Abstract
Recent studies have shown that many molecular mechanisms, such as the EGFR, AKT, STAT3, and beta-catenin pathways, are involved in glioma. However, the prognosis of the disease remains poor. Explorations of the underlying mechanisms of glioma and identification of effective markers for early diagnosis and accurate prognostication remain important today. In this study, we employed survival analysis to determine that TPM3 overexpression was significantly associated with high-grade gliomas and higher mortality. Using microarray combined with Pearson correlation analysis, we found that TPM3 was positively correlated with the expression of MMP family members and EMT-like activators. Reduction of TPM3 (via TPM3-siRNA) inhibited cellular invasion and migration and decreased MMP-9 and SNAI1 levels in glioma cells. To the best of our knowledge, our work is the first to show that TPM3 plays a critical role in the progression of gliomas and provides novel insights into the key roles of MMP family members and EMT-like activators that mediate TPM3 functional signaling for glioma regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High-grade gliomas (HGGs) account for the majority of all gliomas, including glioblastoma (World Health Organization [WHO] grade IV) and anaplasticglioma (WHO grade III). Despite tremendous efforts to develop multimodal treatments, the overall prognosis of the disease remains poor [1]. Given the limitations of current diagnostic tools, including both predictive and prognostic markers, development of improved therapeutic strategies remains a critical need.
Tropomyosins (TPMs) are a series of actin-binding proteins present in the skeletal and smooth muscle and some non-muscular tissues. In the skeletal muscle, TPMs mediate a myosin-actin response to calcium ions and take part in the stabilization of cytoskeletal microfilaments [2]. Several studies have recently suggested that non-muscular TPMs may be involved in the progression of cancer. As a tumor suppressor, TPM1 has been shown to play a role in the suppression of malignant phenotypes [3]. By contrast, TPM3, as an important oncogene, has been reported to be involved in hematopoietic tumorigenesis, migration, and invasion [4]. Miyado et al. [5] observed that TPM3 expression is higher in a highly metastatic mouse melanoma cell line than in a low-metastatic one; this study suggests that TPM3 plays an important role in the invasion or migration of human malignancies. However, the expression and function of TPM3 in gliomas remain poorly understood.
In the present study, TPM3 was shown to be overexpressed in HGGs and involved in the mortality of HGG patients. TPM3 knockdown was demonstrated to inhibit cellular invasion and migration in glioma cells. Using microarray combined with Pearson correlation analysis, we found that a series of matrix metalloproteinase (MMP) family members and epithelial-to-mesenchymal transition (EMT) like markers are overexpressed in HGGs and positively correlated with TPM3 expression. Among these markers, MMP-9 and SNAI1 were selected for functional validation because of their strong association with the malignant phenotype. Consistent with the microarray data, TPM3 knockdown reduced the expression level of MMP-9 and SNAI1. To the best of our knowledge, our study is the first to indicate that TPM3 may be used as a therapeutic target for glioblastoma multiforme (GBM). TPM3 may be an important promoter of glioma cell invasion and migration by induction of the expression of MMP family members and EMT-like activators.
Materials and methods
Microarray analysis
mRNA expression data on gliomas were downloaded from the Chinese Glioma Genome Atlas (CGGA) data portal (http://www.cgga.org.cn/portal.php) [6]. Microarray analysis of the whole genome gene profiles of 220 glioma tissues was performed using Agilent Whole Human Genome Array, and data were normalized using GeneSpring GX 11.0. Retrospective analysis of clinical outcomes was based on CGGA mRNA expression microarray data, including survival information of 89 GBM patients. Overall survival curves were plotted according to the Kaplan–Meier method, and the log-rank test was applied for comparison.
Human glioma tissues and cell lines
Glioma tissue samples were collected from the Chinese Glioma Cooperative group (CGCG). Diagnosis of gliomas was established histologically according to the 2007 WHO classification. Human U87 and U251 glioma cell lines were obtained from the Chinese Academy of Sciences cell bank. Glioma cell lines were cultured in Dulbecco’ s modified Eagle’ s medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), 100 units/ml penicillin, and 100 ng/ml streptomycin. All cells were incubated at 37 °C in an atmosphere of 5 % CO2. The present study was approved by the medical review board of Nanjing Medical University. Written informed consent was obtained from all participating patients.
