Abstract
Chocolate spot (CS), caused by Botrytis fabae Sard., is an important threat to global faba bean production. Growing resistant faba bean cultivars is, therefore, paramount to preventing yield loss. To date, there have been no reported quantitative trait loci (QTL) associated with CS resistance in faba bean. The objective of this study was to identify genomic regions associated with CS resistance using a recombinant inbred line (RIL) population derived from resistant accession ILB 938. A total of 165 RILs from the cross Mélodie/2 × ILB 938/2 were genotyped and evaluated for CS reactions under replicated controlled climate conditions. The RIL population showed significant variation in response to CS resistance. QTL analysis identified five loci contributing to CS resistance on faba bean chromosomes 1 and 6, accounting for 28.4% and 12.5%, respectively, of the total phenotypic variance. The results of this study not only provide insight into disease-resistance QTL, but also can be used as potential targets for marker-assisted breeding in faba bean genetic improvement for CS resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Faba bean (Vicia faba L., 2n = 2x = 12) is a widely grown diploid cool-season legume (pulse) crop with a genome size of approximately 13 Gb (Soltis et al. 2003). It is the fifth most important pulse crop, after common bean (Phaseolus vulgaris L.), chickpea (Cicer arietinum L.), pea (Pisum sativum L.), and lentil (Lens culinaris Medik.), with an annual production of 5.4 Mt (FAOSTAT 2021). It provides an affordable source of dietary protein, fiber, carbohydrates, and other valuable nutrients (Duc et al. 2015; Khazaei and Vandenberg 2020). The faba bean seed contains about 30% protein (Warsame et al. 2018), making it one of the best alternative sources for the plant-based protein industry for food, feed, and extractable protein. The crop plays an important role in a sustainable agriculture system, due to its symbiotic fixation of atmospheric nitrogen and its use as a break crop in cereal-dominated rotations (Angus et al. 2015). Global faba bean production is generally challenged by foliar diseases such as ascochyta blight (Ascochyta fabae Speg.), chocolate spot (Botrytis fabae Sard.), rust (Uromyces viciae fabae Pers.), downy mildew (Peronospora viciae (Berk.) Caspary), and gall disease (Physioderma viciae) (Adhikari et al. 2021; Rubiales and Khazaei 2022).
Chocolate spot (CS), caused by the necrotrophic fungal pathogen Botrytis fabae (Sard.), is one of the most destructive diseases of faba bean plants worldwide and can cause up to 90% yield reduction on susceptible cultivars when conducive environmental conditions prevail (Gorfu and Yaynu 2001; Bouhassan et al. 2004; Tivoli et al. 2006; Beyene et al. 2018). CS can manifest itself on the above-ground parts of the plant at any growth stage, mainly on leaves. It initially appears on leaves as reddish-brown spots that develop into chocolate-colored lesions. These can enlarge into extensive necrotic zones that lead to severe premature defoliation and stem damage. If the disease develops at flowering time, it can cause complete crop failure. The pathogen survives on crop residues or lodged stubble of the previous year’s growing season and on contaminated seeds (Harrison 1988). Its spores are spread primarily between fields by wind and rain. Variation in virulence among B. fabae isolates has been demonstrated (Hutson and Mansfield 1980), although no race classification has been described so far. The strategies used to control chocolate spots on faba bean include crop rotation, reduced planting density, timely fungicide applications, use of clean seeds, and host-plant resistance (Stoddard et al. 2010). Other species of Botrytis contribute to chocolate spot disease, but B. fabae is the main one (Fan et al. 2015).
Genetic resistance is a key part of any integrated disease management approach to prevent yield loss caused by CS. Fungal evolution and migration lead to the need for continuous incorporation of new sources of resistance genes or alleles into elite breeding materials. A few sources of partial genetic resistance to CS have been identified in faba bean germplasm (Hanounik and Robertson 1988; Bouhassan et al. 2004; Villegas-Fernández et al. 2009; Maalouf et al. 2016). Partial resistance to CS was transferred into elite breeding lines and resulted in the release of several cultivars with moderate levels of resistance (Temesgen et al. 2015; Maalouf et al. 2019). Incorporation and pyramiding of CS resistance genes from multiple sources into elite cultivars could be facilitated with marker-assisted selection (MAS). However, the use of MAS in faba bean CS resistance breeding is limited by a lack of knowledge of the genetic basis of the resistance so-far identified, by the presence of pathogen variability in the major faba bean diseases (Hanounik and Robertson 1988; Gorfu 1996), and by the lack of adequate genomic resources.
