cocktail) and left at room temperature for 45 min, followed by 5 cycles of sonication (3 s ON, 30 s OFF, 50 Hz). Samples were then centrifuged at 12,000 g for 15 min at 4 °C and the supernatants were collected in fresh tubes. Protein concentration in each sample was estimated by Bradford assay, using BSA as a protein standard.
Proteomic analysis by tandem mass spectrometry (LC–MS/MS)
Label-free quantitative proteomic analysis was performed as previously described (Chhikara et al. 2022). Briefly, protein samples (50 µg each) were reduced with 5 mM tris(2-carboxyethyl)phosphine (TCEP) followed by alkylation with 50 mM iodoacetamide (IAA). For digestion, samples were incubated overnight in trypsin (1:50, trypsin/sample ratio) at 37 °C. The digested peptides were then cleaned using a C18 silica cartridge, and dried in a speed vac. After drying, samples were resuspended in buffer A (5% acetonitrile, 0.1% formic acid). Mass spectrometric analysis of peptide mixtures was performed using the Ultimate 3000 RSLCnano system coupled with a Thermo QE Plus (Thermo Fisher Scientific). The peptides were resolved on a 3.0 μm EASY-Spray C18 column (50 cm, Thermo Fisher Scientific) and eluted at a flow rate of 300 nL/min with a gradient of buffer B (0–40%, 80% acetonitrile, 0.1% formic acid) for 100 min. MS1 and MS2 spectra were acquired in the Orbitrap at 70,000 and 17,500 resolution, respectively, and dynamic exclusion was employed for 10 s. The raw files were processed and analyzed with Proteome Discoverer™ software v2.2 (Thermo Fisher Scientific) and searched against UniProt’s human proteome database, with both peptide-spectrum match (PSM) and protein false discovery rate set to 0.01. Following parameters were used for Sequest and Amanda search: digesting enzyme—trypsin; maximum missed cleavages value of two; precursor and fragment mass tolerances—10 ppm and 1 Da, respectively; fixed modification—carbamidomethyl on cysteine; and variable modification—oxidation of methionine and N terminal acetylation. The Minora feature detection approach of the Proteome Discoverer framework was employed for label-free quantification of proteins. It detects LC/MS peaks in the raw data files and maps them to identified PSMs. It determines the theoretical isotope pattern of a PSM, identifies the LC/MS peaks that map to it, and then creates a feature signal for a single molecule and charge state throughout the elution profile.
Statistical and bioinformatics analysis
Statistical and bioinformatics analyses were performed as described previously (Naglot et al. 2021). Before analysis, protein abundance data were processed to add missing values by the k-nearest neighbor (knn) imputation method and normalized by Pareto scaling. Statistical analyses, including fold-change and t-test (volcano plot), principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA), orthogonal projections to latent structures discriminant analysis (OPLS-DA), and hierarchical clustering analysis (HCA) were performed by MetaboAnalyst 5.0 (Pang et al. 2021). Enrichment analysis of GO terms and over-represented pathways was performed by DAVID gene enrichment tool v6.8 (Sherman et al. 2022).
Western blot analysis
The expression of selected DEPs was validated in cervical cancer and healthy tissue samples by western blotting. For this, 25 μg protein from each sample was resolved on 10% SDS-PAGE and transferred to a 0.20 μm PVDF membrane (Pall Life Sciences, USA). After completion of protein transfer, the membrane was blocked with 5% (w/v) skim milk in TBST buffer (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Tween-20) for 2 h and incubated overnight in primary antibodies (anti-afamin, MAB8065, R & D systems; anti-stathmin, #3352, cell signaling Technology; GAPDH, sc-47724, Santa Cruz) at 4 °C. Next day, the membrane was washed with TBST and incubated in HRP conjugated secondary antibodies for 2 h. After washing, proteins were detected by enhanced chemiluminescence (ECL, Bio-Rad Labs, USA) and images were acquired on a Syngene G-box (Syngene, UK).
Afamin mRNA expression by quantitative PCR
Total RNA was isolated from tissue samples of cervical cancer (n = 2) and controls (n = 2) by trizol method. Briefly, samples were incubated in trizol for 15 min, and sonicated followed by phase separation in chloroform. The clear aqueous phase obtained after centrifugation (12,000 × g at 4 °C for 15 min) was transferred into a new micro-centrifuge tube and incubated in chilled isopropanol for 10 min. After centrifugation, RNA pellet was washed with 75% ethanol, air-dried, and resuspended in nuclease free water, followed by DNAse I treatment to remove DNA traces, if any. Total RNA was estimated by calculating A260/A280 absorbance ratio and cDNA was prepared by Script cDNA synthesis kit (Bio-Rad) using 1 μg total RNA as the template. Primers for afamin (AFM) and GAPDH (reference gene) were designed using primer3Plus software, followed by optimization of the respective gene. Primer sets used for real-time PCR are presented in Supplementary Table 2. For real-time PCR, 20 μl reaction mixture including 10 μl SsoFast EvaGreen supermix (Biorad), 1 μl cDNA and 10 mM final concentration of each primer was prepared. Quantitative PCR was performed on CFX 96 thermocycler (Bio-Rad) and relative mRNA expression of the AFM was determined by ΔΔCT method. Experiments were performed in triplicates.
