Abstract
Signal peptide is an essential part during protein translocation for extracellular expression in the plants. A secretomics approach was performed to identify secretory proteins from oil palm and predict the presence of the signal peptide in each protein. The transcriptome and secretome profiles were generated from oil palm ramets. A combination of bioinformatics tools including SignalP, TargetP, TMHMM, and SecretomeP was used to predict both classical and leaderless secretory proteins. The secretome analysis from transcriptome data revealed 2259 genes that encoded secretory proteins. The proteome analysis identified a total of 37 proteins from which 10 classical secretory proteins can be distinguished. It was important to note that oil palm ramet’s secretome was dominated by stress or defence related proteins which may act as protection during certain plant developmental stages. We also found that some proteins can be both intracellular and extracellular protein. Among 10 identified classical secretory proteins, the signal peptides of oil palm beta-1,3-glucanase and putative class III chitinase were interesting for further study due to their high cleavage site scores. Overall, this study provided broader view of oil palm secretomes and specific signal peptides for extracellular expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
To carry out its destined function, a certain class of protein needs to be translocated either to specific organelle in cytoplasm or to extracellular spaces. The translocation of protein is usually determined by the existence of specific targeting signal called the signal peptide (SP) which is part of the N-terminus of a protein. SPs are cleaved from its mature peptide by signal peptidases after the targeting process has been completed (Peng et al. 2019). As an integral part of the protein, malfunction SP may lead to a loss of protein activity or even changes in phenotype. The study of maize floury4 (fl4) mutant showed that nucleotide alteration in SP regions resulted in formation of misshapen and aggregated zein protein bodies in endosperm which was due to the uncleaved SP (Wang et al. 2014). This mutations lead to opaque endosperm phenotypes, increased starchy regions, and soft kernel textures. Apart from their translocation function, SPs also have other roles such as controlling protein secretion rates, determining protein folding-state, affecting downstream trans-membrane behaviour and amino terminal glycosylations, gene regulations, and in some cases might act as an antigen (Owji et al. 2018).
Given the fact that SP is an essential part during protein translocations, the information of SPs from a particular plant and its targeting locations may give benefits for certain purposes such as recombinant protein production in plants. By using SP, the protein could be directed to the extracellular pathway and secreted to the cell culture medium through the plant roots. This approach could increase the protein yields and simplify the harvesting and purification method (Pham et al. 2012). The type of SP also determines protein secretion efficiency which directly affected protein yields. It has been demonstrated in various expression system that the type of SP is important for efficient secretion (Jonet et al. 2012; Huang et al. 2015; Jiang et al. 2020). In another example, SP was used for directing beneficial protein such as phytase and acid phosphatase outside of transgenic alfalfa plants (Ma et al. 2012). The combined activities of these extracellular proteins lead to improvement of P acquisition from natural agricultural soils thus allowing reduced P inputs while maintaining high yields.
The information about extracellular proteins from oil palm including its SPs is very limited and needs to be explored. The availability of such information would allow a better understanding of secretome profiles and development of extracellular expression systems in oil palm. In this study, secretomics approach combines the transcriptomics, the bioinformatics, and the proteomics to identify the native oil palm SP candidates which can be used for protein secretions. A list of predicted oil palm root and leaf secretory proteins which contained SP was generated from transcriptome data. The existence of secreted proteins was confirmed by comparing transcriptome data with proteome data generated from oil palm leaf, root, and also root exudates. At last, a list of recommended SP sequences was identified and presented in this study.
Materials and methods
Plant materials
Ramets, that were grown in liquid medium and ready for acclimatization, were used in this research. Ramets were selected under the following criteria: have a minimum of two roots with over than 1 cm length, have a minimum of two leaves, and the height is more than 10 cm. These ramets were subjected to RNA and protein analysis for identification of secretory proteins. In addition, the liquid medium which was used to grow ramets, was also collected and pooled for protein analysis. Three biological replications were applied for all the samples in this research.
