Abstract
Multidrug resistance is one of the worldwide public health concerns. Water represents the most suitable environment, for the exchange of antibiotic resistance genes among pathogenic to non-pathogenic bacteria. Therefore, we aimed to screen the presence of blaNDM-1, blaTEM, blaSHV, blaCTX-M and mcr1–5 genes among water samples from different locations of Lower Himachal Pradesh. We examined the genotypic incidences of blaNDM-1, blaTEM, blaSHV, blaCTXM and mcr1–5 by polymerase chain reaction. Survivability assay, fitness cost assay and biofilm assay were performed for phenotypic characterization. The presence of blaNDM-1 and its related variants were analysed and confirmed by sequencing-based approaches. A total of 73 bacterial strains were identified on M-lauryl sulphate agar medium. Out of 73 colistin-resistant isolates, 34 were E. coli and 39 were K. pneumoniae. Out of 34 samples, 2 (5.8%), 2 (5.8%), 5 (14.7%), 5 (14.7%) and 4 (11.76%) E. coli were blaTEM, blaSHV, blaCTXM-1, blaCTXM-2 and blaCTXM-15 positive, respectively. Among 39 K. pneumoniae, 15 (38.4%), 6 (15.3%), 10 (25.6%), 9 (23.07%) and 10 (25.6%) were blaTEM, blaSHV, blaCTXM-1, blaCTXM-2 and blaCTXM-15 positive, respectively. Interestingly, we observed one E. coli (HG4) isolate with both blaNDM-1 and mcr-1 gene. Further analysis showed HG4 isolate has lesser survivability on the cotton swab, long lag phase and less biofilm production compared to colistin-sensitive isolates. Detection of E. coli with blaNDM-1 and mcr-1 in this geographical region is an alarming signal for tourists, community, health workers and policymakers. Hence, it is utmost important to take appropriate measures to control the dissemination of antibiotic resistance gene in such pristine locations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Overuse of antimicrobial agents promotes the spread of antibiotic resistance genes worldwide [1]. An environment containing bacteria with a trace of antibiotic resistance genes (ARGs) can increase the possibility of acquiring antibiotic resistance in human pathogens [2]. In addition, the population dynamics of natural microbial populations can also be challenged by spreading the ARGs in the natural ecosystem [3]. The increasing incidences of antimicrobial resistance in human pathogens apply an incredible effect on the global human health care system and are assessed to cause thousands of deaths annually [4].
In 2017, a report on major rivers of India showed most of the bacterial population of Indian rivers have high levels of resistance to broad-spectrum antibiotics (i.e. third-generation Cephalosporin [5]). A study on the River Cauvery in Karnataka showed 100% (283/283) of E. coli isolates were resistant to Cephalosporin [6]. In 2016 study, 17.4% (40/230) of Gram-negative bacteria of River Yamuna were found to be extended spectrum β-lactamase producers [7]. Interestingly, 17% of E. coli and 13% of Klebsiella species were found to be resistant to Cephalosporin in potable water sources apart from rivers from Uttar Pradesh, India [8]. Similarly, a report from East Sikkim showed 50% of E. coli and 72% of Klebsiella species were resistant to cephalosporin in natural water bodies [9]. Colistin, a last-resort antibiotic, is used for the treatment of infections caused by carbapenem-resistant E. coli and K. pneumoniae bacterium. In China, mcr-1-mediated colistin resistance among E. coli was isolated from pigs, chickens and humans [10]. Recently, colistin resistance among GNBs has been reported in India [11, 12].