Results
Proteomics analysis
As mentioned in the methodology section, proteins extracted from cervical cancer and healthy tissue samples (n = 10, 5 of each type) were analyzed by LC–MS/MS and quantified by a label-free method. In total, 15,080 peptides corresponding to 2271 proteins were identified (Supplementary File 1). All the proteins identified with one or more unique peptides were processed for label-free quantification, which resulted in the quantification of 2183 proteins in at least one of the samples (out of 10). Among them, 417 and 54 proteins were exclusively quantified in cervical cancer and healthy control tissue, respectively, whilst 1712 proteins were quantified in both (Fig. 2).
Identification and validation of differentially expressed proteins
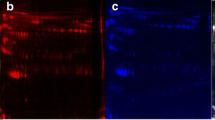
To identify DEPs, protein abundance data were processed to remove the proteins, which were quantified in less than 50% samples followed by imputation of missing values in the remaining, if any. In this way, 1343 proteins were sorted for statistical analysis (Supplementary File 1). To begin with, the volcano plot identified 201 DEPs with fold-change threshold 2 and t-test p-value threshold 0.05, of which 188 proteins were upregulated and 13 downregulated in cervical cancer tissues compared to controls (Fig. 3, Table 1, Supplementary File 2). The list of upregulated proteins included cofilin-1 (CFL1), DnaJ homolog subfamily C member 5 (DNAJC5), syntaxin-7 (STX7), tax1-binding protein 3 (TAX1BP3), PRA1 family protein 2 (PRAF2), syntenin-1 (SDCBP), integrin beta-5 (ITGB5), calponin-3 (CNN3), TPM4, catalase (CAT), malate dehydrogenase, mitochondrial (MDH2), transgelin-2 (TAGLN2), nucleoside diphosphate kinase A (NME1), HLA class I histocompatibility antigen (HLA), transferrin receptor protein 1 (TFRC), 14-3-3 protein beta/alpha (YWHAB), integrin alpha-V (ITGAV), peroxiredoxin-2 (PRDX2), tumor protein D52 (TPD52), glutathione synthetase (GSS), catenin alpha-1 (CTNNA1), RNA-binding protein FUS (FUS), prosaposin (PSAP), nucleobindin-1 (NUCB1), nucleobindin-2 (NUCB2), beta-2-microglobulin (B2M), stathmin (STMN1), tapasin (TAPBP), glucosamine-6-phosphate isomerase 1 (GNPDA1), proliferating cell nuclear antigen (PCNA), growth factor receptor-bound protein 2 (GRB2), tumor-associated calcium signal transducer 2 (TACSTD2), cadherin-1 (CDH1), and prohibitin (PHB). On the other hand, plasminogen (PLG), serum albumin (ALB), serotransferrin (TF), hemopexin (HPX), alpha-2-HS-glycoprotein (AHSG), AFM, apolipoprotein A-II (APOA2), and transmembrane protease serine 13 (TMPRSS13) were found downregulated in cervical cancer. For orthogonal validation, two of the DEPs, STMN1, and AFM were analyzed by western blotting. Similar to LC–MS/MS-based label-free quantitation, STMN1 was found upregulated, while AFM was downregulated in cervical cancer tissue samples (Fig. 3 inset).
Differentially expressed proteins (DEPs) selected by volcano plot analysis. Both fold-change (FC) and p-values were log-transformed. (Insert) Orthogonal validation of stathmin (Uniprot accession P16949) and afamin (Uniprot accession P43652) expression in cervical cancer and control tissue samples by western blotting. GAPDH was used as an internal control; C1 and C2: controls; P1 and P2: cervical cancer
Statistical and bioinformatics analysis
A two-dimensional PCA, a statistical method designed to alleviate the degree of complexity in high-dimensional data was executed to display the abundance variation of proteins within both groups. The PCA scores plot displayed an incomplete separation between cervical cancer and control groups when all the quantified proteins were considered (Fig. 4A); however, separation quality improved when only statistically significant DEPs were considered for PCA analysis (Fig. 4B). The PLS-DA and OPLS-DA score plots showed a clear separation between cervical cancer and control groups (Fig. 4C–E). PLS-DA is a supervised method that evaluates multivariate regression test statistics to assess class discrimination. For each component, PLS-DA calculates Variable Importance in Projection (VIP) scores as a measure of variance; a VIP score greater than 1 denotes a DEP. A total of 218 proteins were identified with VIP score > 1. Similar to PLS-DA, OPLS-DA is also a supervised statistical method, but more powerful for dimension reduction and identifying spectral features that promote group separation. HCA, a popular clustering method that creates clusters of data points with similar characteristics correctly grouped the members of cervical cancer and healthy controls in separate clusters (Fig. 4F). Collectively, these findings signify the statistical power of identified DEPs to classify cervical cancer and control groups.
Statistical analysis of differentially expressed proteins (DEPs). A, B Principal component analysis (PCA) scores plot; C PLS-DA scores plot; D Top 20 DEPs identified by PLS-DA, colored boxes on the right indicate the relative abundance of the corresponding proteins; E OPLS-DA scores plot; F Hierarchical clustering analysis
To understand the biological significance of DEPs, an enrichment analysis of GO terms and pathways was performed. Among the proteins with higher abundance in cervical cancer tissue, a total of 94 GO terms were enriched that included 17 biological processes, 59 cellular components, and 18 molecular functions (Supplementary File 3). Also, 19 KEGG pathways were over-represented in cervical cancer.