RNA isolation and sequencing
The total RNA was extracted from root and leaf samples of oil palm ramets by using Rneasy Plant Mini Kit (QIAgen, Hilden, Germany) according to the manufacture’s procedure. The quantity and quality of extracted RNA were measured using NanoDrop™ 2000c Spectrophotometer (Thermo Scientific, Massachusetts, USA). The RNA was preserved in RNAstable (Biomatrica, California, USA) and the sequencings were performed by Novogene Co., Ltd., Beijing, China. The RNA quantity and integrity was re-checked using the 2100 Bioanalyzer Instrument (Agilent Company, USA) while electrophoresis was used for checking the sample purity. The parameters including RNA concentration, rRNA ratio, and RNA integrity number were calculated. The samples whose RIN or RINe value were more than 5.9 were processed further. RNA was converted into cDNA and then the sequencing adapters were added. The cDNA was sequenced using Illumina HiSeq 4000 System. The raw reads were trimmed to remove adapter and the clean reads was mapped into oil palm transcriptomics reference version NCBI 101 (Singh et al. 2013) using Kallisto version 0.46 software (Bray et al. 2016). A transcript that has a value of more than 40 transcripts per million (TPM) was selected for further analysis.
Isolation of secreted proteins
Secreted proteins were isolated from leaf and root of oil palm ramets and its liquid medium. Root and leaf from ramets were cut into pieces (0.5–0.8 cm) and submerged into 15 mL of sterile ddH2O. The samples were incubated at 4 °C for overnight with 100 rpm agitation and then the supernatant was collected by centrifugation at 1500 × g for 30 min at 4 °C. As for liquid medium, the samples were filtered and concentrated using a freeze dryer until the volume reach 1/3 of the initial volume. Proteins from both of supernatant and concentrated medium were purified by adding 3 volumes of cold acetone followed by incubation at − 20 °C for an hour and centrifugation at 3000 × g for 10 min at 4 °C. The protein pellets were collected and dried using a vacuum centrifuge (Tomy Micro Vac MV-100) for 5 min. Isolated proteins were quantified by the Bradford method (Nouroozi et al. 2015). SDS-PAGE was used for protein quality control before and after the digestion. A qualitative identification of proteins was performed using in-solution digestion followed by high performance liquid chromatography tandem mass spectrometry (LC–MS/MS)(Ngcala et al. 2020).
Bioinformatics analysis of transcriptome and proteome data
The prediction and validation pipeline for genome-wide secretome (Agrawal et al. 2010) was adopted for main analysis of oil pam secretome in this study. In addition, other softwares such as MultiLoc2 and CateGOrizer were used to generate more information for deeper analysis of the secretomes. Each transcript was translated into amino acid sequence and subjected to further analyses. SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP; Armenteros et al. 2019a, b), TMHMM v2 (http://www.cbs.dtu.dk/services/TMHMM; Sonnhammer and Krogh, 2008), TargetP-2.0 (http://www.cbs.dtu.dk/services/TargetP; Armenteros et al. 2019a, b), and SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP; Bendtsen et al. 2004) were employed to identify secretory proteins from both of transcript data and identified proteins. Default software parameters were applied for all bioinformatics analysis. For transcriptome data, analysis using SignalP, TargetP, and TMHMM were performed simultaneously to generate predicted classical secretory proteins (CSP). Proteins with SP and without transmembrane region were taken as secreted protein. The excluded proteins were analyzed by SecretomeP (NN-score > 0.6) for predicting leaderless secretory proteins (LSP). MultiLoc2 (https://abi-services.informatik.uni-tuebingen.de/multiloc2/webloc.cgi; Blum et al. 2009) was used to predict the subcellular location of protein while CateGOrizer (https://www.animalgenome.org/tools/catego; Hu et al. 2008) was used to classify proteins based on the gene ontology. GO-Slim was used as classification method for GO terms in CateGOrizer.