The Indian pharmaceutical industry has an estimated US$15 billion in revenue in 2014 which is approximately 20% of the global demand for generic drugs (Nordea Asset Management 2015). Eighty per cent of antibiotics sold by multinational pharmaceutical companies are being manufactured in India or China. The effluents from manufacturing units contain significant levels of antibiotic residues, leading to waterways and lakes in India being contaminated [1, 13]. The Central Pollution Control Board (CPCB) sets effluent standards for the pharmaceutical industry in India, which is also applicable to all state pollution control boards. Unfortunately, antibiotics are not included in the current standards and therefore they are not monitored in pharmaceuticals effluent discharge (CPCB Effluent Standards 2013). Himachal Pradesh is a major touristic place in India; its economy is majorly dependent on tourism and agriculture. The Baddi town is one of the Asia's largest pharmaceutical hubs; this is home of around 270 pharmaceutical companies. These manufacturing units are discharging their effluent with residual antibiotics in local water bodies and contaminating them in the absence of any strict guidelines. Traces of antibodies can change the microbial ecosystem of the surrounding environment by putting selection pressure on the microbial population for antibiotic resistance. Therefore, we aimed for the presence of blaNDM-1, blaTEM, blaSHV, blaCTX-M and mcr1–5 genes among water samples from different locations of Lower Himachal Pradesh.
2 Materials and methods
2.1 Sampling sites
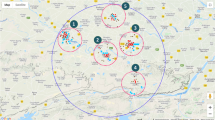
Water samples were collected from different locations of Manali to Baddi (Himachal Pradesh, India) in sterile water collection bottles (Fig. 1) following the standard method in the month of January 2019 [1]. Briefly, 500 mL of water sample was collected in autoclaved polypropylene bottles from 60 cm beneath the water surface and bottles were immediately kept on ice and stored overnight at 4 °C, and bacteriological analysis was undertaken on the following day.
2.2 Bacteria isolation
Screening of colistin-resistant bacteria was done on M-lauryl sulphate agar media (Hi-Media, Mumbai, India) with 2 µg per mL colistin. Bluish-green coloured colonies on the media were E. coli and yellowish colonies on media were K. pneumoniae. We further characterized these isolates by standard biochemical tests by using Hi25™ Enterobacteriaceae Identification Kit (Hi-Media, India). Bacterial isolates were preserved in 20% glycerol stocks and stored at −80 °C.
2.3 Antimicrobial susceptibility testing
The overnight grown culture of E. coli and K. pneumoniae was adjusted to the McFarland standard of 0.5 (equal to 1.5 × 108 CFU/mL) (Hi-Media, Mumbai, India). Then, both E. coli and K. pneumoniae were subjected to antibiotic susceptibility test (AST) by E test and broth microdilution test for colistin, using Mueller Hinton media in micro-titter plate (Hi-media, Mumbai, India) (Bauer, et al. 1966: 493-6). The detection of colistin-resistant isolates by micro-broth dilution method was done as per CLSI guidelines M07-A9 [14] The E. coli (ATCC25922) (Pan sensitive) was taken as negative control [15]. ESBL’s presence was confirmed by a double-disc synergy test utilizing ceftazidime (30 µg) and a mix of ceftazidime (30 µg) and clavulanic acid (10 µg) as per standard guideline [16]. Phenotypic detection of Metallo beta-lactamases was done by using Imipenem with & without EDTA Ezy MIC™ Strips test (IPM+EDTA/IPM (Hi-Media India).
2.4 Molecular characterization by PCR
Overnight culture of colistin-resistant E. coli and Klebsiella was centrifuged at 10,000 × g for 5 min. DNA was extracted from pellet using PureLink® Genomic DNA Kit (Invitrogen, CA, USA). The DNA integrity was confirmed by 1% agarose gel electrophoresis and quantitated using Nano-Drop 2000 (Thermo Fisher Scientific, MA, USA) at 260 and 280 nm. Primers for blaNDM-1, blaTEM, blaSHV, blaCTXM and mcr1–5, blaCTXM-1, blaCTXM-2, blaCTXM-8 and blaCTXM-15 were used; the sequence of these primers is given in Supplementary File: Table S1. Polymerase chain reaction (PCR) was performed using Master Cycler Gradient (Bio-Rad, USA) and amplified products were analysed on 1.5% agarose gel electrophoresis (Figure 2a, b) stained with EtBr and analysed on gel doc Bio-Plex 200TM (Bio-Rad, USA).