Afamin mRNA expression analysis
Relative expression of AFM mRNA was determined in tissue samples of cervical cancer with respect to controls after normalization with the reference gene. Similar to protein profile, AFM mRNA expression was found downregulated in cervical cancer compared to controls (Supplementary Fig. 1). Fold-change (FC) analysis revealed that AFM mRNA expression was downregulated by ~ fourfold in cervical cancer tissue samples (log2 FC = − 2.02).
Discussion
The prognosis of cervical cancer is greatly associated with its grade/stage. If detected in the early stage, treatment strategies are capable of eliminating the disease resulting in a high survival rate among the patients; however, with detection in advanced stages chances of survival reduce significantly. Despite significant advancements in the detection methods for cervical cancer and multimodality management strategies, treatment response in high-grade cervical cancer patients is still unremarkable. This highlights an essential need to discover novel diagnostic markers and therapeutic targets of cervical cancer. This study is another step in our quest to enhance our molecular understanding of cervical cancer and for searching prospective diagnostic markers and therapeutic targets. The study of protein changes becomes important due to their dynamic nature in various stages of cancer progression. Proteins are the main functional biomolecules that participate in all kinds of biological processes and regulate many physiological activities including receiving and sending chemical signals, synthesizing and repairing DNA, etc. (Pan et al. 2013). Failure of the quality control mechanisms that maintain the cellular interplay of biomolecules causes disturbance in proteostasis leading to dysregulation in protein synthesis, folding, trafficking, and degradation of proteins. These abnormalities, in turn, cause several diseases including cancers (Powers et al. 2009).
Identification and validation of differentially expressed proteins
A total of 2271 proteins were identified and 2183 proteins were quantified by high-throughput proteomics analysis. Further, 201 proteins were differentially expressed, of which 188 and 13 proteins were found upregulated and downregulated, respectively in cervical cancer tissue samples. CFL1, DNAJC5, STX7, TAX1BP3, PRAF2, SDCBP, ITGB5, CNN3, TPM4, CAT, MDH2, TAGLN2, NME1, HLA, TFRC, ITGAV, PRDX2, TPD52, GSS, CTNNA1, FUS, PSAP, NUCB1, B2M, STMN1, TAPBP, GNPDA1, PCNA, GRB2, CDH1, and PHB were identified as key upregulated proteins, whilst PLG, ALB, TF, HPX, AHSG, AFM, APOA2, and TMPRSS13 were found downregulated in cervical cancer. Many of these upregulated proteins have previously been shown to possess tumor-promoting properties. For example, CFL1 plays a key role in the invasion and metastasis of solid tumors (Wang et al. 2006). It is a member of cofilin family and is involved in normal cell functions, particularly the mechanisms that promote cell motility (Sequeda-JuArez et al. 2020). The overexpression of CFL1 in pancreatic cancer has recently been associated with higher invasiveness and poor survival (Werle et al. 2021). Similarly, STX7 also aids in cancer cell invasion (Parveen et al. 2022). TAX1BP3 has also been found overexpressed in cancer and its overexpression was linked to cancer cell adhesion, migration, and metastasis (Wang et al. 2014, 2023). PRAF2 is an oncogene that helps in the proliferation and invasion of breast cancer cells (Wang et al. 2022). In esophageal squamous cell carcinoma, PRAF2 overexpression predicts a poor prognosis and encourages carcinogenesis (Qian et al. 2019). Similarly, overexpression of SDCBP and other upregulated proteins has been reported to be associated with cancer progression, metastasis, and poor survival (Ban et al. 2016; Talukdar et al. 2019; Li et al. 2021; Liu et al. 2021; Mir et al. 2021). Upregulated expression of few proteins, such as CNN3, ITGB5, and TDP52 has also been reported to promote cervical cancer (Shi et al. 2020; Xia et al. 2020). In our study, similar expression trends in cervical cancer highlight tumor-promoting nature of these proteins. On the other hand, downregulated proteins identified in this study are known to play crucial roles in inhibiting tumor growth (Ahmed et al. 2005; Nojiri and Joh 2014; Garcia-Martinez et al. 2015; Zhang et al. 2019).
The expression of two proteins, STMN1 and AFM was also validated by western blotting. STMN1, also known as oncoprotein 18 is a cytosolic phosphoprotein that plays an important role in microtubule dynamics that regulate mitosis and is essential for cell division and proliferation. Higher expression of STMN1 has been marked in cancer tissue samples of the ovary, endometrium, breast, and cervix (Curmi et al. 2000; Wei et al. 2008; Su et al. 2009Trovik et al. 2011). It is known to cause mitotic arrest enough to persuade tumorigenesis, tumor invasion, metastasis, and chemoresistance (Howitt et al. 2013). Associated with poor outcomes, overexpression of STMN1 in high-grade cervical cancer has been correlated with its involvement in the progression of disease with more aggressive behavior due to its oncogenic properties (Xi et al. 2009; Howitt et al. 2013). Thus, it can be explored further as a potential diagnostic marker as well as a therapeutic target for cervical cancer, specifically in high-grade malignancies. Other protein, AFM is a highly glycosylated member of the albumin gene family abundantly present in human plasma and other biological fluids (Dieplinger and Dieplinger 2015). It is a multi-functional vitamin E transporter that interacts with fatty-acylated Wnt proteins and helps them in retaining their biological activity during intracellular trafficking (Nusse and Varmus 2012; Wilson 2017). Wnt proteins are signaling molecules involved in the regulation of various cell development processes like proliferation, differentiation, migration, invasion, apoptosis, and tissue homeostasis (Wilson 2017). AFM expression has been reported in different cancers like gastric, bladder, colorectal, breast, and thyroid cancer and found significantly downregulated in ovarian cancer (Jackson et al. 2007; Dieplinger and Dieplinger 2015). A reduced expression of AFM in cervical cancer tissue samples correlates with previous reports of a negative correlation between AFM expression and cancer progression. However, not much information is available about the functional aspects of AFM. The targeted studies exploring the physiological role of this protein can uncover several unknown facets of cervical cancer pathogenesis, which can help us in designing improved therapeutics.