SignalP and TargetP are machine learning method-based tools which are usually used to identify N-terminal SP on protein sequences (Emanuelsson et al. 2007; Nielsen 2017). Both softwares can predict the presence of SP in protein sequence but TargetP can further distinguish between secretory SP and transit peptide for subcellular localization (mitochondria, chloroplast, and thylakoid luminal). TMHMM can be used to determine transmembrane proteins by predicting the number of transmembrane helices in proteins. Proteins with predicted SPs but did not contain predicted transmembrane region are selected as secreted protein candidates (Chen et al. 2003). By combining the analysis of this software, we could get a shortlist of CSPs from root and leaf of oil palm ramets. On the other hand, it was known that around 50% of plant secretory proteins do not possess SP (LSPs) (Agrawal et al. 2010; Alexandersson et al. 2013; Krause et al. 2013). The LSPs are mainly identified under biotic and biotic stress conditions, suggesting their roles in defense or stress responses (Lum and Min 2011). This type of secretory protein was predicted by SecretomeP software based on sequence-derived features such as composition, size, charge, post-translational modification, predicted structure and degradation signal (Bendtsen et al. 2004). In addition, Multiloc2 software, which incorporates overall amino acids composition and sorting signals with phylogenetic profiles and GO terms, was used for predicting subcellular location of protein within eukaryotic cells (Blum et al. 2009). This series of bioinformatics analysis allowed better precision in predicting the secretome profiles of targeted samples.
Results
Broad view of oil palm secretome profiles
Based on NCBI Reference Sequence database (GenBank Accession No. PRJNA268357), oil palm has a total of 41,801 protein-coding genes. Transcriptomics analysis showed that 4402 genes were expressed in roots while 4377 genes were expressed in leaves of oil palm ramets (data not shown). Parallel bioinformatics analysis was carried out towards transcriptomics data using SignalP, TargetP and TMHMM for prediction of CSPs (Supplementary file). The analysis of root’s transcriptome data showed that 771 genes (17.51%) contained SP sequence but only 292 genes (6.63%) were predicted to encode CSPs (Fig. 1). As for leaf transcriptome, 856 genes (19.56%) contained SP sequence but only 188 genes (4.29%) were predicted to encode CSPs. Overall, our analysis generated the total of 347 CSPs, 159 proteins were found in roots and 55 proteins were found in leaves while 133 proteins were found in both organs. In addition, SecretomeP software was used to predict LSPs from transcriptome data. We found that 1,462 root genes (33.21%) and 1497 leaf genes (34.2%) were belong to this group. A large portion of LSPs (1067 genes) was found in both organs.
Identification and characterization of secretory proteins from extracted proteins
Proteomics analysis was used to validate the predicted secretomes. A total of 37 proteins were successfully identified from the medium, leaf, and root samples (Table 1). Among them, 12 proteins were found across the samples while the rest could only be found in its respective sources. Once the proteins were identified, the full amino sequences of the protein were derived from database and used for predicting the presence of secretory signals and transit peptides, function in biological processes, and the subcellular locations within the cells.
Based on the result of combined bioinformatics analysis, identified proteins were categorized into 4 predicted protein types: classical secretory, leaderless secretory, mitochondrial, and non-secretory protein. Out of 37 identified proteins, ten proteins were predicted to contain secretory SP which could direct the proteins to extracellular compartment (CSP) while only one protein contained mitochondrial transit peptide. Further analysis using SecretomeP showed that 13 proteins were secretory proteins without signal sequences (LSP). The remaining proteins belong to non-secretory protein class. Overall, 24 identified proteins were predicted as secretory proteins.
We used GO terms to determine the protein function and then GO-Slim classification for grouping the proteins based on its function. A large portion of identified secretory proteins involved in cellular processes (38%) while the others play a role in protein metabolic process (13%), lipid metabolic process (13%), and other miscellaneous functions (Fig. 2). These data suggest that secretory proteins involve in wide range of cellular processes. For subcellular locations, analysis using Multiloc2 showed that most of identified proteins were located in cytoplasm (40%) while the rest were divided between extracellular (38%), chloroplast (14%), mitochondria (5%), and nuclear (3%).
Determination of SP candidates
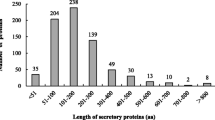
From identified CSPs, we could generate 10 SPs candidates. The length of SPs ranges from 20 to 28 amino acids (Fig. 3) regardless of the total protein lengths which vary between 118 and 525 amino acids. The SP length prediction was based on the probability of cleavage site position. The higher the probability value, the more likely that the SP would be cleavaged in the predicted position.
