Abstract
Breeding for disease resistance in plants is an integral part of integrated disease management, which helps develop resistance against devastating plant diseases and reduces yield losses. The present study focuses on characterizing the Cercospora leaf spot (CLS) resistant genotypes through simple sequence repeats (SSR) markers linked to CLS resistance. Two hundred sixty-one mung bean genotypes were screened against CLS resistance from 2018 to 2020 under field conditions with a completely randomized block design. Further, six susceptible and six resistant genotypes were tested along with a susceptible check, artificially inoculated with the pathogen to induce disease, and screened through SSR markers to identify the CLS-resistant genotypes. Out of 10 reported CLS-linked SSR markers, seven markers i.e. CEDAAG002, CEDC050, CEDG037, CEDG084, CEDG150, VR108, and VR393, have been validated and confirmed to show polymorphism with different DNA banding patterns in resistant and susceptible genotypes. The extent of polymorphism analyzed, SSR markers such as CEDG084 (71.42%), CEDG150 (50%), VR108 (85.71%), and VR393 (50%) showed higher polymorphism than other markers, which may serve as promising markers for identification of CLS resistance source.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mung bean (Vigna radiata) is a versatile, short-duration legume crop cultivated worldwide, mainly in South East Asian countries. Its production in India is remarkable, contributing around 54% of the total mung bean production globally (Singh and Singh 2011). It is widely cultivated for its rich protein content and many vitamins and minerals. Along with the high nutritional components, the leaves and stems are utilized as green manure and forage. Various adverse factors like biotic and abiotic stresses have drastic effects on the production of mungbean. Cercospora leaf spot (CLS) is one of the biotic stresses in mung bean crops, which seriously threatens crop production and yield (Chand et al., 2015). CLS disease needs high humidity for its incidence and spread, so it occurs in the Kharif or rainy season (Chankaew et al. 2011). The initial phase of the disease is marked by spots on the leaves, which further increase in number and size at the flowering and pod-filling stage. In the presence of favorable environmental conditions like high relative humidity of about 90%, intermittent rainfall, and warm temperatures, the pathogen infects the plants. It forms many spots, which further coalesce into blight-like symptoms. In severe cases in highly susceptible mung bean cultivars, CLS results in premature defoliation with the smaller size of pods and seeds, thus hampering the seed yield.
Cercospora canescens is a genetically highly diverse fungal pathogen isolated from different locations, and cultivars showed significant variation in resistance to the pathogen (Joshi et al. 2006). The plant-pathogen interaction in the mung bean - CLS system is analyzed in the field by the phenotypic screening to the disease based on the disease reactions. The yield loss is often proportional to the disease progress curve and higher in susceptible cultivars than the resistant one concerning CLS (Bhat et al. 2008). Deploying resistant varieties is the most efficient and durable strategy for managing this disease (Pink 2002). So, screening for CLS disease resistance is essential for identifying resistant mung bean genotypes and using them in breeding programs. According to Chankaew et al. (2011), although the phenotypic disease screening and analysis of disease progress is vital for breeders to develop resistant cultivars, it is still associated with many limitations. Even though the mung bean is a short-duration crop and cultivated multiple times a year, CLS is predominant in the kharif season. Hence, the evaluation of the disease severity is possible in only one season of the year. The repeatability of the screening experiments makes the task a lot more time-consuming. Molecular markers are used to identify genetic differences between individuals of different or the same species by tagging the gene of interest (Vidak et al., 2017). Exploitation of CLS resistance markers for the identification of promising mung bean cultivars carrying these resistance genes can be helpful to combat the shortcomings of seasonal disease screening (Papan et al. 2021) and aid marker-assisted selection (MAS) for the development of novel resistant varieties round the year (Collard and Mackill 2008). Therefore, several CLS resistance-linked DNA markers have been reported to identify resistant sources and develop improved resistant mungbean varieties in response to CLS (Chankaew et al., 2011). Papanet al. (2021) reported a QTL associated with CLS resistance using SSR markers system in CN72 × V4718 F2 population between VR393 and CEDG084 markers. Inter-simple sequence repeat (ISSR) marker I16274 linked to the CLS resistance gene was identified (Tantasawat et al. 2020). Previous reports have identified various sources of resistance and proved that resistance to CLS in mungbean is controlled by a single dominant gene, which can benefit breeding programs for developing resistant cultivars (Chankaew et al. 2011). The present study highlights the efforts to screen and characterize the CLS resistance genotypes through CLS-linked SSR markers.