a Gel Image of NDM-1 Positive HG4 isolate (M: 50 bp ladder, HG4 sample, P positive control, N negative control. b Gel Image of mcr-1 positive 5 isolates (M 50 bp ladder, P positive control, HG4, HG2, BY8.1, LG8, EY2 mcr-1 positive isolates, N negative control). c The evolutionary history was inferred using the neighbour-joining method. The optimal tree with the sum of branch length = 0.02409398 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. The analysis involved 19 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There was a total of 813 positions in the final dataset. Evolutionary analyses were conducted in MEGA6
2.5 Sanger sequencing and sequence alignment
Sanger sequencing was performed by using an ABI 3730xl DNA Analyzer (Applied Biosystems, India) at the Eurofins Genomics India Pvt. Ltd, Bengaluru. Bio-edit software was used for sequence alignment and cluster analysis; 100% similarity with referral gene was deemed as gene match. Mega software was used for the construction of the phylogenetic tree.
2.6 Biofilm assay
Biofilm assay was performed as previously published protocol [17]. Biofilm assay was performed in 3 isolates: HG4 (environmental isolate), positive control of colistin-resistant isolate (Gifted by Prof. Kashi Nath Prasad, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow) and sensitive strain E. coli ATCC25922.
2.7 Survivability assay on a dry cotton swab
Survivability assay was done as our previously published protocol [18].
2.8 Fitness cost
The persistence of an ARG mutation in bacterial populations is determined in part by its fitness; mutations with little or no fitness cost are considered to be more likely to persist if there is no antibiotic selection pressure. The fitness cost of hVISA isolates was studied by comparing the length of the lag phase during the growth curve [18]. The lag phase duration was taken to be the beginning of the maximum growth rate.
3 Results
Water samples were collected from the low pharma waste region (Lower Himalayas) to the high pharma waste region (plains). Total, 73 bacterial strains were identified on the M-lauryl sulphate agar medium. Out of 73 colistin-resistant isolates, 34 were E. coli and 39 were K. pneumoniae. Five isolates: 2/34 (5.8%) E. coli and 3/39 (7.6%) K. pneumoniae were mcr-1 positive, but most interestingly, one E. coli had both blaNDM-1 and mcr-1 gene. The E test result showed 11 (32.35%) E. coli and 13 (33.33%) K. pneumoniae had MIC more than 2µg/ml (Table 1). Similarly, BMD (broth microdilution assay) result showed 8 (23.5%) of E. coli and 12 (30.7%) K. pneumoniae have MIC more than 2µg/ml (Table 1). No colistin-resistant E. coli or K. pneumoniae were found in Kullu and Manali valley and underground water (Near Baddi) during the screen (Table 2). ESBL detection kit showed a total of 14.7% (5/34) E. coli and 38.46% (15/39) K. pneumoniae were ESBL positive phenotypically (Table 3). The molecular analysis for the mcr-1 gene by PCR showed 5 isolates: 2 (5.8%) E. coli and 3 (7.6%) K. pneumoniae were mcr-1 positive (Table 4). In this study, we noticed the presence of both mcr-1 and NDM-1 in an isolate of E. coli (HG4); this was further confirmed by DNA sequencing and the sequence of NDM-1 was submitted to the gene bank (accession number MT367572). This is the first report from Himachal Pradesh where an environmental sample has both mcr-1 and NDM-1 positive gene in an isolate. The antibiotic sensitivity pattern of the HG4 isolate showed that this is resistant to all selected antibiotics except meropenem (Table 4). Out of 34 isolates, 2 (5.8%), 2 (5.8%), 5 (14.7%), 5 (14.7%) and 4 (11.76%) E. coli were blaTEM, blaSHV, blaCTXM-1, blaCTXM-2 and blaCTXM-15 positive, respectively. Among 39 K. pneumoniae, 15 (38.4%), 6 (15.3%), 10 (25.6%), 9 (23.07%) and 10 (25.6%) were blaTEM, blaSHV, blaCTXM-1, blaCTXM-2 and blaCTXM-15 positive, respectively (Table 4).