Oligonucleotides and cell transfection
The TPM3-siRNA, MMP9-siRNA, and SNAI1-siRNA oligonucleotides were designed and synthesized by GenePharma and Gene chem (Shanghai, China). A siRNA that was unrelated to any human sequence was used as a negative control (NC). The plasmid containing the ORF of TPM3 (TPM3), was generated from Gene chem. Blank vector was used as an NC. For transfection, transfection complexes were formed from oligonucleotides using Lipofectamine 2000 (Invitrogen). Transfection complexes were added to glioma cells and incubated for 6–8 h before the medium was changed.
Western blot analysis
Western blot was performed as previously described [7]. Immunoblot was performed using appropriate primary antibodies: TPM3 (1:800; Abcam), MMP-9 (1:800; Abcam), SNAI1 (1:500; Abcam), and GAPDH (1:1,000; Santa Cruz).
Invasion assay
Cell invasion assays were performed using transwell membranes coated with Matrigel (BD Biosciences). Transfected cells were plated at a density of 3 × 104 cells per well in the upper chamber and in serum-free medium. The lower chamber was filled with 20 % FBS as a chemo-attractant. After 24 h of incubation, cells remaining in the upper chamber of each well were carefully removed with cotton swabs, and invading cells were fixed with 3 % paraformaldehyde, stained with crystal violet, and counted from three independent fields (×100 magnification).
Wound healing assay
Cells were cultured until reached 90 % confluence in 6-well plates. Cell layers were scratched using a 20-μl tip to form wound gaps, washed twice with PBS and cultured. The wound healing was photographed at different time points and wounded gaps were analyzed by measuring the distance of migrating cells for three different areas for each wound.
Cell proliferation assay
Cells in the logarithmic phase of growth were seeded at 2,000 per well in 96-well plates and cultured. Cell proliferation was assayed at the indicated time points using a CCK8 kit (Beyotime, China) according to the manufacturer’s instructions.
Statistical analysis
Each value was obtained from at least three independent experiments and presented as means ± SD. Significant differences were calculated using one-way ANOVA for three-group comparisons and t-test for two-group comparisons. The SPSS 13.0 software package was employed. Pearson correlation analysis was performed using MATLAB software. A probability value of <0.05 was considered statistically significant.
Results
TPM3 expression in glioma tissues and association with GBM patient prognosis
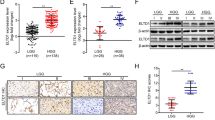
Using CGGA microarray data, we initially analyzed TPM family expression patterns in different grades of glioma tissues and found that TPM3 expression is elevated in HGG tissues compared with low-grade glioma ones (Fig. 1a and b). The mean overall survival time of GBM patients with high level of TPM3 was 287.0 days, significantly shorter than that of patients with low levels of TPM3 (506.0 days; P = 0.02, log-rank test; Fig. 1c).
TPM3 expression in glioma tissues and Kaplan–Meier plots of overall survival duration in patients with GBM. a A heat map showing the expression profiles TPM family members in 220 glioma tissues. b Microarray analysis of TPM3 expression in different grades of glioma. ***P < 0.0001. c Kaplan–Meier survival analysis of overall survival duration in 89 GBM patients according to TPM3 expression. The log-rank test was used to calculate the P value. Results are presented as mean ± SD from three independent experiments
Effects of TPM3 on cellular invasion and migration in glioma cells
To explore the potential role of TPM3 on the invasiveness of glioma cells, TPM3 was downregulated using small interfering TPM3 (si-TPM3). In vitro invasion assay was employed. As shown in Fig. 2a and b, TPM3 knockdown led to a 43.84 % reduction in invasion of the basement membrane (Matrigel) by U87 cells and 44.19 % reduction in invasion by U251 cells compared with the scramble group. Subsequently, the effect of si-TPM3 on cell migration was tested. We found that down-expression of TPM3 obviously decreased the wound healing effect of glioma cells (Fig. 2c and d). To exclude the interference of cell proliferation, we did CCK8 assay and captured relative cell number at two time points. As shown in supplementary figure 1, CCK-8 assays showed that there was no significant difference between si-TPM3-transfected U87 and U251 cells and NC group within 24 h.