Materials and methods
Plant materials and evaluation of disease resistance under field conditions
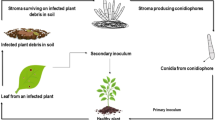
Screening for CLS disease resistance was carried out using 261mung bean genotypes under field conditions during the Kharif crop seasons 2018-19 and 2019 -20 at the agricultural farm of Odisha University of Agricultural Technology, Bhubaneswar (20.26° N, 85.81° E) in a randomized complete block design (Mahapatra et al. 2022). Since the incidence of the disease is negligible in rabi season due to lack of adequate moisture, the experiment was carried out in kharif only. The plants were subjected to artificial inoculation of the pathogen at 50% flowering stage and characterized based on disease incidence (Fig. 1). The relative humidity was maintained in the experimental field at 85–90% by flooding through the channels in the area along with a regular spray of water with the atomizer in the absence of natural rainfall, and the temperature ranged from 25 to 28 °C throughout the test period. The fertilizers were applied per the recommended agronomic dose of 20:60:40 kg N–P–K ha − 1 as basal dose during the field preparation. The observations were carried out on initiatiation of the disease and appearance of specific symptoms to CLS. The condition was scored on a scale of 0–9 (Umer et al. 2009), where 9 implies the most severe infection with more than 90% leaf area damage and 0 being the least with no visible symptoms. The observation was made at seven days intervals after the disease initiation.
Based on the CLS disease reaction, the mung bean genotypes were distributed into resistant, moderately resistant, moderately susceptible, and susceptible on the AUDPC value. AUDPC = Area under disease progress curve, R = Resistance, MR = Moderately resistant, MS = Moderately susceptible, S = Susceptible
Further, the disease severity percentage was calculated in the Area Under Disease Progress Curve (AUDPC) using the following formula given by Shaner and Finney (1977). The genotypes were categorized based on the extent of the disease reaction. The genotypes with higher AUDPC values were classified as susceptible, whereas those with lower AUDPC values were resistant.
Where, Yi = disease level/severity at time ti.
t (i + 1) – ti= Time (days) between two disease scores.
n = number of dates at which CLS was recorded.
Out of the screened population of mungbean genotypes, six susceptible and six resistant genotypes were selected based on the AUDPC value for the present study.
Polyhouse experiment
Based on AUDPC value, resistant mung bean genotypes (CPR BAM GP 316, Kamdev, CPR BAM GP 325, CPR BAM GP 345, OBGG-102, ANGUL Mung GR) and susceptible genotypes (CPR BAM GP 267, CPR BAM GP 301, KVK Jagatsinghpur-1, KVK Dhenkanal-1, KVK Sambalpur-3 and CPR BAM GP 265) were selected. These selected genotypes were grown in the polyhouse along with a susceptible check i.e., Kopergaon (Maurya et al. 2018), and artificially inoculated to induce disease with the pathogen mass cultures of C. canescens isolate BBS1 (Accession no- MZ475049) isolated from diseased plant samples collected from Bhubaneswar (20.26° N, 85.81°), Odisha. Optimum irrigation is provided to maintain the humidity for the pathogen infection and disease occurrence. Observation were recorded on the yield components and extent of infection in the selected genotypes.
Molecular analysis
The genomic DNA of the tested genotypes was isolated using CTAB method (Murray and Thompson 1980). About 1gm of the freshly harvested leaf tissues were ground in liquid nitrogen, and pre-warmed DNA extraction buffer was added to the powdered plant material and incubated at 65 °C for 1 h. Chloroform: isoamyl alcohol (24:1 v/v) solution of equal volume was added to the plant mixture and centrifuged at 12,000 rpm for 10 min. The supernatant was collected, and chilled ethanol and sodium acetate were added for DNA precipitation. The DNA pellet was washed with chilled 70% (v/v) ethanol and dried at room temperature. The well-dried DNA pellet was re-suspended in Tris-EDTA buffer and stored at -20 °C.