Phylogenetic analysis of NDM sequences showed maximum similarity with NDM type 1 isolates by using MEGA6 software (Fig. 2c). Fitness cost result showed in the presence with half MIC of ColR HG4 isolate have a higher lag phase (3 hours) compared to the control strain (1 hour) (Fig. 4a). Survivability study showed environmental isolate ColR HG4 has lesser survivability (14 days) on dry cotton swab compared to positive (28 days) and negative control (35 days) (Fig. 4b). Results of biofilm assay also showed HG4 has laser biofilm production compared to colistin-sensitive E. coli ATCC 25922 strain (Fig. 4c).
a Growth kinetics of HG4, positive control and E. coli ATCC 25,922 sensitive strain to check the lag phase duration. b Box and whisker graph shows survivability of Hg4, positive control and E. coli ATCC 21,922 sensitive strain (survivability is given in number of days). c Biofilm production by HG4, positive control and E. coli ATCC 25,922 sensitive strain
4 Discussion
The presence of (ARBs) in natural water sources is a matter of serious concern for a healthy community as they may transfer ARGs to human commensals [19, 20]. The presence of ARBs and ARGs in water sources, water treatment plants and drinking water distribution systems and their effects on community and environment have been previously reported [21, 22]. The discharge of pharmaceuticals waste (mainly antibiotics) in natural water sources creates a selection pressure for the spread of antibiotic resistance [23]. The overuse of antibiotics among livestock animals also favours the distribution of antibiotic resistance in the aquatic environment [24, 25]. In this study, we have reported the first case of colistin-resistant E. coli with mcr-1 and blaNDM-1 gene from the water samples of Himachal Pradesh, India. Himachal Pradesh is situated in the Western Himalaya part of India, and tourism and agriculture are two important constituents of this state’s economy. Baddi is the biggest and only industrial town in this state (Fig. 3).
The mcr-1 and NDM-1 genes are mostly located on plasmids of bacteria and can be easily transferred to sensitive strains by horizontal transfer of genes. The presence of such superbugs (ColR HG4) in environment is of serious concern for the general community, agriculture and human health. A 2013 study described the presence of antimicrobial resistance microbes as “serious threats to the environment” to low-labour cost Asian countries like India, Bangladesh, China and Pakistan. Most of these manufacturing sites did not follow standard environmental regulation and guidelines, and effluent is directly discharged into the domestic sewage network without any prior treatment, thereby exposing microbes to drugs and drug-resistant microbes to humans and animals subsequently [26]. In this study, we have reported that most of the colistin-resistant isolates and NDM-1 and mcr-1 positive E. coli isolates were found to be from Baddi area, compared to other non-industrial sites (Kullu and Mandi) (Fig. 5). Probably, significant quantitative levels of antibiotics released from manufacturing units, that combine with runoff from farms, human wastewater, give an ideal rearing ground for drug-resistant bacteria. Bacteria in these environments are ready to share or exchange genetic material within intra- or inter-species.
The genotypic characterization showed ColRHG4 isolate was blaNDM-1, blaTEM, blaSHV and mcr-1 positive. The Sanger sequencing and sequence alignment further confirmed that this isolate was blaNDM-1. The presence of mcr-1 positive isolate in animal and human-borne Enterobacteriaceae with co-occurrence of other resistance genes such as ESBL and MBL has been reported worldwide [10, 27, 28]. Like our study, a report from Switzerland showed co-occurrence of mcr-1 gene with ESBLs in Enterobacteriaceae from rivers and lakes [29]. In 2017, another report from China had identified 23 mcr-1 positive isolates from the environmental waters in Hangzhou, China, which is another industrial city [30]. Similarly, in 2018, blaKPC, blaNDM and vanA gene along with 18 mcr-1 positive strains and six mcr-3 positive strains were found from Fauna River in China. They concluded it might be due to the urban activities and presence of antibiotic-resistant bacteria in water [31]. In another study, the first case of the colistin-resistant mcr-1 gene in two E. coli strain from seawater of Algerian coasts was reported by Drali R. et al. 2018 [32]. Noteworthy, ARGs such as blaCTXM, blaNDM-1 and the mcr-1 genes that provide resistance to broad-spectrum antibiotics, including last-resort agents, were reported in all major rivers of India [33, 34]. But it was not reported from this geographical region of India.