Selection of biomarker candidates for cervical cancer
For the selection of cervical cancer biomarker candidates, the previously described two-step selection method was employed (Saraswat et al. 2020; Naglot et al. 2021). The first step was course selection, which included the identification of DEPs by volcano plot, PLS-DA, and OPLS-DA. In the second step, regarded as fine selection, overlapping DEPs with a p-value threshold of 0.05 were selected. This resulted in the selection of 46 DEPs as candidate biomarkers of cervical cancer, including stomatin-like protein 3 (STOML 3), ITGB2, STMN1, CAT, TF, HLA class II histocompatibility antigen (HLA-DRA), HPX, tropomyosin (TMP3/TMP4), ras GTPase-activating-like protein IQGAP1 (IQGAP1), YWHAB, ALB, putative heat shock 70 kDa protein 7 (HAPA7), FUS, PRDX2, CFL1, MDH2, NME1, TFRC, lysosome-associated membrane glycoprotein 1 (LAMP1), and TAGLN2 (Supplementary File 4). Many of these proteins have been studied previously for their role in cancers and their altered expressions were found in conjunction with cancer progression and/or therapy resistance. For example, ITGB2, which is a cell surface adhesion glycoprotein associated with autophagy and PI3K–Akt signaling pathways overexpresses in various cancers, including cervical cancer, and is involved in cell adhesion and migration (Peng et al. 2020; Sun et al. 2021). CAT, which is a key enzyme of H2O2 metabolism was found upregulated in many cancers and correlated with resistance to therapy (Sander et al. 2003; Hwang et al. 2007; Rainis et al. 2007; Oliva et al. 2015; Flor et al. 2021). Similarly, overexpression of TMP3 in cancers such as glioma, colon cancer, and liver cancer has been linked with the development of tumors and statistically correlated with tumor prognosis (Tao et al. 2014; Chen et al. 2021). IQGAP1, a PI3K scaffolding protein, is required for optimal induction of the PI3K/AKT/mTOR pathway and promotes the progression of papillomavirus-induced carcinogenesis (Wei et al. 2021). ALB plays a vital role in tumorigenesis by exerting antioxidant effect against carcinogens and modulating the immune response, and a low level of ALB might correlate with reduced immune competency, and poor anti-cancer response (Nojiri and Joh 2014; Garcia-Martinez et al. 2015). It has shown significant potential as a prognostic marker of cervical cancer (Zheng et al. 2016; Zhang et al. 2019). Conclusively, altered expression of these proteins, in coherence with previous studies, highlights their potential to serve as prognostic markers of cervical cancer. However, further studies are required in a large cohort to validate them as potential markers.
Bioinformatics analysis
The biological processes enriched among overexpressed proteins in cervical cancer were cell cycle, cell adhesion, protein folding, small GTPase mediated signal transduction, inflammatory disorders, post-translational protein modification, and tumorigenesis. Interestingly, the ferroptosis pathway was over-represented in cervical cancer, in addition to several other KEGG pathways, such as antigen processing and presentation, cell adhesion molecules, spliceosome, and adherens junction. The DEPs associated with ferroptosis were voltage-dependent anion-selective channel proteins 2 and 3 (VDAC2/VDAC3), GSS, TFRC, and PCBP2. Ferroptosis is a non-apoptotic, iron-dependent regulated cell death due to the accumulation of lipid-based reactive oxygen species (ROS). It is an adaptive process to eliminate the nutrient-deficient cancer cells, which helps in maintaining the death balance and plays an important role in inhibiting tumor growth (Zhao et al. 2022). Thus, ferroptosis is targeted for developing strategies for promising cancer therapy (Dixon and Stockwell 2019; Li et al. 2020). During chronic inflammation and tissue injury transferrin, an iron-binding serum protein is transported into the cells via TFRC-mediated endocytosis and induces ferroptotic cell death (Lu et al. 2017). In agreement with previous reports, TFRC was also found elevated in cervical cancer in our study (Shen et al. 2018; Xu et al. 2019; Huang et al. 2022). However, TF was found downregulated in cervical cancer samples. TF also helps in stimulating cell proliferation and has been found downregulated in ovarian cancer and other chronic inflammations (Baumann and Gauldie 1994; Ahmed et al. 2005). This emphasizes on exploring the interplay between these proteins to determine their precise involvement in ferroptosis and consequently, in the regulation of cervical cancer progression. In line with a number of earlier research studies, these biomolecules can be targeted for developing effective anti-cancer strategies for cervical cancer.
Conclusively, this quantitative proteomics study identified a number of candidate biomarkers of cervical cancer. The DEPs, specifically STMN1, AFM, TF, and those associated with the ferroptosis pathway, such as TFRC can further be explored to validate the possibility to use these proteins as diagnostic markers and/or therapeutic targets.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary information files.