In recent years, progress has been made in developing single nucleotide polymorphism (SNP)–based genetic maps of faba bean (e.g., Ellwood et al. 2008; Cruz-Izquierdo et al. 2012; Kaur et al. 2014; Webb et al. 2016; Carrillo-Perdomo et al. 2020). Among these maps, the densest SNP-based consensus maps are reported by Webb et al. (2016), constructed from six mapping populations using 687 SNP markers, and Carrillo-Perdomo et al. (2020), a consensus map of three mapping populations consisting of 1728 SNP markers distributed in six linkage groups. Concurrently, several quantitative trait loci (QTL) studies for various faba bean improvement traits have been carried out using different genetic maps (see reviews by O’Sullivan and Angra 2016; Maalouf et al. 2019; Khazaei et al. 2021a) for major foliar diseases such as ascochyta blight (e.g., Avila et al. 2004; Kaur et al. 2014; Atienza et al. 2016; Sudheesh et al. 2019) and rust (e.g., Adhikari et al. 2016; Ijaz et al. 2021). Despite the importance and widespread global nature of CS, no QTL studies have been published to our knowledge. In addition, breeding for CS resistance has been difficult because screening for the disease under field conditions is unreliable, especially in dry environments where faba bean is grown mostly as a rain-fed crop (Adhikari et al. 2021). Humid conditions, which are uncommon in many faba bean-growing areas, provide the opportunity to screen in the field. Identification of reliable molecular markers for use in MAS systems for improving CS resistance is essential for future faba bean breeding. The objective of this study was to identify QTL associated with CS resistance using an advanced RIL mapping population developed from accession ILB 938 which has partial resistance to CS (Khazaei et al. 2018) under climate-controlled conditions.
Material and methods
Plant material
Genetic resistance to B. fabae was evaluated using 165 recombinant inbred lines (RILs) from the Mélodie/2 × ILB 938/2 cross (Khazaei et al. 2014a). The RILs were advanced using single seed descent to the F8 generation. Then the F8-derived bulked seeds of the RILs were selfed for at least two additional generations before disease phenotyping. Mélodie/2 was bred at INRA (France) as a source of low vicine-convicine (Khazaei et al. 2014a). ILB 938 (IG 12,132) originates from the Andean region of Colombia and Ecuador and has resistance to several biotic and abiotic stresses, including CS (Maalouf et al. 2016; Khazaei et al. 2018). It has high water use efficiency (Link et al. 1999; Khazaei et al. 2014a) and carries a gene that decouples pigmentation in flowers from that in stipules (Khazaei et al. 2014b). It was the first identified source of resistance for CS in faba bean (Hanounik 1982; Khalil et al. 1984; Robertson 1984). ILB 938 resulted from mass selection within ILB 438 (IG 11,632). ICARDA’s registered BPL (pure line) derivatives of ILB 438 and ILB 938 are BPL 710 and BPL 1179, respectively.
Inoculation and phenotyping for chocolate spot reactions
A culture stock of B. fabae isolate FB-7 was obtained from the pulse pathology laboratory, University of Saskatchewan for mass spore production. Conidia were cultured on faba bean extract media plates (18 g of agar, 20 g of dextrose, 20 g of NaCl, and 1 L of faba bean extract which is prepared by boiling 400 g of faba bean seeds in 1 L of distilled water for 45 min and diluting with 1 L of distilled water after removing the faba bean seeds) and incubated for 14 d at room temperature in a cycle of 12 h of darkness and 12 h of light to induce sporulation. Plates were then flooded with sterile-deionized water and conidia were harvested by scraping the colonies with the edge of a sterile glass microscope slide. The suspension was collected and filtered through one layer of cheesecloth into a clean Erlenmeyer flask. The concentration of the conidial suspension was adjusted to 5 × 105 spores mL−1 using a hemocytometer. The surfactant Tween 20 (polyoxyethylene sorbitan monolaurate) was added at the rate of 1 to 2 drops per 1000 mL of suspension, and the suspension was shaken well before inoculation.