Polymerase chain reaction (PCR) analysis
Ten pairs of SSR primers reported to be linked to CLS resistance (Chankaew et al. 2011) were selected and used to screen the mung bean genotypes for disease resistance (Table 1). This molecular screening used the selected resistant and susceptible genotypes and susceptible CLS checks with the ten sets of primer. The reaction volume of 12.5 µl consisted of 1U Taq DNA polymerase, 1×Taq buffer,2 mM MgCl2, 0.2 mM dNTPs, 2 ng template DNA, and forward and reverse primers (5 pmol each). The amplification was carried out in Thermocycler (BIORAD, USA) with PCR parameters programmed as follows: initial denaturation at 94 °C for 3 min, primer annealing at 48–60 °C for 50 s depending on primer, chain extension, and final extension at 72 °C for 50 s and 7 min, respectively.
Electrophoresis and detection of amplified products by polyacrylamide gel electrophoresis (PAGE)
The PCR amplification products were checked and characterized by gel electrophoresis on 10% denaturing polyacrylamide gel in 1 × TBE buffer at 90 V for 150 min. The amplicons were visualized under UV with ethidium bromide as the staining agent. A low molecular weight (50 bp) ladder was used as a reference standard marker.
Results
The mungbean genotypes were categorised into four groups i.e. resistant, moderately resistant, moderately susceptible and susceptible to CLS based on the AUDPC value. Out of 261 genotypes, 89 genotypes were moderately resistant to CLS and the AUDPC value varied from 251 to 500 (Fig. 1). Genotypes exhibited very narrow range of variation between moderately susceptible to susceptible screened under natural conditions. Both selected resistant and susceptible genotypes along with check variety subjected to artificial inoculation of the CLS at 50% flowering period and were characterised on the basis of disease incidence (Fig. 1). Both disease severity and yield characteristics of the infected mungbean plants grown in the polyhouse condition were recorded (Table 2). The number of sporulating lesions varied from 0.67 to 10.21 in resistant and susceptible genotypes. Average number of pods per plant and number of seeds per plant ranged from 15.00 to 27.00 and 8.75 to 10.68 in both resistant and susceptible genotypes, respectively. The highest (32.00) number of pods/plant was observed in genotype OBGG-102, which was resistant to CLS and lowest (15.00) in the genotype KVK Jagasinghpur-1.
There was a significant yield variation between susceptible and resistant genotypes. Molecular analysis of both genotypes using 10 SSR primers linkage with CLS resistance showed polymorphism with known CLS reactions. Of the 10 SSR markers, CEDC031, CEDG117, and CEDG305 did not show consistent results among the genotypes. CEDAAG002, CEDC050, CEDG037, CEDG084, CEDG150, VR108, and VR393 showed polymorphism among the susceptible and resistant genotypes (Fig. 2a, 2b, 2c, 2d, 2e, 2f and 2g). The targeted fragment range of these seven amplifiable markers was between 100 and 300 bp. CEDAAG002 primer produced multiple amplified bands between 100 and 400 bp. In contrast, the six susceptible genotypes along with the susceptible check showed bands of 140 bp, which was not present in any of the resistant genotypes (Fig. 2a). Amplicons at 192 bp and 80 bp found to be linked to CLS resistance were amplified by CEDC050 marker in the resistant genotypes and absent in the susceptible genotypes (Fig. 2b; Table 3). CEDG037 amplified a 142 bp amplicon that linked to CLS resistance in the six resistant genotypes (Fig. 2c). CEDG084 primer showed polymorphism in the selected resistant and susceptible genotypes with the differential banding pattern for both the groups (Fig. 2d). CEDG150 amplified a 190 bp amplicon in the resistant genotypes and not found in the susceptible genotypes which implied to link with disease resistance (Fig. 2e). The VR108 marker showed polymorphism for multiple bands in resistant genotypes (Fig. 2f). The amplicons at 200 bp, 240 bp, 270 bp, 400 and 500 bp were absent in the susceptible genotypes along with the susceptible check. A 150 bp amplicon amplified by VR393 marker in all the six resistant genotypes. In contrast, it was located at 160 bp in six susceptible genotypes (Fig. 2g). Based on the distinct arrangement of DNA banding pattern and polymorphism percent, CEDG150, CEDG084, VR108, and VR393 exhibited 50%, 71.42%, 85.71%, and 50% polymorphism, respectively can be considered as promising markers for identification of CLS resistance source (Table 3).