References
Ahmed N, Oliva KT, Barker G, Hoffmann P, Reeve S, Smith IA, Quinn MA, Rice GE (2005) Proteomic tracking of serum protein isoforms as screening biomarkers of ovarian cancer. Proteomics 5(17):4625–4636. https://doi.org/10.1002/pmic.200401321
Aljawad MF, Faisal A, Alqanbar MF, Wilmarth PA, Hassan BQ (2022) Tandem mass tag-based quantitative proteomic analysis of cervical cancer. Proteom Clin Appl 17(1):e2100105. https://doi.org/10.1002/prca.202100105
Bae SM, Lee CH, Cho YL, Nam KH, Kim YW, Kim CK, Han BD, Lee YJ, Chun HJ, Ahn WS (2005) Two-dimensional gel analysis of protein expression profile in squamous cervical cancer patients. Gynecol Oncol 99(1):26–35. https://doi.org/10.1016/j.ygyno.2005.05.041
Ban HS, Xu X, Jang K, Kim I, Kim BK, Lee K, Won M (2016) A novel malate dehydrogenase 2 inhibitor suppresses hypoxia-inducible factor-1 by regulating mitochondrial respiration. PLoS ONE 11(9):e0162568. https://doi.org/10.1371/journal.pone.0162568
Baumann H, Gauldie J (1994) The acute phase response. Immunol Today 15(2):74–80. https://doi.org/10.1016/0167-5699(94)90137-6
Chen S, Shen Z, Gao L, Yu S, Zhang P, Han Z, Kang M (2021) TPM3 mediates epithelial-mesenchymal transition in esophageal cancer via MMP2/MMP9. Ann Transl Med 9(16):1338. https://doi.org/10.21037/atm-21-4043
Chhikara N, Tomar AK, Datta SK, Yadav S (2022) Proteomic changes in human spermatozoa during in vitro capacitation and acrosome reaction in normozoospermia and asthenozoospermia. Andrology. https://doi.org/10.1111/andr.13289
Clifford GM, Smith JS, Aguado T, Franceschi S (2003) Comparison of HPV type distribution in high-grade cervical lesions and cervical cancer: a meta-analysis. Br J Cancer 89(1):101–105. https://doi.org/10.1038/sj.bjc.6601024
Curmi PA, Nogues C, Lachkar S, Carelle N, Gonthier MP, Sobel A, Lidereau R, Bieche I (2000) Overexpression of stathmin in breast carcinomas points out to highly proliferative tumours. Br J Cancer 82(1):142–150. https://doi.org/10.1054/bjoc.1999.0891
Demarco M, Hyun N, Carter-Pokras O, Raine-Bennett TR, Cheung L, Chen X, Hammer A, Campos N, Kinney W, Gage JC, Befano B, Perkins RB, He X, Dallal C, Chen J, Poitras N, Mayrand MH, Coutlee F, Burk RD, Lorey T, Castle PE, Wentzensen N, Schiffman M (2020) A study of type-specific HPV natural history and implications for contemporary cervical cancer screening programs. EClinicalMedicine 22:100293. https://doi.org/10.1016/j.eclinm.2020.100293
Dieplinger H, Dieplinger B (2015) Afamin–a pleiotropic glycoprotein involved in various disease states. Clin Chim Acta 446:105–110. https://doi.org/10.1016/j.cca.2015.04.010
Ding Y, Yang M, She S, Min H, Xv X, Ran X, Wu Y, Wang W, Wang L, Yi L, Yang Y, Gao Q (2015) iTRAQ-based quantitative proteomic analysis of cervical cancer. Int J Oncol 46(4):1748–1758. https://doi.org/10.3892/ijo.2015.2859
Dixon SJ, Stockwell BR (2019) The Hallmarks of Ferroptosis. Annu Rev of Cancer Biol 3(1):35–54. https://doi.org/10.1146/annurev-cancerbio-030518-055844
Einstein MH, Schiller JT, Viscidi RP, Strickler HD, Coursaget P, Tan T, Halsey N, Jenkins D (2009) Clinician’s guide to human papillomavirus immunology: knowns and unknowns. Lancet Infect Dis 9(6):347–356. https://doi.org/10.1016/s1473-3099(09)70108-2
Feng Q, Yu M, Kiviat NB (2006) Molecular biomarkers for cancer detection in blood and bodily fluids. Crit Rev Clin Lab Sci 43(5–6):497–560. https://doi.org/10.1080/10408360600922632
Flor S, Oliva CR, Ali MY, Coleman KL, Greenlee JD, Jones KA, Monga V, Griguer CE (2021) Catalase overexpression drives an aggressive phenotype in glioblastoma. Antioxidants 10(12):1988. https://doi.org/10.3390/antiox10121988
Garcia-Martinez R, Andreola F, Mehta G, Poulton K, Oria M, Jover M, Soeda J, Macnaughtan J, De Chiara F, Habtesion A, Mookerjee RP, Davies N, Jalan R (2015) Immunomodulatory and antioxidant function of albumin stabilises the endothelium and improves survival in a rodent model of chronic liver failure. J Hepatol 62(4):799–806. https://doi.org/10.1016/j.jhep.2014.10.031
Grulich AE, van Leeuwen MT, Falster MO, Vajdic CM (2007) Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis. Lancet 370(9581):59–67. https://doi.org/10.1016/S0140-6736(07)61050-2
Howitt BE, Nucci MR, Drapkin R, Crum CP, Hirsch MS (2013) Stathmin-1 expression as a complement to p16 helps identify high-grade cervical intraepithelial neoplasia with increased specificity. Am J Surg Pathol 37(1):89–97. https://doi.org/10.1097/PAS.0b013e3182753f5a