The parents and 165 RILs were grown in a climate-controlled growth chamber (GR48, Conviron, Winnipeg, Canada) at the University of Saskatchewan’s College of Agriculture and Bioresources phytotron facility, Saskatoon, Canada. The growth chamber conditions were adjusted to 18 h light and 6 h dark, with the temperatures maintained at 21 °C (day) and 18 °C (night), and the photosynthetic photon flux density was set to 300 μmol m−2 s−1 during the light period at the crop canopy level. Three seeds of each genotype were sown in individual 3.8 L pots (15.5 cm in diameter) containing a soil-less mixture (Sunshine Mix No. 3, Sun Grow Horticulture® Ltd., Vancouver, BC, Canada) and fertilized once per week using 3 g L−1 of soluble N:P:K (20:20:20) PlantProd® fertilizer (Nu-Gro Inc., Brantford, ON, Canada). Three weeks after seeding, the plants were inoculated with a spore suspension (5 × 105 spores mL−1) at ~ 3 mL per plant until runoff using a pressurized knapsack sprayer. Plants were placed in an incubation chamber for 7 days. Two humidifiers (Vicks Fabrique Paz Canada, Inc., Milton, ON, Canada) were placed in the incubation chamber to ensure 90–100% relative humidity for infection and disease development. The disease severity data were collected per individual plant at 7 days post-inoculation (dpi) using a 0 to 10 rating scale with 10% increments. The experiment was repeated four times under the same growing conditions during 2020–2021, and each time disease scores were recorded from three replicates (one plant per replicate). Data were converted to percentage disease severity using the class midpoints for data analysis and analyzed as a randomized complete block design.
Statistical analysis
Disease score data were analyzed using SAS v.9.4 (SAS Institute 2015, Cary, USA). Normality and variance homogeneity of the residuals were analyzed using the Shapiro–Wilk normality test and Levene’s test for homogeneity, respectively. Genotypes were treated as fixed effects and blocks as random effects; the significance of variances was declared at 5% significance level. Least square means were estimated for genotype using LSMEANS statements and used for QTL analysis. Multivariate relationships between RILs and experimental repeats were investigated by principal component analysis (PCA) and biplots of autoscaled (i.e., each repeat mean and grand mean centered at the origin) data in R (R Core Team 2020) with the ‘factoextra’ package.
Genotyping
Genomic DNA was extracted from 3-day-old, germinated embryo axes of the parents and 188 RILs from the mapping population using the CTAB method, as described previously (Björnsdotter et al. 2021). DNA quality was assessed by agarose gel electrophoresis; concentrations were determined with a Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, UK) following the manufacturer’s guidelines. DNA samples were brought to a concentration of 35 ng/µL in 40 µL aliquots and genotyped using the Axiom ‘Vfaba_v2′ 60 K array that was developed from metatranscriptome data (Personal communication D. M. O’Sullivan; O’Sullivan et al. 2019).
Linkage map construction
Genotypic data for the RIL population was filtered against markers showing significant segregation distortion (deviating from the expected 1:1 ratio) using a chi-square (χ2) test. Markers missing in 1% or more of the data were removed from the analysis. Draft maps were generated from the remaining 180 RILs using ASMap software (Taylor and Butler 2017) with a p-value of 1E−10 and a maximum distance between SNP markers of 15.0 cM for grouping them into linkage groups. This linkage map was refined using MapDisto v. 1.7.7.0.1 (Lorieux 2012) with a logarithm of odds (LOD) score of 3.0 and a cutoff recombination value of 0.35. The best marker order was estimated using both “AutoCheckInversions” and “AutoRipple” commands in MapDisto. Distances between markers were calculated using the Kosambi function (Kosambi 1943). Linkage groups (LGs) were scanned and corrected for double recombinants using MapDisto v. 1.7.7.0.1 (Lorieux 2012). The final LGs were assigned to chromosomes according to the NV644 × NV153 genetic map (unpublished) developed at the University of Reading, UK, and the Webb et al. (2016) faba bena consensus genetic map.
QTL mapping of disease resistance
Windows QTL Cartographer 2.5 software (Wang et al. 2012) and multiple QTL mapping (MQM, Manichaikul et al. 2009), run in R/qtl software (Broman et al. 2003), were used to detect QTL. For Windows QTL Cartographer, composite interval mapping (CIM) was implemented using the Kosambi map function with Ri1 cross-type (recombinant inbred line, derived by selfing) at a 1.0 cM interval walk speed. Cofactor selection was performed using forward and backward regression in the standard CIM model with a probability of in and out of 0.1 and a window size of 5 cM. QTL significance thresholds were determined by permutation tests (1000 permutations) at a significance level of P = 0.05. Multiple QTL mapping was completed with the “stepwiseqtl” function (Broman et al. 2003), using the Haley-Knott regression (Haley and Knott 1992) methods. The optimal QTL model was chosen based on the highest penalized LOD score (Manichaikul et al. 2009) after forward and the backward selection and elimination modeling. Penalties for model selection and the genome-wide significance threshold (= 0.05) were determined through 1000 permutations of the “scantwo” function for two-dimensional QTL scanning. The percentage of the phenotypic variance explained (PVE) and effects of QTL were obtained by fitting a mixed linear model using the “fitqtl” function. The confidence intervals for each QTL were estimated using the “lodint” function that calculates the 2-LOD support intervals. The QTL was declared to be independent when two peaks on one chromosome were separated by a minimum of 20 cM (Ravi et al. 2011). QTL positions on the linkage map were drawn by MapChart v. 2.2 (Voorrips 2002).