Effects of TPM3 on glioma cell invasion and migration as determined by in vitro assay. a, b Effect of si-TPM3 on the invasion of glioma cell lines as determined by in vitro transwell invasion assay. **P < 0.01. c, d Effect of si-TPM3 on the migration of glioma cell lines as determined by in vitro wound-healing assay. **P < 0.01. Results are presented as mean ± SD from three independent experiments
Correlation of MMP family genes and EMT-like activators with TPM3
To determine the potential mechanisms of the aberrant expression of TPM3 and malignant progression of glioma, we used Pearson correlation to analyze the relationships between TPM3 and all known human protein-coding genes. MMP family genes and a set of classical EMT-like activators were found to positively correlate with TPM3 expression (Fig. 3 and Table 1). Among these genes, MMP-9 and SNAI1, two factors known to promote cell invasion and migration in cancers and positively correlate with glioma grade (Fig. 4), were selected for validation studies on glioma cells.
MMP-9 and SNAI1 involved in TPM3 functional signaling for glioma regulation
To determine whether or not TPM3 regulates MMP-9 and SNAI1 in glioma cells, MMP-9 and SNAI1 expression was determined in blank, scrambled, and si-TPM3-treated cells by Western blot analysis. Compared with the blank and scrambled control cells, si-TPM3 cells showed decreased MMP-9 and SNAI1 expression (Fig. 5). Further, transwell and wound healing assays showed that overexpression of TPM3 promoted glioma cell invasion and migration. Introduction of si-MMP9 or si-SNAI1 abrogated TPM3 overexpression-induced cellular invasion and migration. These results suggest that TPM3 modulates malignant progression at least partially through MMP9 and SNAI1 (Supplementary Fig2).
Discussion
Glioma is the most common lethal intracranial tumor in adults. Even after years of efforts in developing and improving therapeutic strategies, the survival times of glioma patients remain short. Local invasion is the hallmark of malignant gliomas and the major cause of recurrence and morbidity [1]. Given the identification of novel biomarkers and new molecular classification systems, a new era of diagnostics is expected to emerge.
TPM3, an important member of the TPM family, is known to play a major role in the pathological processes of hepatocellular carcinoma, specifically cell migration and invasion [8]. Expression levels of TPM3 are reportedly higher in a highly metastatic mouse melanoma cell line than in a low metastatic one [5]. However, the effect of TPM3 on gliomas and the associated regulatory networks of TPM3 have yet to be elucidated. In the present study, we found that TPM3 is overexpressed in HGGs and that TPM3 overexpression is significantly associated with higher mortality in GBM patients. Hence, TPM3 shows potential as a prognostic marker and therapeutic target. We subsequently conducted a study to elucidate the biological effects of TPM3 overexpression in gliomas using RNA interference (RNAi) technology to knock-down TPM3 expression. We found that TPM3 knockdown profoundly represses the invasion and migration potential of glioma cells compared with identical cell lines without siTPM3 treatment. We also explored the mechanisms behind the reduced invasion and migration observed in TPM3 knockdown glioma cells. Using microarray combined with Pearson correlation analysis, we found that several MMP family members and EMT-like markers are overexpressed in HGGs and positively correlated with TPM3 expression. Among these markers, MMP-9 and SNAI1, which are genes strongly associated with malignancy in glioma, were selected for functional validation. In vitro studies reveal that TPM3 knockdown reduces the levels of MMP-9 and SNAI1 expression in glioma cells. Restoration experiments suggest that TPM3 modulates malignant progression at least partially through MMP9 and SNAI1in glioma.