Discussion
A plant was known to secrete many proteins to extracellular spaces which played an important role during stress responses, maintenance of cell structures, communications, and development processes (Agrawal et al. 2010; Alexandersson et al. 2013; Krause et al. 2013). In this study, we focused on identification of extracellular proteins of oil palm ramets to get a broader view of oil palm secretome profiles and obtain a list of SP candidates. Transcriptome data from leaf and roots of oil palm ramets were used for predicting the secretory proteins then a comparison with proteomics data was made to validate the result.
Analysis on transcriptome data showed that in total, oil palm ramet roots had higher number of expressed secretory proteins compared to leaf. It was known that plants are continuously secreting an enormous range of compounds to their surrounding environment including soils (Badri and Vivanco 2009; Vives-Peris et al. 2020). The root exudates which consist of antimicrobial compounds, signal molecules, and nutrients, facilitates wide range of interactions between plant and its surrounding rhizosphere (Haichar et al. 2014). In addition, the data also showed that a large number of proteins were present in both organs, especially for LSPs group.
Plant as a sessile organism developed unique and advanced biological mechanisms to deal with environmental challenges. Cell secretion is one of the critical biological systems in this adaptation process. It was interesting to see that stress/defence related genes were highly expressed in root and leaf (Supplementary file). Pathogenesis-related protein, germin, and protease were among those genes in root. Germin and germin-like protein which are expressed during specific periods of plant growth and development, involve in plant protection toward various environmental stresses (Dunwell et al. 2008). On a different manner, proteases play a role as regulator in plant immunity system and a large number of them act in apoplast (Balakireva and Zamyatnin 2018). BURP domain-containing protein, non-specific lipid-transfer protein, and phylloplanin were some of highly expressed stress/defence related-genes in leaf. BURP-domain-containing is a protein class that unique to plant and involves in development and metabolism. Many of these proteins are stress related gene as shown in Medicago truntula plant where all of its BURP genes were regulated by drought stress (Li et al. 2016). Non-specific lipid-transfer protein present in abundance in higher plant and involves in various cell processes including resistance to biotic and abiotic stress (Liu et al. 2015). On the other hand, phylloplanins acts as leaf surface defence protein in tobacco where pathogen spore germination and leaf infection were inhibited in the presence of this proteins (Shepherd et al. 2005).
Multiloc2 software was used to further strengthen the analysis of sample-derived proteins. All proteins that belong to CSP were predicted to enter the secretory pathway (Table 1); however, most of LSPs were predicted to be located in other organelles such as mitochondria, cytoplasm, or chloroplast. Putative G-D-S-l family lipolytic protein which contained a mitochondrial transit peptide was also predicted to enter the secretory pathway. The difference in analysis method between used softwares might cause this kind of disagreement. We found that some intracellular proteins such as ribosomal protein, histone, or ribulose bisphosphate carboxylase were predicted as LSP by SecretomeP. The location for these proteins could be easily clarified because its biological roles are already well-defined. As for other proteins, the function and location should be determined individually by literature studies. It should also be noted that SecretomeP were designed to predict LSPs from bacteria and mammals therefore the analysis may not be suitable for some plant proteins.
In the case of non-secretory proteins, no disagreement was found between software outputs. We also found that some particular proteins can be both intracellular and extracellular protein. Enolase (Didiasova et al. 2019) and peptidyl-prolyl cis–trans isomerase (Xue et al. 2018) are two examples of protein that can be found in both locations. These proteins are predicted as intracellular protein but actually they are also secreted to extracellular compartment. Intracellular proteins contamination could be found in our proteomics analysis output. The cytoplasmic leakage from broken plant cells which occurred naturally or were resulted from protein extraction processes may be the cause of this contamination (Alexandersson et al. 2013).
Comparison between transcriptomics and proteomics data showed that almost all sample-derived CSPs could be found in the transcript data. Only pathogenesis-related protein that was absent in leaf transcripts even though the protein was identified in extracted proteins. This might happen due to different analysed samples where individual sample was used for transcriptome analysis while pooled sample was used for protein identification. The amount of required samples for proteomics analysis was a limiting factor in our experiment thus the isolation method should be further improved in the future research.