Results
Phenotypic analysis of the parental lines and RIL population
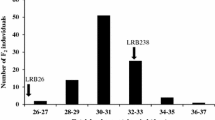
Individuals in the RIL population showed significant variation in CS resistance (F-value = 2.21, P = 0.0001). The parental lines Mélodie/2 and ILB 938/2 exhibited significant differences in resistance to CS under growth chamber conditions (Fig. 1). ILB 938/2 had a partial resistant reaction with mean disease severity of 30.4%, while Mélodie/2 showed a moderately susceptible reaction with a mean of 53.3%. The distribution of disease severity as a measure of CS response for RILs showed continuous variation ranging from 20.8 to 62.9%, with a mean of 44.5% (Fig. 1), suggesting polygenic regulation of CS severity. Similarly, the autoscaled PCA indicated that the data had no underlying structure for the first two principal components (PC1 vs PC2), which explained approximately 66.1% of the variance. The PCA-biplot graph also displayed a strong positive correlation among the four screening repeats, which were represented well by the graph (i.e., the vector with equal length), demonstrating that the data could be used for accurate mapping of CS resistance QTL (Fig. 2).
Frequency distributions of chocolate spot disease scores in 165 RILs derived from cross Mélodie/2 × ILB 938/2 evaluated under growth chamber conditions. Each value is the mean of four screenings, each with three replicates. The arrows indicate the average disease severity of the parents. Disease severity was rated on a 0–10 scale, where the disease severity score increased in 10% increments
PCA-biplot graph of PC1 vs PC2 for autoscaled and centered data for chocolate spot severity for each repeat (rep) collected from a 165 recombinant inbred lines (RILs) population of a cross Mélodie/2 × ILB 938/2 cross. Each RIL score, which is a deviation from its rep average value, was displayed as a point on the graph and reps as a blue arrow (vector). The parental lines are circled with black dots. The origin of the graph represents the average value for both each rep and across all repeats. The angle between any two arrows indicates the correlation between repeats (if the angle is 90 degrees, it means both variables show a lack of correlation), and the length of the arrow represents the variance explained by two components (all equal length means a perfect fit). The color bar indicator on the right shows the deviation of each score from the averages
Linkage map construction
A total of 35,363 SNP markers were filtered for polymorphism between the parents, significant segregation distortion, and missing data. A final genetic map was constructed from 4089 SNP markers, which mapped to six linkage groups (LGs) representing the six chromosomes of faba bean (Table 1). The LGs were numbered to match the respective faba bean consensus map based on where the markers lie (Webb et al. 2016). The linkage map spanned a total genetic distance of 1229.5 cM with an average marker interval of 0.3 cM. The genetic distance within LGs varied from 124.2 cM for LG 4 to 396.1 cM for LG 1. LG 4 contained the fewest SNP markers, whereas LG 1 contained the most SNP markers (Table 1; Fig. S1).
QTL analysis
Five QTL associated with chocolate spot resistance were detected in the RIL population (Table 2 and Fig. 3). Among these, two QTL (qBF1.1 and qBF1.2) were identified on LG 1 by both QTL mapping methods, CIM and MQM, implemented in WinQTLCart and R/qtl software, respectively. The QTL qBF1.1 accounted for 8.9–10.7% of PVE, whereas qBF1.2 explained 12.2–12.8% of the PVE (Table 2). These QTL were derived from the resistant line ILB 938/2. On the other hand, the remaining three QTL, one on LG 1 (qBF1.3), explained 7.3% of the PVE, and two on LG 6 (qBF6.1 and qBF6.2), accounted for 6.2 and 6.3% of the PVE, respectively, were detected solely by CIM methods. The resistance allele for qBF6.1 was contributed by ILB 938/2, whereas the moderately susceptible parent Mélodie/2 was the donor of qBF1.3 and qBF6.2 (Table 2).