Huang N, Wei Y, Cheng Y, Wang X, Wang Q, Chen D, Li W (2022) Iron metabolism protein transferrin receptor 1 involves in cervical cancer progression by affecting gene expression and alternative splicing in HeLa cells. Genes Genom 44(6):637–650. https://doi.org/10.1007/s13258-021-01205-w
Hwang TS, Choi HK, Han HS (2007) Differential expression of manganese superoxide dismutase, copper/zinc superoxide dismutase, and catalase in gastric adenocarcinoma and normal gastric mucosa. Eur J Surg Oncol 33(4):474–479. https://doi.org/10.1016/j.ejso.2006.10.024
Jackson D, Craven RA, Hutson RC, Graze I, Lueth P, Tonge RP, Hartley JL, Nickson JA, Rayner SJ, Johnston C, Dieplinger B, Hubalek M, Wilkinson N, Perren TJ, Kehoe S, Hall GD, Daxenbichler G, Dieplinger H, Selby PJ, Banks RE (2007) Proteomic profiling identifies afamin as a potential biomarker for ovarian cancer. Clin Cancer Res 13(24):7370–7379. https://doi.org/10.1158/1078-0432.CCR-07-0747
Li Z, Chen L, Chen C, Zhou Y, Hu D, Yang J, Chen Y, Zhuo W, Mao M, Zhang X, Xu L, Wang L, Zhou J (2020) Targeting ferroptosis in breast cancer. Biomark Res 8(1):58. https://doi.org/10.1186/s40364-020-00230-3
Li L, Ye T, Zhang Q, Li X, Ma L, Yan J (2021) The expression and clinical significance of TPM4 in hepatocellular carcinoma. Int J Med Sci 18(1):169–175. https://doi.org/10.7150/ijms.49906
Liu D, Liu S, Fang Y, Liu L, Hu K (2021) Comprehensive analysis of the expression and prognosis for ITGBs: identification of ITGB5 as a biomarker of poor prognosis and correlated with immune infiltrates in gastric cancer. Front Cell Dev Biol 9:816230. https://doi.org/10.3389/fcell.2021.816230
Lu B, Chen XB, Ying MD, He QJ, Cao J, Yang B (2017) The role of ferroptosis in cancer development and treatment response. Front Pharmacol 8:992. https://doi.org/10.3389/fphar.2017.00992
Mir C, Garcia-Mayea Y, Garcia L, Herrero P, Canela N, Tabernero R, Lorente J, Castellvi J, Allonca E, Garcia-Pedrero J, Rodrigo JP, Carracedo A, EstherLLeonart M (2021) SDCBP modulates stemness and chemoresistance in head and neck squamous cell carcinoma through Src activation. Cancers (basel) 13(19):4952. https://doi.org/10.3390/cancers13194952
Naglot S, Tomar AK, Singh N, Yadav S (2021) Label-free proteomics of spermatozoa identifies candidate protein markers of idiopathic recurrent pregnancy loss. Reprod Biol 21(3):100539. https://doi.org/10.1016/j.repbio.2021.100539
Nguyen HH, Broker TR, Chow LT, Alvarez RD, Vu HL, Andrasi J, Brewer LR, Jin G, Mestecky J (2005) Immune responses to human papillomavirus in genital tract of women with cervical cancer. Gynecol Oncol 96(2):452–461. https://doi.org/10.1016/j.ygyno.2004.10.019
Nojiri S, Joh T (2014) Albumin suppresses human hepatocellular carcinoma proliferation and the cell cycle. Int J Mol Sci 15(3):5163–5174. https://doi.org/10.3390/ijms15035163
Nusse R, Varmus H (2012) Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J 31(12):2670–2684. https://doi.org/10.1038/emboj.2012.146
Oliva CR, Markert T, Gillespie GY, Griguer CE (2015) Nuclear-encoded cytochrome c oxidase subunit 4 regulates BMI1 expression and determines proliferative capacity of high-grade gliomas. Oncotarget 6(6):4330–4344. https://doi.org/10.18632/oncotarget.3015
Pan S, Brentnall TA, Kelly K, Chen R (2013) Tissue proteomics in pancreatic cancer study: discovery, emerging technologies, and challenges. Proteomics 13(3–4):710–721. https://doi.org/10.1002/pmic.201200319
Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques PE, Li S, Xia J (2021) MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucl Acids Res 49(W1):W388–W396. https://doi.org/10.1093/nar/gkab382
Parveen S, Khamari A, Raju J, Coppolino MG, Datta S (2022) Syntaxin 7 contributes to breast cancer cell invasion by promoting invadopodia formation. J Cell Sci. https://doi.org/10.1242/jcs.259576
Peng Y, Wu D, Li F, Zhang P, Feng Y, He A (2020) Identification of key biomarkers associated with cell adhesion in multiple myeloma by integrated bioinformatics analysis. Cancer Cell Int 20:262. https://doi.org/10.1186/s12935-020-01355-z
Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78:959–991. https://doi.org/10.1146/annurev.biochem.052308.114844
Qian Z, Wei B, Zhou Y, Wang Q, Wang J, Sun Y, Gao Y, Chen X (2019) PRAF2 overexpression predicts poor prognosis and promotes tumorigenesis in esophageal squamous cell carcinoma. BMC Cancer 19(1):585. https://doi.org/10.1186/s12885-019-5818-7