EMT is a complex cellular process that reflects a high level of phenotypic plasticity. Milestone studies in colon cancer have led to the construction of a classic EMT-mediated model for epithelial solid tumor metastasis [9]. Interestingly, recent publications in the field of brain tumor research indicate an apparent link between EMT-like processes and GBM progression and invasion [10]. In recent years, extensive experimental work has yielded an improved understanding of the molecular mechanisms of these processes and resulted in the elucidation of core components of EMT-like activators in glioma [11]. Initial reports have implicated TWIST1overexpression in malignant glioma development and GBM invasion promotion in vitro [12]. The same authors reported that shRNA-mediated knockdown of TWIST1 significantly impairs the in vitro migration and invasion of GBM cells and reduces their sphere formation capacity. Higher ZEB2 levels have been detected in 90 clinicopathologically characterized specimens derived from glioblastoma patients who experienced early relapse and rapid tumor progression [13]. In the same study, knockdown of ZEB2 in GBM-derived cell cultures resulted in impaired migration and invasion, decreased proliferation, and increased cell death. The SNAI family represents a third group of transcriptional activators that play a significant role in the EMT process in cancer cells [14]. SNAI1 and SNAI2 are reportedly closely associated with the elevated invasion and migration of malignant gliomas [15–17]. Members of the MMP family are involved in the breakdown of the extracellular matrix in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in several diseases [18, 19]. MMP-9 is overexpressed in various human cancers and acts as an important oncogene that promotes the invasiveness of cancer cells [20, 21]. Many studies have attempted to investigate the predictive value of MMP-9 in various malignancies. In one study, MMP-9 conferred a poor prognosis in patients with GBM [22]. Similarly, increased expression of MMP-9 was correlated with poor prognosis in gastric cancer, bladder cancer, and nasopharyngeal carcinoma patients [23–25]. In the present study, MMP-9 and SNAI1 show significantly higher expression in HGG tissues than in low-grade gliomas and are positively regulated by TPM3 in in vitro assays.
In summary, we confirmed that TPM3 is correlated with poor prognosis in GBM patients and showed for the first time that TPM3 expression profile in different grades of gliomas. In addition, we demonstrated that TPM3 knockdown profoundly represses the invasion and migration of glioma cell lines as well as the expression of MMP-9 and SNAI1. To the best of our knowledge, our work is the first to show that TPM3 is involved in the malignant phenotype of glioma by activating MMP and EMT-like signaling pathways. Considering the key role of TPM3 in cell invasion and migration, TPM3 may participate in glioma progression and thus indicate poor prognosis. As such, TPM3 may be a potential target for GBM therapy.
References
Wang Y, Jiang T. Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013;331:139–46.
Pieples K, Arteaga G, Solaro RJ, et al. Tropomyosin 3 expression leads to hypercontractility and attenuates myofilament length-dependent Ca(2+) activation. Am J Physiol Heart Circ Physiol. 2002;283:H1344–53.
Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328–36.
Kim TM, Yim SH, Shin SH, et al. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer. 2008;123:2808–15.
Miyado K, Kimura M, Taniguchi S. Decreased expression of a single tropomyosin isoform, TM5/TM30nm, results in reduction in motility of highly metastatic B16-F10 mouse melanoma cells. Biochem Biophys Res Commun. 1996;225:427–35.
Yan W, Zhang W, You G, et al. Molecular classification of gliomas based on whole genome gene expression: a systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neuro Oncol. 2012;14:1432–40.
Zhang C, Zhang J, Hao J, et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J Transl Med. 2012;10:119.
Choi HS, Yim SH, Xu HD, et al. Tropomyosin3 overexpression and a potential link to epithelial–mesenchymal transition in human hepatocellular carcinoma. BMC Cancer. 2010;10:122.
Brabletz T, Jung A, Reu S, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A. 2001;98:10356–61.
Kahlert UD, Nikkhah G, Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–8.
Brabletz T. To differentiate or not—routes towards metastasis. Nat Rev Cancer. 2012;12:425–36.
Elias MC, Tozer KR, Silber JR, et al. TWIST is expressed in human gliomas and promotes invasion. Neoplasia. 2005;7:824–37.