Both of transcriptomics and proteomics data showed that secretome of oil palm ramet was dominated by stress/defence related proteins. The proteins may directly involve in stress response or play a part on activation of more complex system. The presence of stress/defence related protein did not directly indicate that the oil palm ramets were infected by pathogens or under a stress condition. The expression of such proteins may be related to a certain phase of plant lifecycle. In Arabidopsis thaliana, the root secretion of defence-related proteins (e.g. chitinase, glucanases, and myrosinases) was enhanced during flowering. This event suggested that plant shifted to defence mode in order to prevent pathogen attack during important stage of developmental cycle (De-la-Peña et al. 2010). Similar event also found in Pisum sativum roots where newly generated root tip was resistance to pathogen infection. The secretory proteins were released when border cell separation occurred and appeared as protection of the root tip (Wen et al. 2006). Oil palm ramet is still at early development stage towards mature plant and it seems that stress related genes were expressed as innate defence strategy for surviving this developmental phase. The expression of stress or defence related genes may also be caused by overlapping roles of the regulator genes. Major transcription factor families in higher plants which are closely related to defence signaling also have overlapping roles in development processes (Ng et al. 2018). It was also reported that growth regulating factors (GRFs) which involves in tissue differentiation and organ development may also regulate various biological processes related to defence response (Liu et al. 2014). These reports suggest that defence and development process are closely related thus may explain the presence of defence or stress related genes in our secretome data.
Although SPs have a low degree of sequence conservation, its structure are usually composed by three typical domains: an amino terminal positively charged and hydrophilic region (n-region), a central hydrophobic region (h-region), and a more polar carboxy region (c-region) which specifies cleavage site (Owji et al. 2018). Based on this information, the existence of SPs can be predicted from the amino sequences of proteins. We identified 10 CSPs from the samples and further analyzed the SPs. Non-specific lipid-transfer protein (A0A1D5AIU7) had the highest cleavage site score while glucan endo-1,3-beta-glucosidase had the lowest one. Higher score means higher probability of SP cleavage in the predicted position. Considering the cleavage site score and the presence of protein in all samples, the SP sequence from beta-1,3-glucanase (B3TLW8; MANRSKVSRAAVALLIGLLVAIPTGVKS) and putative class III chitinase (K4MQC1; MATNQLLPLLLLALVAGSHA) were recommended for further use. The SP from pathogenesis-related protein may also be used but it had relatively low cleavage site score. These SPs should be tested first using a reporter gene such as beta glucuronidase or fluorescent protein in the future study.
Conclusion
This study generated a broader view of secretomes from oil palm ramets and specific SP candidates for future applications. The existence of some secretory proteins was confirmed by identification of proteins from root, leaf, and growth medium samples. We found that secretome of oil palm ramet was dominated by stress or defence related proteins which may act as protection during development process. In addition, the expression of stress or defence related genes in development process may indicate the overlapping roles of regulator genes. Furthermore, SPs from identified proteins were selected based on cleavage site score and the presence of protein in various samples. The combination of different bioinformatics software greatly increased the level of confidence when determining secretory proteins but some result disagreement between software may emerge especially for the LSPs. If possible, additional information from related references may be used to solve this problem. On the other hand, protein isolation methods should be optimized further to increase the number of identified proteins while minimizing the presence of contaminant proteins. To our knowledge, this is the first report about oil palm secretome and the provided information should be useful for wide range of research. In the future, similar approach may be used to identify oil palm secretome under certain conditions such as abiotic stress or pathogen infection. The performance of recommended signal sequences should also be assessed with reporter gene first before it can be used for extracellular expression.
Availability of data and materials
Not applicable.
Code availability
Not applicable.