Biofilm formation is one of the most important virulence factors exhibited by E. coli among other virulence factors. In this study, we found ColRHG4 as less biofilm former than ColS E. coli ATCC 25922. This suggests that the ColS-type E. coli tend to form more biofilm than resistant ones. In 2016, Qi et al. studied the relationship between biofilm production and resistance among Acinetobacter baumannii and they also found that MDR and XDR isolates tend to form thin biofilms than non-MDR strains [35]. However, in contrast to our results, other previous studies had shown a direct relationship between antimicrobial resistance and biofilm production [36].
The HG4 also has less survivability than the clinical isolates and ColS E. coli ATCC 25922, which suggests that the presence of resistance gene in resistant strain may lead to less survival. Similar to our study, Pettibone et al. also observed antibiotic sensitive bacteria can survive longer than resistant bacteria [36]. The growth kinetic analysis showed a higher lag phase of ColRHG4 (3 hours) compared to ColS E. coli ATCC 25922(1 hour). This result indicates fitness cost associated with ColR HG4 isolate but for a conclusion validation study on more samples is required.
5 Conclusions
In conclusion, incidences of blaNDM-1 and mcr-1 isolate in single bacteria in this geographical region are an alarming signal for tourists, the community, health workers and policymakers. Hence, it is utmost important to think about strict regulation and monitoring of pharma waste management in such an industrial area. Further research is needed on a bigger sample size to know the relationship between anthropogenic activities and antibiotic resistance and the underlying genetic basis of colistin resistance and the accompanying biological features including fitness, growth rate, biofilm formation ability and other virulence characteristics.
References
Lubbert C, Baars C, Dayakar A, Lippmann N, Rodloff AC, Kinzig M et al (2017) Environmental pollution with antimicrobial agents from bulk drug manufacturing industries in Hyderabad, South India, is associated with dissemination of extended-spectrum beta-lactamase and carbapenemase-producing pathogens. Infection 45:479–491
Pauwels B, Verstraete W (2006) The treatment of hospital wastewater: an appraisal. J Water Health 4:405–416
Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M (2008) Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol Rev 32:386–408
WHO (2014) Antimicrobial resistance: global report on surveillance. Geneva. World Health Organization. ISBN 978 92 4 156474 8
Gandra S, Joshi J, Trett A, Lamkang AS, Laxminarayan R (2017) Scoping report on antimicrobial resistance in India. Center for Disease Dynamics. Economics & Policy, Washington
Skariyachan S, Mahajanakatti AB, Grandhi NJ, Prasanna A, Sen B, Sharma N et al (2015) Environmental monitoring of bacterial contamination and antibiotic resistance patterns of the fecal coliforms isolated from Cauvery River, a major drinking water source in Karnataka. India Environ Monit Assess 187:279
Azam M, Jan AT, Haq QM (2016) blaCTX-M-152, a Novel Variant of CTX-M-group-25, Identified in a study performed on the prevalence of multidrug resistance among natural inhabitants of River Yamuna. India Front Microbiol 7:176
Kumar S, Tripathi VR, Garg SK (2012) Antibiotic resistance and genetic diversity in water-borne Enterobacteriaceae isolates from recreational and drinking water sources. Int J Environ Science Technol 10(4):789–798
Poonia S, Singh TS, Tsering DC (2014) Antibiotic susceptibility profile of bacteria isolated from natural sources of water from rural areas of East Sikkim. Indian J Commun Med 39:156–160
Liu X, Li R, Zheng Z, Chen K, Xie M, Chan W-C et al (2017) Molecular characterization of Escherichia coli isolates carrying mcr-1, fosA3, and extended-spectrum-beta-lactamase genes from food samples in China. Antimicrob Agents Chemother 61:e00064-17