Location of chocolate spot resistance QTL identified on chromosome 1 and 6 using 165 RILs derived from cross Mélodie/2 × ILB 938/2. The QTL positions are shown with a red bar, and the loci within the QTL regions are shown on the right of the bar. Only portions of the linkage map related to the QTL positions are displayed. a, b, and, c represents the position of qBF1.1, qBF1.2, and qBF1.3, respectively on chromosome 1. d and e represents the position of qFB6.1 and qFB6.2 respectively on chromosome 6
Discussion
Resistance breeding is the most cost-effective and environmentally friendly strategy for controlling CS. Many studies have reported ILB 938 as a source of resistance to CS (e.g., Hanounik and Robertson 1988; Rhaïem et al. 2002; Villegas-Fernández et al. 2009; Maalouf et al. 2016). The introduction of CS resistance from this source into elite cultivars could be accelerated using MAS. To implement MAS in faba bean breeding programs, the identification of markers linked to QTL or genes conferring resistance to CS is the first step. In this study, the analysis of QTL was conducted using a RIL population derived from ILB 938/2. The RIL population showed significant variation in pathogen response, as expected from the presence of resistance genes or alleles. The frequency distribution of the CS disease score in RILs showed a continuous distribution, indicating polygenic inheritance of CS resistance in ILB 938/2. To our knowledge, this study provides the first identification of genomic regions linked to CS resistance in this species.
Here, we report a linkage map, comprised of 4089 SNP markers in six linkage groups that correspond to the six chromosomes of faba bean, generated for the ILB938-derived RIL population. The genetic linkage map spanned 1229.5 cM with an average marker density of 0.3 cM. According to Stange et al. (2013), an increase in marker density from 5 to 1 cM could increase the power sufficiently to precisely localize and resolve closely linked QTL. Therefore, the genetic map developed in this study has sufficient marker density to provide adequate power for QTL mapping. The total map size and the marker distributions along the LGs of the current genetic map are consistent with the consensus map previously published by Webb et al. (2016). It is shorter than the recently published SNP-based consensus map (1547.71 cM) by Carrillo-Perrillo et al. (2020). Like previous marker-based genetic maps (Webb et al. 2016; Carrillo-Perrillo et al. 2020) and cytogenetic studies (Lucretti et al. 1993; Doležel and Lucretti 1995), LG 1, corresponding to chromosome 1, contained the highest number of SNP markers and a larger size.
Using this genetic map and the CS phenotypic data for the RIL population, we mapped five QTL associated with CS resistance on LG 1 and LG 6. Two QTL on LG 1 (qBF1.1 and qBF1.2) with high LOD scores were detected by both WinQTLCart and R/qtl. These QTL were derived from ILB 938/2 and explained a total phenotypic variation of 22%. The other three QTL, one on LG 1 (qBF1.3) and two on LG 6 (qBF6.1 and qBF6.2), passed the threshold after permutation and were detected only by WinQTLCart, collectively accounting for 19.8% of the phenotypic variance. Interestingly, two of these three QTL were inherited from the moderately susceptible parent Mélodie/2. In QTL studies for various traits in different crops, the inheritance of the resistance allele from a susceptible parent is not unusual, and a good example is the large effect QTL for improved yield in rice under drought conditions derived from the susceptible parent (Bernier et al. 2007).