Abbreviations
- SP:
-
Signal peptide
- CSP:
-
Classical secretory protein
- LSP:
-
Leaderless secretory protein
References
Agrawal GK, Jwa NS, Lebrun MH et al (2010) Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10:799–827. https://doi.org/10.1002/pmic.200900514
Alexandersson E, Ali A, Resjö S, Andreasson E (2013) Plant secretome proteomics. Front Plant Sci 4:1–6. https://doi.org/10.3389/fpls.2013.00009
Armenteros JJA, Salvatore M, Emanuelsson O et al (2019a) Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance 2:1–14. https://doi.org/10.26508/lsa.201900429
Armenteros JJA, Tsirigos KD, Sønderby CK et al (2019b) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. https://doi.org/10.1038/s41587-019-0036-z
Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32:666–681. https://doi.org/10.1111/j.1365-3040.2009.01926.x
Balakireva AV, Zamyatnin AA (2018) Indispensable role of proteases in plant innate immunity. Int J Mol Sci. https://doi.org/10.3390/ijms19020629
Bendtsen JD, Jensen LJ, Blom N et al (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17:349–356. https://doi.org/10.1093/protein/gzh037
Blum T, Briesemeister S, Kohlbacher O (2009) MultiLoc2: Integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinform 10:274. https://doi.org/10.1186/1471-2105-10-274
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. https://doi.org/10.1038/nbt.3519
Chen Y, Yu P, Luo J, Jiang Y (2003) Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm Genome 14:859–865. https://doi.org/10.1007/s00335-003-2296-6
De-la-Peña C, Badri DV, Lei Z et al (2010) Root secretion of defense-related proteins is development-dependent and correlated with flowering time. J Biol Chem 285:30654–30665. https://doi.org/10.1074/jbc.M110.119040
Didiasova M, Schaefer L, Wygrecka M (2019) When place matters: shuttling of enolase-1 across cellular compartments. Front Cell Dev Biol 7:1–11. https://doi.org/10.3389/fcell.2019.00061
Dunwell JM, Gibbings JG, Mahmood T, Saqlan Naqvi SM (2008) Germin and germin-like proteins: evolution, structure, and function. CRC Crit Rev Plant Sci 27:342–375. https://doi.org/10.1080/07352680802333938
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971. https://doi.org/10.1038/nprot.2007.131
Haichar FZ, Santaella C, Heulin T, Achouak W (2014) Root exudates mediated interactions belowground. Soil Biol Biochem 77:69–80. https://doi.org/10.1016/j.soilbio.2014.06.017
Hu Z-L, Bao J, Reecy J (2008) CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J Bioinform 9:108–112
Huang LF, Tan CC, Yeh JF et al (2015) Efficient secretion of recombinant proteins from rice suspension-cultured cells modulated by the choice of signal peptide. PLoS ONE 10:1–16. https://doi.org/10.1371/journal.pone.0140812
Jiang MC, Hu CC, Hsu WL et al (2020) Fusion of a novel native signal peptide enhanced the secretion and solubility of bioactive human interferon gamma glycoproteins in Nicotiana benthamiana using the bamboo mosaic virus-based expression system. Front Plant Sci 11:1–18. https://doi.org/10.3389/fpls.2020.594758
Jonet MA, Mahadi NM, Murad AMA et al (2012) Optimization of a heterologous signal peptide by site-directed mutagenesis for improved secretion of recombinant proteins in Escherichia coli. J Mol Microbiol Biotechnol 22:48–58. https://doi.org/10.1159/000336524
Krause C, Richter S, Knöll C, Jürgens G (2013) Plant secretome—from cellular process to biological activity. Biochim Biophys Acta - Proteins Proteomics 1834:2429–2441. https://doi.org/10.1016/j.bbapap.2013.03.024
Li Y, Chen X, Chen Z et al (2016) Identification and expression analysis of BURP domain-containing genes in Medicago truncatula. Front Plant Sci 7:1–16. https://doi.org/10.3389/fpls.2016.00485
Liu J, Rice JH, Chen N et al (2014) Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS ONE 9:1–14. https://doi.org/10.1371/journal.pone.0098477
Liu F, Zhang X, Lu C et al (2015) Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J Exp Bot 66:5663–5681. https://doi.org/10.1093/jxb/erv313