Qi S, Song Y, Peng Y, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS One. 2012;7:e38842.
Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28.
Bhat-Nakshatri P, Appaiah H, Ballas C, et al. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24− phenotype. BMC Cancer. 2010;10:411.
Han SP, Kim JH, Han ME, et al. SNAI1 is involved in the proliferation and migration of glioblastoma cells. Cell Mol Neurobiol. 2011;31:489–96.
Weissenberger J, Priester M, Bernreuther C, et al. Dietary curcumin attenuates glioma growth in a syngeneic mouse model by inhibition of the JAK1,2/STAT3 signaling pathway. Clin Cancer Res. 2010;16:5781–95.
Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–73.
Surgucheva I, Chidambaram K, Willoughby DA, Surguchov A. Matrix metalloproteinase 9 expression: new regulatory elements. J Ocul Biol Dis Infor. 2010;3:41–52.
Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74.
Comincini S, Paolillo M, Barbieri G, et al. Gene expression analysis of an EGFR indirectly related pathway identified PTEN and MMP9 as reliable diagnostic markers for human glial tumor specimens. J Biomed Biotechnol. 2009;2009:924565.
Yan W, Zhang W, Sun L, et al. Identification of MMP-9 specific microRNA expression profile as potential targets of anti-invasion therapy in glioblastoma multiforme. Brain Res. 2011;1411:108–15.
Liu Z, Li L, Yang Z, et al. Increased expression of MMP9 is correlated with poor prognosis of nasopharyngeal carcinoma. BMC Cancer. 2010;10:270.
Chu D, Zhang Z, Li Y, et al. Matrix metalloproteinase-9 is associated with disease-free survival and overall survival in patients with gastric cancer. Int J Cancer. 2011;129:887–95.
Offersen BV, Knap MM, Horsman MR, Verheijen J, Hanemaaijer R, Overgaard J. Matrix metalloproteinase-9 measured in urine from bladder cancer patients is an independent prognostic marker of poor survival. Acta Oncol. 2010;49:1283–7.
Acknowledgments
This work was supported by grants from the National High Technology Research and Development Program of China (863) (2012AA02A508), International Cooperation Program (2012DFA30470), National Natural Science Foundation of China (91229121, 81272792, 81172389,81372709, 81302185, 81101901,81302184), Jiangsu Province’s Natural Science Foundation (BK2011847 and 20131019), Jiangsu Province’s Key Provincial Talents Program (RC2011051), Jiangsu Province’s Key Discipline of Medicine (XK201117), Jiangsu Provincial Special Program of Medical Science (BL2012028), and Program for Development of Innovative Research Team in the First Affiliated Hospital of NJMU, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflicts of interest
None
Author information
Authors and Affiliations
Consortia
Corresponding authors
Additional information
T. Tao, Y. Shi and D. Han contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig.1
Cell proliferation determined by CCK-8 assay. (A and B) CCK-8 assays showed that there was no significant difference between si-TPM3-transfected cells and negative control group within 24 hours. #P > 0.05. Results were presented as mean±SD from three independent experiments. (GIF 12 kb)
Supplementary Fig.2
MMP-9 and SNAI1 involved in TPM3 functional signaling for glioma invasion and migration. (A and B) si-MMP9 and si-SNAI1 were introduced into TPM3-treated cells. Decreased MMP9 or SNAI1 in TPM3-treated cells rescued the invasion phenotype induced by TPM3. (C and D) Introduction of si-MMP9 or si-SNAI1 abrogated TPM3 overexpression-induced cellular migration. (*P < 0.05, **P < 0.01). Results were presented as mean±SD from three independent experiments. (GIF 93 kb)
Rights and permissions
About this article
Cite this article
Tao, T., Shi, Y., Han, D. et al. TPM3, a strong prognosis predictor, is involved in malignant progression through MMP family members and EMT-like activators in gliomas. Tumor Biol. 35, 9053–9059 (2014). https://doi.org/10.1007/s13277-014-1974-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-1974-1