Despite the fact that CS is one of the most damaging faba bean diseases worldwide, progress remains slow in breeding for CS resistance. Among the challenges related to faba bean improvement, the restricted appearance of the disease only during the humid climate cropping season makes traditional field-based phenotypic selection unreliable, particularly in dry areas like the Western Canadian prairies and parts of Australia (Adhikari et al. 2021; Khazaei et al. 2021b). This might explain why there were no QTL studies of CS previously reported. Therefore, conventional faba bean breeding could be augmented with molecular marker technologies to develop CS-resistant cultivars that are suitable for fulfilling the growing global demand for faba bean generated by its high seed protein content and great ecological service in cropping systems. Our study provides the first QTL identification for CS resistance using high-resolution SNP markers. Some of the identified QTL in this work can be used as potential targets for further studies, and the linked markers can enable the possibility of using MAS in faba bean CS resistance breeding after validation in appropriate germplasm.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
References
Adhikari KN, Zhang P, Sadeque A, Hoxha S, Trethowan R (2016) Single independent genes confer resistance to faba bean rust (Uromyces viciae-fabae) in the current Australian cultivar Doza and a central European line Ac1655. Crop Pasture Sci 67:649–654
Adhikari KN, Khazaei H, Ghaouti L, Maalouf F, Vandenberg A, Link W, O’Sullivan DM (2021) Conventional and molecular breeding tools for accelerating genetic gain in faba bean (Vicia faba L.). Front Plant Sci 12:744259
Angus JF, Kirkegaard JA, Hunt JR, Ryan MH, Ohlander L, Peoples MB (2015) Break crops and rotations for wheat. Crop Pasture Sci 66:523–552
Atienza SG, Palomino C, Gutiérrez N, Alfaro CM, Rubiales D, Torres AM, Ávila CM (2016) QTLs for ascochyta blight resistance in faba bean (Vicia faba L.): validation in field and controlled conditions. Crop Pasture Sci 67:216–224
Avila CM, Satovic Z, Sillero JC, Rubiales D, Moreno MT, Torres AM (2004) Isolate and organ-specific QTLs for ascochyta blight resistance in faba bean. Theor Appl Genet 108:1071–1078
Bernier J, Kumar A, Ramaiah V, Spaner D, Atlin G (2007) A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci 47:507–516
Beyene AT, Derera J, Sibiya J (2018) Genetic variability of faba bean genotypes for chocolate spot (Botrytis fabae) resistance and yield. Euphytica 214:132
Björnsdotter E, Nadzieja M, Chang W, Escobar-Herrera L, Mancinotti D, Angra D, Xia X, Tacke R, Khazaei H, Crocoll C, Vandenberg A, Link W, Stoddard FL, O’Sullivan DM, Stougaard J, Schulman AH, Andersen SU, Geu-Flores F (2021) VC1 catalyses a key step in the biosynthesis of vicine in faba bean. Nat Plants 7:923–931
Bouhassan A, Sadiki M, Tivoli B (2004) Evaluation of a collection of faba bean (Vicia faba L.) genotypes originating from the Maghreb for resistance to chocolate spot (Botrytis fabae) by assessment in the field and laboratory. Euphytica 135:55–62
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Carrillo-Perdomo E, Vidal A, Kreplak J, Duborjal H, Leveugle M, Duarte J, Desmetz C, Deulvot C, Raffiot B, Marget P, Tayeh N, Pichon JP, Falque M, Martin OC, Burstin J, Aubert G (2020) Development of new genetic resources for faba bean (Vicia faba L.) breeding through the discovery of gene-based SNP markers and the construction of a high-density consensus map. Sci Rep 10:6790
Cruz-Izquierdo S, Avila CM, Satovic Z, Palomino C, Gutierrez N, Ellwood SR, Phan HTT, Cubero JI, Torres AM (2012) Comparative genomics to bridge Vicia faba with model and closely-related legume species: stability of QTLs for flowering and yield-related traits. Theor Appl Genet 125:1767–1782
Doležel J, Lucretti S (1995) High-resolution flow karyotyping and chromosome sorting in Vicia faba lines with standard and reconstructed karyotypes. Theor Appl Genet 90:797–802
Duc G, Aleksic JM, Marget P, Mikic A, Paull J, Redden RJ, Sass O, Stoddard FL, Vandenberg A, Vishnyakova M, Torres AM (2015) Faba bean. In: DeRon A (ed) Grain Legumes Handbook of Plant Breeding, vol 10. Springer, New York, pp 141–178
Ellwood SR, Phan HTT, Jordan M, Hane J, Torres AM, Avila CM, Cruz-Izquierdo S, Oliver RP (2008) Construction of a comparative genetic map in faba bean (Vicia faba L.); conservation of genome structure with Lens culinaris. BMC Genom 9:380