Kaur A, Gandra S, Gupta P et al (2017) Clinical outcome of dual colistin- and carbapenem-resistant Klebsiella pneumoniae bloodstream infections: a single-center retrospective study of 75 cases in India. Am J Infect Control 45:1289–1291
Manohar P, Shanthini T, Ayyanar R, Bozdogan B, Wilson A, Tamhankar AJ et al (2017) The distribution of carbapenem- and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India. J Med Microbiol 66:874–883
Larsson DG, de Pedro C, Paxeus N (2007) Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J Hazard Mater 148:751–755
Graham DW, Olivares-Rieumont S, Knapp CW, Lima L, Werner D, Bowen E (2010) Antibiotic resistance gene abundances associated with waste discharges to the Almendares River near Havana. Cuba Environ Sci Technol 45:418–424
CLSI (2012) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard, 9th edn, CLSI document M07-A9. In: Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA
Gales AC, Reis AO, Jones RN (2001) Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol 39:183–190
Jacoby GA (2009) AmpC beta-lactamases. Clin Microbiol Rev 22:161–182
Singh A, Ahmed A, Prasad KN, Khanduja S, Singh SK, Srivastawa JK et al (2015) Antibiofilm and membrane-damaging potential of cuprous oxide nanoparticles against Staphylococcus aureus with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 59:6882–6890
Singh A, Singh S, Singh J, Rahman M, Pathak A, Prasad KN (2017) Survivability and fitness cost of heterogeneous vancomycin-intermediate Staphylococcus aureus. Indian J Med Microbiol 35:415–416
Sanganyado E, Gwenzi W (2019) Antibiotic resistance in drinking water systems: occurrence, removal, and human health risks. Sci Total Environ 669:785–797
Huijbers PM, Blaak H, de Jong MC, Graat EAM, Vandenbroucke-Grauls CMJE, Husman AMdR (2015) Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol 49:11993–12004
Abera B, Kibret M, Mulu W (2016) Extended-spectrum beta (beta)-lactamases and antibiogram in enterobacteriaceae from clinical and drinking water sources from Bahir Dar City, Ethiopia. PLoS ONE 11:e0166519
Gao H, Zhang L, Lu Z, He C, Li Q, Na G (2017) Complex migration of antibiotic resistance in natural aquatic environments. Environ Pollut 232:1–9
Devarajan N, Laffite A, Mulaji CK, Otamonga J, Mpiana PT, Mubedi JI et al (2016) Occurrence of antibiotic resistance genes and bacterial markers in a tropical river receiving hospital and urban wastewaters. PLoS ONE 11:e0149211
Aubertheau E, Stalder T, Mondamert L, Ploy M, Dagot C, Labanowski J (2016) Impact of wastewater treatment plant discharge on the contamination of river biofilms by pharmaceuticals and antibiotic resistance. Sci Total Environ 579:1387–1398
Chen B, Hao L, Guo X, Wang N, Ye B (2015) Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province China. Environ Sci Pollut Res Int. 22:13950–13959
Rehman MS, Rashid N, Ashfaq M, Saif A, Ahmad N, Han J-I (2013) Global risk of pharmaceutical contamination from highly populated developing countries. Chemosphere 138:1045–1055
Du H, Chen L, Tang YW, Kreiwirth BN (2016) Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287–288
Hasman H, Hammerum AM, Hansen F, Handriksen RS, Olesen B, AgersØ Y et al (2015) Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:30085
Zurfuh K, Poirel L, Nordmann P, Nuesch-Inderbinen M, Hachler H, Stephan R (2016) Occurrence of the plasmid-borne mcr-1 Colistin ResiSTANCE gene in extended-spectrum-beta-lactamase-producing Enterobacteriaceae in River Water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother 60:2594–2595
Zhou HW, Zhang T, Ma JH, Fang Y, Wang HY, Huang ZX et al (2017) Occurrence of plasmid- and chromosome-carried mcr-1 in waterborne Enterobacteriaceae in China. Antimicrob Agents Chemother 61(8):e00017-17
Tuo H, Yang Y, Tao X, Liu D, Li Y, Xie X et al (2019) The prevalence of colistin resistant strains and antibiotic resistance gene profiles in Funan River. China Front Microbiol 9:3094
Drali R, Berrazeg M, Zidouni LL, Hamitouche F, Abbas AA, Deriet A et al (2018) Emergence of mcr-1 plasmid-mediated colistin-resistant Escherichia coli isolates from seawater. Sci Total Environ 642:90–94