Rainis T, Maor I, Lanir A, Shnizer S, Lavy A (2007) Enhanced oxidative stress and leucocyte activation in neoplastic tissues of the colon. Dig Dis Sci 52(2):526–530. https://doi.org/10.1007/s10620-006-9177-2
Ramirez-Torres A, Gil J, Contreras S, Ramirez G, Valencia-Gonzalez HA, Salazar-Bustamante E, Gomez-Caudillo L, Garcia-Carranca A, Encarnacion-Guevara S (2022) Quantitative proteomic analysis of cervical cancer tissues identifies proteins associated with cancer progression. Cancer Genom Proteom 19(2):241–258. https://doi.org/10.21873/cgp.20317
Sander CS, Hamm F, Elsner P, Thiele JJ (2003) Oxidative stress in malignant melanoma and non-melanoma skin cancer. Br J Dermatol 148(5):913–922. https://doi.org/10.1046/j.1365-2133.2003.05303.x
Saraswat M, Joenväärä S, Tohmola T, Sutinen E, Vartiainen V, Koli K, Myllärniemi M, Renkonen R (2020) Label-free plasma proteomics identifies haptoglobin-related protein as candidate marker of idiopathic pulmonary fibrosis and dysregulation of complement and oxidative pathways. Sci Rep 10(1):7787. https://doi.org/10.1038/s41598-020-64759-x
Sequeda-JuArez A, JimEnez A, Espinosa-Montesinos A, Del Carmen C-AM, RamOn-Gallegos E (2020) Use of AKR1C1 and TKTL1 in the diagnosis of low-grade squamous intraepithelial lesions from Mexican Women. Anticancer Res 40(11):6273–6284. https://doi.org/10.21873/anticanres.14648
Shen Y, Li X, Dong D, Zhang B, Xue Y, Shang P (2018) Transferrin receptor 1 in cancer: a new sight for cancer therapy. Am J Cancer Res 8(6):916–931
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W (2022) DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucl Acids Res. https://doi.org/10.1093/nar/gkac194
Shi P, Zhang X, Lou C, Xue Y, Guo R, Chen S (2020) Hsa_circ_0084927 regulates cervical cancer advancement via regulation of the miR-634/TPD52 Axis. Cancer Manag Res 12:9435–9448. https://doi.org/10.2147/CMAR.S272478
Singh M, Jha RP, Shri N, Bhattacharyya K, Patel P, Dhamnetiya D (2022) Secular trends in incidence and mortality of cervical cancer in India and its states, 1990–2019: data from the Global Burden of Disease 2019 Study. BMC Cancer 22(1):149. https://doi.org/10.1186/s12885-022-09232-w
Su D, Smith SM, Preti M, Schwartz P, Rutherford TJ, Menato G, Danese S, Ma S, Yu H, Katsaros D (2009) Stathmin and tubulin expression and survival of ovarian cancer patients receiving platinum treatment with and without paclitaxel. Cancer 115(11):2453–2463. https://doi.org/10.1002/cncr.24282
Suh KS, Park SW, Castro A, Patel H, Blake P, Liang M, Goy A (2010) Ovarian cancer biomarkers for molecular biosensors and translational medicine. Expert Rev Mol Diagn 10(8):1069–1083. https://doi.org/10.1586/erm.10.87
Sun Z, Dang Q, Liu Z, Shao B, Chen C, Guo Y, Chen Z, Zhou Q, Hu S, Liu J, Yuan W (2021) LINC01272/miR-876/ITGB2 axis facilitates the metastasis of colorectal cancer via epithelial-mesenchymal transition. J Cancer 12(13):3909–3919. https://doi.org/10.7150/jca.55666
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Talukdar S, Das SK, Pradhan AK, Emdad L, Windle JJ, Sarkar D, Fisher PB (2019) MDA-9/Syntenin (SDCBP) Is a critical regulator of chemoresistance, survival and stemness in prostate cancer stem cells. Cancers (basel) 12(1):53. https://doi.org/10.3390/cancers12010053
Tao T, Shi Y, Han D, Luan W, Qian J, Zhang J, Wang Y, You Y, Chinese Glioma Cooperative G (2014) TPM3, a strong prognosis predictor, is involved in malignant progression through MMP family members and EMT-like activators in gliomas. Tumour Biol 35(9):9053–9059. https://doi.org/10.1007/s13277-014-1974-1
Trovik J, Wik E, Stefansson IM, Marcickiewicz J, Tingulstad S, Staff AC, Njolstad TS, Vandenput I, Amant F, Akslen LA, Salvesen HB, MoMaTec Study G (2011) Stathmin overexpression identifies high-risk patients and lymph node metastasis in endometrial cancer. Clin Cancer Res 17(10):3368–3377. https://doi.org/10.1158/1078-0432.CCR-10-2412
Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS (2006) The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol 173(3):395–404. https://doi.org/10.1083/jcb.200510115
Wang H, Han M, Whetsell W Jr, Wang J, Rich J, Hallahan D, Han Z (2014) Tax-interacting protein 1 coordinates the spatiotemporal activation of Rho GTPases and regulates the infiltrative growth of human glioblastoma. Oncogene 33(12):1558–1569. https://doi.org/10.1038/onc.2013.97