Fan X, Zhang J, Yang L, Wu M, Chen W, Li G (2015) Development of PCR-based assays for detecting and differentiating three species of Botrytis infecting broad bean. Plant Dis 99(5):691–698
FAOSTAT (2021) Food and Agriculture Organization of the United Nations. Available online at: http://faostat.fao.org. Accessed 15 Oct 2021
Gorfu D (1996) Morphological, cultural and pathogenic variability among nine isolates of Botrytis fabae from Ethiopia. FABIS Newsl 38–39:37–41
Gorfu D, Yaynu H (2001) Yield loss of crops due to plant diseases in Ethiopia. Pest Manag J Ethiopia 5:55–67
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324
Hanounik SB (1982) Resistance in faba beans to chocolate spot. FABIS Newsl 5:24–26
Hanounik SB, Robertson LD (1988) New sources of resistance in Vicia faba to chocolate spot caused by Botrytis fabae. Plant Dis 72:696–698
Harrison JG (1988) The biology of Botrytis spp. on Vicia beans and chocolate spot disease - a review. Plant Pathol 37:168–201
Hutson RA, Mansfield JW (1980) A genetical approach to the analysis of mechanisms of pathogenicity in Botrytis/Vicia faba interactions. Physiol Plant Pathol 17:309–317
Ijaz U, Sudheesh S, Kaur S, Sadeque A, Bariana H, Bansal U, Adhikari K (2021) Mapping of two new rust resistance genes Uvf-2 and Uvf-3 in faba bean. Agronomy 11:1370
Kaur S, Kimber RBE, Cogan NOI, Materne M, Forster JW, Paull JG (2014) SNP discovery and high-density genetic mapping in faba bean (Vicia faba L.) permits identification of QTLs for ascochyta blight resistance. Plant Sci 217(218):47–55
Khalil SA, Nassib AM (1984) Identification of some sources of resistance to disease in faba bean. I. Chocolate spot (Botrytis fabae Sard.). FABIS Newsl 10:18–21
Khazaei H, Vandenberg A (2020) Seed mineral composition and protein content of faba beans (Vicia faba L.) with contrasting tannin contents. Agronomy 10:511
Khazaei H, O’Sullivan DM, Sillanpää MJ, Stoddard FL (2014) Use of synteny to identify candidate genes underlying QTL controlling stomatal traits in faba bean (Vicia faba L.). Theor Appl Genet 127:2371–2385
Khazaei H, O’Sullivan DM, Sillanpää MJ, Stoddard FL (2014) Genetic analysis reveals a novel locus in Vicia faba decoupling pigmentation in the flower from that in the extra-floral nectaries. Mol Breed 34:1507–1513
Khazaei H, Link W, Street K, Stoddard FL (2018) ILB 938, a valuable faba bean (Vicia faba L.) accession. Plant Genet Resour 16:478–482
Khazaei H, O’Sullivan DM, Stoddard FL, Adhikari KN, Paull JG, Schulman AH, Andersen SU, Vandenberg A (2021) Recent advances in faba bean genetic and genomic tools for crop improvement. Legume Sci 3:e75
Khazaei H, Hawkins G, Vandenberg A (2021) Historical review of faba bean improvement in western Canada. Legume Sci 2021:e92
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Link W, Abdelmula AA, Kittlitz EV, Bruns S, Riemer H, Stelling D (1999) Genotypic variation for drought tolerance in Vicia faba. Plant Breed 118:477–484
Lorieux M (2012) MapDisto: fast and efficient computation of genetic linkage map. Mol Breed 30:1231–1235
Lucretti S, Doležel J, Schubert I, Fuchs J (1993) Flow karyotyping and sorting of Vicia faba chromosomes. Theor Appl Genet 85:665–672
Maalouf F, Ahmed S, Shaaban K, Bassam B, Nawar F, Singh M, Amri A (2016) New faba bean germplasm with multiple resistances to ascochyta blight, chocolate spot and rust diseases. Euphytica 211:157–167
Maalouf F, Hu J, O’Sullivan DM, Zong Z, Hamwieh A, Kumar S, Baum M (2019) Breeding and genomics status in faba bean (Vicia faba). Plant Breed 138:465–473
Manichaikul A, Moon JY, Sen S, Yandell BS, Broman KW (2009) A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics 181:1077–1086
O’Sullivan DM, Angra D (2016) Advances in faba bean genetics and genomics. Front Genet 7:150
O’Sullivan DM, Angra D, Harvie T, Tagkouli V, Warsame A (2019) A genetic toolbox for Vicia faba improvement. In: International conference on legume genetics and genomics, May 13–17, 2019. Dijon, France
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org
Ravi K, Vadez V, Isobe S, Mir RR, Guo Y, Nigam SN, Gowda MVC, Radhakrishnan T, Bertioli DJ, Knapp SJ, Varshney RK (2011) Identification of several small main-effect QTLs and a large number of epistatic QTLs for drought tolerance related traits in groundnut (Arachis hypogaea L.). Theor Appl Genet 6:1119–1132