Lum G, Min XJ (2011) Plant secretomes: current status and future perspectives. Plant Omics 4:114–119
Ma XF, Tudor S, Butler T et al (2012) Transgenic expression of phytase and acid phosphatase genes in alfalfa (Medicago sativa) leads to improved phosphate uptake in natural soils. Mol Breed 30:377–391. https://doi.org/10.1007/s11032-011-9628-0
Ng DWK, Abeysinghe JK, Kamali M (2018) Regulating the regulators: the control of transcription factors in plant defense signaling. Int J Mol Sci. https://doi.org/10.3390/ijms19123737
Ngcala MG, Goche T, Brown AP et al (2020) Heat stress triggers differential protein accumulation in the extracellular matrix of sorghum cell suspension cultures. Proteomes 8:1–19. https://doi.org/10.3390/proteomes8040029
Nielsen H (2017) Predicting secretory proteins with signaIP. Methods Mol Biol 1611:59–73. https://doi.org/10.1007/978-1-4939-7015-5_6
Nouroozi RV, Noroozi MV, Ahmadizadeh M (2015) Determination of protein concentration using bradford microplate protein quantification assay. Int Electron J Med 4:11–17. https://doi.org/10.31661/iejm158
Owji H, Nezafat N, Negahdaripour M et al (2018) A comprehensive review of signal peptides: structure, roles, and applications. Eur J Cell Biol 97:422–441. https://doi.org/10.1016/j.ejcb.2018.06.003
Peng C, Shi C, Cao X et al (2019) Factors influencing recombinant protein secretion efficiency in gram-positive bacteria: signal peptide and beyond. Front Bioeng Biotechnol 7:1–9. https://doi.org/10.3389/fbioe.2019.00139
Pham NB, Schäfer H, Wink M (2012) Production and secretion of recombinant thaumatin in tobacco hairy root cultures. Biotechnol J 7:537–545. https://doi.org/10.1002/biot.201100430
Shepherd RW, Bass WT, Houtz RL, Wagner GJ (2005) Phylloplanins of tobacco are defensive proteins deployed on aerial surfaces by short glandular trichomes. Plant Cell 17:1851–1861. https://doi.org/10.1105/tpc.105.031559
Singh R, Ong-Abdullah M, Low ETL et al (2013) Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500:335–339. https://doi.org/10.1038/nature12309
Sonnhammer ELL, Krogh A (2008) A hidden Markov model for predicting transmembrane helices in protein sequence. Sixth Int Conf Intell Syst Mol Biol 8
Vives-Peris V, de Ollas C, Gómez-Cadenas A, Pérez-Clemente RM (2020) Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep 39:3–17. https://doi.org/10.1007/s00299-019-02447-5
Wang G, Qi W, Wu Q et al (2014) Identification and characterization of maize floury4 as a novel semidominant opaque mutant that disrupts protein body assembly. Plant Physiol 165:582–594. https://doi.org/10.1104/pp.114.238030
Wen F, VanEtten HD, Tsaprailis G, Hawes MC (2006) Extracellular proteins in pea root tip and border cell exudates. Plant Physiol 143:773–783. https://doi.org/10.1104/pp.106.091637
Xue C, Sowden MP, Berk BC (2018) Extracellular and intracellular cyclophilin A, native and post-translationally modified, show diverse and specific pathological roles in diseases. Arterioscler Thromb Vasc Biol 38:986–993. https://doi.org/10.1161/ATVBAHA.117.310661
Acknowledgements
The authors would like to express their great appreciation to the director of Plant Production and Biotechnology Division for permission to publish this article.
Funding
This project was fully funded by PT SMART Tbk.
Author information
Authors and Affiliations
Contributions
CD: conceptualization, methodology, writing—original draft and visualization. RAR: conceptualization, methodology and data curation. ZAT: methodology, software, data curation and vizualization. WA: resources. CU: writing—review, editing and supervision. TL: writing—review, editing, supervision and funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflicts of interest that could influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Darmawan, C., Rohman, R.A., Tanjung, Z.A. et al. The prediction of specific oil palm extracellular signal peptides using plant secretomics approach. J Proteins Proteom 13, 29–38 (2022). https://doi.org/10.1007/s42485-021-00081-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-021-00081-y