Wang Y, Zhao Z, Jiao W, Yin Z, Zhao W, Bo H, Bi Z, Dong B, Chen B, Wang Z (2022) PRAF2 is an oncogene acting to promote the proliferation and invasion of breast cancer cells. Exp Ther Med 24(6):738. https://doi.org/10.3892/etm.2022.11674
Wang Y, Gan Y, Dong Y, Zhou J, Zhu E, Yuan H, Li X, Wang B (2023) Tax1 binding protein 3 regulates osteogenic and adipogenic differentiation through inactivating Wnt/beta-catenin signalling. J Cell Mol Med 27(7):950–961. https://doi.org/10.1111/jcmm.17702
Wei SH, Lin F, Wang X, Gao P, Zhang HZ (2008) Prognostic significance of stathmin expression in correlation with metastasis and clinicopathological characteristics in human ovarian carcinoma. Acta Histochem 110(1):59–65. https://doi.org/10.1016/j.acthis.2007.06.002
Wei T, Choi S, Buehler D, Lee D, Ward-Shaw E, Anderson RA, Lambert PF (2021) Role of IQGAP1 in Papillomavirus-Associated Head and Neck Tumorigenesis. Cancers (basel) 13(9):2276. https://doi.org/10.3390/cancers13092276
Werle SD, Schwab JD, Tatura M, Kirchhoff S, Szekely R, Diels R, Ikonomi N, Sipos B, Sperveslage J, Gress TM, Buchholz M, Kestler HA (2021) Unraveling the molecular tumor-promoting regulation of Cofilin-1 in pancreatic cancer. Cancers (basel) 13(4):725. https://doi.org/10.3390/cancers13040725
Wilson MA (2017) Structural Insight into a Fatty-Acyl chaperone for Wnt proteins. Structure 25(12):1781–1782. https://doi.org/10.1016/j.str.2017.11.009
Xi W, Rui W, Fang L, Ke D, Ping G, Hui-Zhong Z (2009) Expression of stathmin/op18 as a significant prognostic factor for cervical carcinoma patients. J Cancer Res Clin Oncol 135(6):837–846. https://doi.org/10.1007/s00432-008-0520-1
Xia L, Yue Y, Li M, Zhang YN, Zhao L, Lu W, Wang X, Xie X (2020) CNN3 acts as a potential oncogene in cervical cancer by affecting RPLP1 mRNA expression. Sci Rep 10(1):2427. https://doi.org/10.1038/s41598-020-58947-y
Xu X, Liu T, Wu J, Wang Y, Hong Y, Zhou H (2019) Transferrin receptor-involved HIF-1 signaling pathway in cervical cancer. Cancer Gene Ther 26(11–12):356–365. https://doi.org/10.1038/s41417-019-0078-x
Zhang C, Li Y, Ji R, Zhang W, Zhang C, Dan Y, Qian H, He A (2019) The prognostic significance of pretreatment albumin/alkaline phosphatase ratio in patients with stage IB-IIA cervical cancer. Onco Targets Ther 12:9559–9568. https://doi.org/10.2147/OTT.S225294
Zhao L, Zhou X, Xie F, Zhang L, Yan H, Huang J, Zhang C, Zhou F, Chen J, Zhang L (2022) Ferroptosis in cancer and cancer immunotherapy. Cancer Commun (lond) 42(2):88–116. https://doi.org/10.1002/cac2.12250
Zheng RR, Huang M, Jin C, Wang HC, Yu JT, Zeng LC, Zheng FY, Lin F (2016) Cervical cancer systemic inflammation score: a novel predictor of prognosis. Oncotarget 7(12):15230–15242. https://doi.org/10.18632/oncotarget.7378
Acknowledgements
The authors thank AIIMS, New Delhi for intramural financial support and Vproteomics, New Delhi for their support in performing LC-MS/MS experiments. AT also thanks University Grants Commission (UGC), New Delhi, India for her Senior Research Fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization, SY, AKT, and KBC; Project administration, and supervision, SY, JBS, and NB; Recruitment of study participants, sample collection, analysis, and processing, KBC, AT, NB, and JBS; Methodology, KBC, AT, and AKT; Data analysis, AT, and AKT; Writing—original draft preparation, KBC, AT, and AKT; Writing—review and editing, AKT. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Informed consent
Yes, tissue samples were collected after the completion of the standard questionnaire and written informed consent.
Research involving human participants and/or animals
Yes, research involved human participants and this study was approved by the Institutional Ethics Committee, All India Institute of Medical Sciences (AIIMS), New Delhi (IECPG-119/24.02.2021, RT-09/24.03.2021).
Supplementary Information
Below is the link to the electronic supplementary material.
42485_2023_114_MOESM2_ESM.xlsx
Supplementary file2 List of proteins identified by LC-MS/MS analysis (sheet 1) and abundance data of 1343 proteins sorted for statistical analysis (XLSX 632 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chandra, K.B., Tomar, A.K., Thapliyal, A. et al. Differential proteomics reveals overexpression of ferroptosis-related proteins in cervical cancer tissue. J Proteins Proteom 14, 163–174 (2023). https://doi.org/10.1007/s42485-023-00114-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-023-00114-8