Discussion
In the present study, phenotyping for CLS resistance in both field and polyhouse conditions to categorized the mungbean genotypes based on disease reaction with respect to CLS and identified the resistant genotypes. The yield potential of resistant and susceptible genotypes under field conditions was used as selection criteria for the breeding program (Kumar and Reena, 2007). But, molecular analysis provides further new insights into identifying genotypes for breeding. Very limited research on molecular identification is available in selected mungbean genotypes linked to CLS disease. The results revealed that the distinct banding pattern of the PCR amplicons of the selected resistant and susceptible genotypes helped to confirm the morphological data, which paves the way to choose promising genotypes for further use in breeding programs. The CEDAAG002, CEDC050, CEDG037, CEDG084, CEDG150, VR108, and VR393 primers have shown differentiation in banding pattern among the genotypes and compared to the disease reaction. Chankaew et al. (2011) reported similar results by using the CLS-linked SSR markers in KPS1 × V4718 cross. But they also found distinct polymorphism among the F2 and BC1F1 mungbean population using CEDC031, CEDG117, and CEDG305 primers, which was not similar to our result as these primers did not show polymorphism and consistency among all the genotypes under consideration. This observation is attributed to the fact that these three primers linked to CLS resistance were specific to the population generated from KPS1 × V4718 cross. But, the other seven selected primers have successfully shown polymorphism and helped to distinguish the resistant and susceptible genotypes in the population considered in the previous studies and the diverse genotypes used in this study. These markers can establish the mapping of CLS resistance in various mungbean genotypes and the genetic distances. Tantasawat et al. (2010) have estimated the genetic distance between VR393 and one ISSR marker I16274 and used it for CLS resistance mapping using 143 recombinant inbred lines (RIL) from CN72 × V4718 cross and have located a major QTL qCLSC72V18-1 between both the markers. The level of polymorphism exhibited by the seven SSR markers was compared based on band differentiation to understand the markers’ credibility in identifying the resistance sources. These markers showing polymorphism in case of resistance and susceptible genotypes used in the present study are validated to identify the relevant QTL for the selected mungbean genotypes. Inheritance of CLS resistance is identified as qualitative and governed by a single dominant gene; the resistance gene can be transferred into susceptible cultivars to develop resistant varieties (Tantasawat et al. 2020). The susceptible genotypes lacking the resistance amplicons can be identified, and thereby, it can transfer the CLS resistance genes from the resistant lines into them. Identifying promising CLS resistance-linked molecular markers shall help us to select the genotypes with genes of interest for future breeding programs and deploy varieties with sustained resistance against the disease. Since Cercospora canescens is a slow-growing fungus in artificial medium, its incidence in natural conditions occurs in the presence of favourable environmental factors, which hinders timely screening for disease resistance.
Conclusion
The present study concluded that the 261 mung bean genotypes screened against CLS were categorized into four groups i.e. resistant, moderately resistant, moderately susceptible, and susceptible. There was significant yield variation between susceptible and resistant genotypes. Molecular analysis indicated that the 192 bp and 80 bp amplicons were linked to CLS resistance using CEDC050 marker. The VR108 marker showed polymorphism for multiple bands in resistant genotypes. The amplicons at 200 bp, 240 bp, 270 bp, 400 bp, and 500 bp were absent in the susceptible genotypes along with the susceptible check. Based on the distinct arrangement of DNA banding pattern and polymorphism percent, CEDG150, CEDG084, VR108, and VR393 with 50%, 71.42%, 85.71%, and 50% polymorphism, respectively, can be considered as promising markers for the identification of CLS resistant genotypes in a population.