Akiba M, Senba H, Otagiri H, Prabhasankar VP, Taniyasu S, Yamashita N et al (2015) Impact of wastewater from different sources on the prevalence of antimicrobial-resistant Escherichia coli in sewage treatment plants in South India. Ecotoxicol Environ Saf 115:203–208
Marathe NP, Pal C, Gaikwad SS, Jonsson V, Kristiansson E, Larsson DGJ (2017) Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. Water Res 124:388–397
Qi L, Li H, Zhang C, Liang B, Li J, Wang L et al (2016) Relationship between Antibiotic Resistance, Biofilm Formation, and Biofilm-Specific Resistance in Acinetobacter baumannii. Front Microbiol 7:483
Rao RS, Karthika RU, Singh SP, Shshikala P, Kanungo R, Jayachandran S et al (2008) Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol 26:333–337
Pettibone GW, Sullivan SA, Shiaris MP (1987) Comparative survival of antibiotic-resistant and -sensitive fecal indicator bacteria in estuarine water. Appl Environ Microbiol 53:1241–1245
Farajzadeh Sheikh A, Rostami S, Jolodar A, Tabatabaiefar MA, Khorvash F, Saki A, Shoja S, Sheikhi R (2014) Detection of Metallo-Beta Lactamases among Carbapenem-Resistant Pseudomonas aeruginosa. Jundishapur J Microbiol 7(11):e12289. https://doi.org/10.5812/jjm.12289
Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, Guerra B, Malorny B, Borowiak M, Hammerl JA, Battisti A, Franco A, Alba P, Perrin-Guyomard A, Granier SA, De Frutos EC, Malhotra-Kumar S, Villa L, Carattoli A, Hendriksen RS (2018) Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 for surveillance purposes. Euro Surveill 23(6):17. https://doi.org/10.2807/1560-7917.ES.2018.23.6.17-00672
Rasheed JK, Anderson GJ, Yigit H, Queenan AM, Domenech-Sanchez A, Swenson JM, Biddle JW, Ferraro MJ, Jacoby GA, Tenover FC (2000) Characterization of the extended-spectrum beta-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob Agents Chemother 44(9):2382–2388. https://doi.org/10.1128/aac.44.9.2382-2388.2000
Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S (2019) Distribution of blaCTX -M, blaTEM, blaSHV and blaOXA genes in Extended-spectrum-beta-lactamase-producing clinical isolates: a three-year multi-center study from Lahore. Pakistan Antimicrob Resist Infect Control 8:80. https://doi.org/10.1186/s13756-019-0536-0
Tamta S, Kumar ORV, Singh SV, Pruthvishree BS, Karthikeyan R, Rupner R, Sinha DK, Singh BR (2020) Antimicrobial resistance pattern of extended-spectrum beta-lactamase-producing Escherichia coli isolated from fecal samples of piglets and pig farm workers of selected organized farms of India. Vet World 13(2):360–363. https://doi.org/10.14202/vetworld.2020.360-363
Ramadan AA, Abdelaziz NA, Amin MA, Aziz RK (2019) Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci Rep 9(1):4224. https://doi.org/10.1038/s41598-019-39730-0
Acknowledgements
Dr. Avinash Singh acknowledged DST SERB (PDF/2017/000654) for providing the funds for doing this work. We also thank Professor Kashi Nath Prasad, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, for providing the positive controls for NDM-1 and mcr-1 positive isolates. AP and SSR acknowledge financial support from the Ramalingaswami Fellowship and Department of Biotechnology, Government of India, New Delhi. AP is supported through research grants BT/PR26841/MED/122/121/2017 Department of Biotechnology, Government of India, New Delhi, and ECR/2016/000817/LS from SERB, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the funders has any role in deciding/influencing the outcome of this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Avinash Singh and Anand Kumar Keshri have contributed equally to this work
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, A., Keshri, A.K., Rawat, S.S. et al. Identification and characterization of colistin-resistant E. coli and K. pneumoniae isolated from Lower Himalayan Region of India. SN Appl. Sci. 3, 615 (2021). https://doi.org/10.1007/s42452-021-04596-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-021-04596-3