Rhaïem A, Cherif M, Kharrat M, Cherif M, Harrabi M (2002) New faba bean genotypes resistant to chocolate spot caused by Botrytis fabae. Phytopathol Mediterr 41:99–108
Robertson LD (1984) A note on the I.L.B. Source of Botrytis fabae resistance. In: Chapman GP, Tarawali SA (Eds.), Systems for Cytogenetic Analysis in Vicia faba L. Adv Agri Bio. Dordrecht: Springer, vol. 11
Rubiales D, Khazaei H (2022) Advances in disease and pest resistance in faba bean. Theor Appl Genet, in Press. https://doi.org/10.1007/s00122-021-04022-7
SAS Institute (2015) The SAS system for windows. Version 9.3. SAS Institute, Inc., Cary
Soltis DE, Soltis PS, Bennett MD, Leitch IJ (2003) Evolution of genome size in the angiosperms. Am J Bot 90:1596–1603
Stange M, Utz HF, Schrag TA, Melchinger AE, Würschum T (2013) High-density genotyping: an overkill for QTL mapping? Lessons learned from a case study in maize and simulations. Theor Appl Genet 126:2563–2574
Stoddard FL, Nicholas AH, Rubiales D, Thomas J, Villegas-Fernández AM (2010) Integrated pest management in faba bean. Field Crops Res 115:308–318
Sudheesh S, Kimber RBE, Braich S, Forster JW, Paull JG, Kaur S (2019) Construction of an integrated genetic linkage map and detection of quantitative trait loci for ascochyta blight resistance in faba bean (Vicia faba L.). Euphytica 215:42
Taylor J, Butler D (2017) R package ASMap: efficient genetic linkage map construction and diagnosis. J Stat Softw 79(6):1–29
Temesgen T, Keneni G, Sefera T, Jarso M (2015) Yield stability and relationships among stability parameters in faba bean (Vicia faba L.) genotypes. Crop J 3:258–269
Tivoli B, Baranger A, Avila CM, Banniza S, Barbetti MCW, Davidson J, Lindeck K, Kharrat M, Rubiales D, Sadiki M, Sillero JC, Sweetingham M, Muehlbauer FJ (2006) Screening techniques and sources of resistance to foliar diseases caused by major necrotrophic fungi in grain legumes. Euphytica 147:223–253
Villegas-Fernández AM, Sillero JC, Emeran AA, Winkler J, Raffiot B, Tay J, Flores F, Rubiales D (2009) Identification and multi-environment validation of resistance to Botrytis fabae in Vicia faba. Field Crops Res 114:84–90
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL Cartographer 25. North Carolina State University, Raleigh, USA, Department of Statistics
Warsame AO, O’Sullivan DM, Tosi P (2018) Seed storage proteins of faba bean (Vicia faba L): current status and prospects for genetic improvement. J Agric Food Chem 66:12617–12626
Webb A, Cottage A, Wood T, Khamassi K, Hobbs D, Gostkiewicz K, White M, Khazaei H, Ali M, Street D, Duc G, Stoddard FL, Maalouf F, Ogbonnaya FC, Link W, Thomas J, O’Sullivan DM (2016) A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba L.). Plant Biotechnol J 14:177–185
Acknowledgements
The authors thank Prof. Donal M. O’Sullivan and Dr. Deepti Angra from the University Reading, UK, for providing SNP genotyping results. We also thank Dr. Sabine Banniza (Pulse pathology lab, University of Saskatchewan) for technical advice and the staff of the Pulse Crop Breeding Programs for technical support at the University of Saskatchewan.
Funding
This work was supported by the Danish Innovation Fund for funding the NORFAB project (Innovation Fund Denmark grant number 5158-00004B); ADF (Agriculture Development Fund – Government of Saskatchewan, Canada); the Western Grains Research Foundation, Canada; the Saskatchewan Pulse Growers, Canada; and the Academy of Finland (decision 298314, “Papugeno”).
Author information
Authors and Affiliations
Contributions
Hamid Khazaei, Albert Vandenberg, Frederick L. Stoddard, and Alan H. Schulman conceived and designed the experiments. Tadesse S. Gela and Margaret Bruce performed the experiments. Wei Chang performed DNA extraction and library preparation. Tadesse S. Gela and Hamid Khazaei analyzed the data. Hamid Khazaei, Albert Vandenberg, Frederick L. Stoddard, and Alan H. Schulman provided resources and acquired the funding. Tadesse S. Gela wrote the first draft of the manuscript and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gela, T.S., Bruce, M., Chang, W. et al. Genomic regions associated with chocolate spot (Botrytis fabae Sard.) resistance in faba bean (Vicia faba L.). Mol Breeding 42, 35 (2022). https://doi.org/10.1007/s11032-022-01307-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-022-01307-7