References
Bhat NA, Maheshwari SK, Ahmad S, Beig MA, Masoodi SD (2008) Field evaluation of mungbean accessions against Cercospora leaf spot. Annl Biol 24:1–9. http://www.scimagojr.com
Chand R, Pal C, Singh V, Kumar M, Singh VK Chowdappa P (2015) draft genome sequence of Cercospora canescens: a leaf spot causing pathogen. Curr Sci 109:2103–2110. https://www.jstor.org/stable/24906710
Chankaew S, Somta P, Sorajjapinun W, Srinives P (2011) Quantitative trait loci mapping of Cercospora leaf spot resistance in mungbean, Vigna radiata (L.) Wilczek. Mol Breed 28:255–264. https://springerlink.bibliotecabuap.elogim.com/article/https://doi.org/10.1007/s11032-010-9478-1
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philosophical Trans Royal Soc B: Biol Sci 363:557–572. https://doi.org/10.1098/rstb.2007.2170
Joshi A, Souframanien J, Chand R, Pawar SE (2006) Genetic diversity study of Cercospora canescens (Ellis & Martin) isolates, the pathogen of Cercospora leaf spot in legumes. Curr Sci 90:564–568 Corpus ID:89151133
Kumar A, Reena S (2007) Assessment of grain yield potential of mungbean in Shivalaik foothills of India. Jour of Agron 6:476–479. https://doi.org/10.3923/ja.2007.476.479
Mahapatra SS, Swain D, Beura SK, Rout GR (2022) Identification of mungbean germplasm for resistance against Cercospora canescens (CLS) and to study the association of biochemical parameters with defence mechanisms. Agron Jour 114:1184–1199. https://doi.org/10.1002/agj2.20987
Maurya AK, Navathe S, Mohapatra C, Chand R (2018) Antioxidants elevates the resistance to Cercosporacanescens in interspecific cross of Vigna radiata (Kopergaon)× Vigna mungo (Pant Urd 31). Indian Phytopathol 71:519–528. https://doi.org/10.1007/s42360-018-0101-4
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res 8:4321–4326
Papan P, Chueakhunthod W, Poolsawat O, Arsakit K, Tharapreuksapong A, Tantasawat, PA (2021) Validation of molecular markers linked to cercospora leaf spot Disease resistance in mungbean (Vigna radiata [L.] Wilczek). SABRAO Jour Breed & Genetic 53:749–758. https://doi.org/10.54910/sabrao2021.53.4.16
Pink DA (2002) Strategies using genes for non-durable Disease resistance. Euphytica 124:227–236. https://doi.org/10.1023/A:1015638718242
Shaner G, Finney RE (1977) The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathol 67:1051–1056
Singh DP, Singh BB (2011) Breeding for tolerance to abiotic stresses in mungbean. Jour Food Legumes 24:83–90 Print ISSN:0970–6380, online ISSN
Tantasawat PA, Poolsawat O, Arsakit K, Papan P (2020) Identification of ISSR, ISSR-RGA and SSR markers associated with Cercospora leaf spot resistance gene in mungbean. Int Jour Agricul & Biol 23:447–453
Tantasawat PA, Trongchuen J, Prajongjai T, Thongpae T, Petkhum C, Seehalak W, Machikowa T (2010) Variety identification and genetic relationships of mungbean and blackgram in Thailand based on morphological characters and ISSR analysis. African Jour Biotech 9(27):4452–4454. http://www.academicjournals.org/AJB
Umer I, Iqbal SM, Zahid MA, Khan SH (2009) Screening of local mungbean genotype against Cercospora leaf spot Disease. Pakistan Jour Phytopathol 21:123–125
Vidak M, Carović-Stanko K, Barešić A, Grdiša M,Šatović, Liber Z (2017) Microsatellite markers in common bean (Phaseolus vulgaris L). Jour Cent Europ Agricul 18:902–917. https://doi.org/10.5513/JCEA01/18.4.1983
Acknowledgements
The authors wish to acknowledge to the Department of Biotechnology, Govt. of India for providing the financial support under the R & D project (BT/IN/UK/PORI/04/GRR/2018-19) to carry out the experiment. SSM is thankful to the Department of Science and Technology, Government of India for financial support in the form of INSPIRE fellowship (IF180456).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mahapatra, S.S., Senapati, A.K., Swain, D. et al. Identification and characterisation of Cercospora leaf spot(CLS) resistance in mungbean (Vigna radiata L.) genotypes through SSR markers. Indian Phytopathology 76, 969–977 (2023). https://doi.org/10.1007/s42360-023-00682-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-023-00682-1