Abstract
Sugarcane viral diseases are most challenging because of their systemic infection, rapid spread through vegetative propagative stem cuttings (setts) and vector transmissions. About 23 virus species are known to infect sugarcane across the countries of which yellow leaf disease (YLD) is caused by sugarcane yellow leaf virus (ScYLV), mosaic caused by sugarcane mosaic virus (SCMV) and sugarcane streak mosaic virus (SCSMV), and leaf fleck caused by sugarcane bacilliform virus (SCBV) are widespread in India in all the high yielding and high sugar varieties. Sorghum, maize, and sugarcane are all C4 plants that belong to the Poaceae family and are grown in many countries under the same crop ecosystem. SCMV infection was reported earlier in these genetically related crops of Saccharum in China, USA, Argentina, Brazil, and other countries. In India, sugarcane viruses are managed effectively by planting virus-free planting material derived through tissue culture. However, tissue culture-derived plants pick up the viruses within one or two seasons under natural field conditions thus necessitating new seed cane requirements at regular intervals adding to the cost of cultivation. YLD is primarily spread through infected planting materials and secondarily spread through phloem sap-feeding aphid vectors from neighboring fields. Hence, we have strongly suspected sorghum and maize crops grown in adjacent field areas as potential reservoirs for sugarcane viruses. Based on the detailed research work carried out with symptomatic and asymptomatic sorghum plants adjacent to sugarcane fields from different places in Tamil Nadu showed 17.14, 5.71 and 11.42% infections of SCMV, SCSMV, and ScYLV, respectively. All the sugarcane viruses of the sorghum host had the highest nucleotide homology of 99–100% in the highly conserved coat protein region with Saccharum sequences from India and other countries. The results clearly revealed that the sorghum-infecting viruses were as identical as those diagnosed in the nearby sugarcane cultivars and they have quickly adapted to the new host environment without any changes in their genomic regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum spp hybrid) is a major field crop grown in the tropics and subtropics around the world for sugar and bioethanol production. In biomass production, sugarcane ranks first among cultivated crops and is one of the top ten most widely cultivated crops worldwide. In 2021, ~ 1.9 billion tons of sugarcane was produced worldwide from an area of 26.3 million ha grown in ~ 100 countries. Brazil and India are the world’s largest producers of sugarcane and sugar and both account for nearly 64.0% of global output. China, Thailand, Mexico, Pakistan, the United States, Colombia, Australia, Cuba, and the Philippines are the other major sugarcane producing countries [http://www.fao.org/faostat (accessed on 22 April 2023)]. Among the disease-causing agents, viruses are most challenging because of their systemic infection, inconspicuous symptoms, rapid spread through vegetative propagative stem cuttings (known as setts) and natural vector transmissions. Advent of next generation sequencing (NGS) based omics technologies and advanced quick diagnosis methods led to report of new viruses and this helps to detect new host spectrum of known viruses in the crop ecology. In India, sugarcane is affected by yellow leaf disease (YLD) caused by sugarcane yellow leaf virus (ScYLV) (Polerovirus, Solemoviridae), mosaic caused by sugarcane mosaic virus (SCMV) (Potyvirus, Potyviridae) and sugarcane streak mosaic virus (SCSMV) (Poacevirus, Potyviridae) and leaf fleck caused by sugarcane bacilliform virus (SCBV) (Badnavirus, Caulimoviridae) (Viswanathan et al. 2018). About 23 virus species are known to infect sugarcane across the countries of which these four viruses are widespread in India in all the high yielding and high sugar varieties, cause more than 50% yield losses, 38.9–42.3% reduction in plant growth traits and 34.15% reduction in juice yield in the susceptible varieties (Viswanathan 2016; Viswanathan et al. 2014, 2018; Bagyalakshmi et al. 2019; Holkar et al. 2020). Recently, infection of maize yellow mosaic virus (MaYMV) (Polerovirus, Solemoviridae) was reported from India (Nithya et al. 2021).
Mosaic causative viruses SCMV, SCSMV, and sorghum mosaic virus (SrMV) are distributed worldwide (Grisham et al. 2011; Perera et al. 2012) in which SCSMV is mostly prevalent in India, Indonesia, Vietnam, Sri Lanka, and Thailand (Chatenet et al. 2005; Parameswari et al. 2013; Putra et al. 2014; Liang et al. 2016; Feng et al. 2018) and recently reported from Ivory Coast in West Africa (Daugrois et al. 2020; Sorho et al. 2021) while SrMV in the USA, China, and Vietnam (Yang and Mirkov 1997; Grisham and Pan 2007; Chen and Chen 2002; Ha et al. 2008). In India, SCMV and SCSMV are the causal agents either as single or mixed infections (Viswanathan et al. 2007, 2008, 2013), whereas in the neighboring country China, mosaic was reported with single or mixed infections of SCMV, SrMV, and/or SCSMV (Lu et al. 2021) and in other countries mixed infections with different combinations of SCMV, SrMV, and SCSMV (Wang et al. 2017). But, SrMV is not diagnosed from sugarcane mosaic and sorghum samples in India till date. Sorghum (Sorghum bicolor), maize (Zea mays), and sugarcane are C4 plants belonging to the Poaceae family and are grown under the same crop ecosystem in many countries. SCMV infection was reported earlier in these genetically related crops of Saccharum in China, USA, Argentina, Brazil, and other countries (Yang and Mirkov 1997; Wu et al. 2012; Braidwood et al. 2019). ScYLV is an important sugarcane virus primarily spread through infected planting materials and secondarily spread through phloem sap-feeding aphid vector Melanaphis sacchari, in a circulative, and non-propagative manner in India (Chinnaraja and Viswanathan 2015), however, Ceratovacuna lanigera, Rhopalosiphum maidis and R. rufiabdominalis were also reported as vectors in other countries (Scagliusi and Lockhart 2000; Zhou et al. 2006).
Earlier, low to moderate level of ScYLV infections were reported in maize, rice and sorghum seedlings by artificial inoculations with viruliferous aphids (Schenck and Lehrer 2000), Erianthus sp under controlled conditions (Comstock et al. 2001) and recently, in barley (Hordeum vulgare) in Tunisia, North Africa (Bouallegue et al. 2014) and as a natural infection in Sorghum almum (Columbus grass) and S. bicolor (grain sorghum) in Florida, USA (Delgado et al. 2015; Wei et al. 2016; El-Sayed et al. 2018). The Columbus grass grown in nearby sugarcane fields harboured M. sacchari and was identified as potential reservoir and secondary host for ScYLV transmission to sugarcane.
In the absence of resistant varieties, the sugarcane viruses are manged effectively by planting virus free planting materials derived through tissue culture (Viswanathan 2016, 2018, 2021; Viswanathan et al. 2018). However, such virus free materials acquire new virus infections under field conditions. Hence, replanting of virus free seed canes is required at regular intervals adding to the cost of cultivation. Therefore, we have suspected sorghum and maize crops grown nearby field areas as potential reservoir for sugarcane viruses and carried out detailed research work to establish host specificity and their other possible host ranges in the sugarcane ecosystem. This study also elaborated sugarcane viral epidemiology in India and brought a new information to formulate crop management advisory to prevent further spread into new areas and healthy seed production.
Materials and methods
Plant material
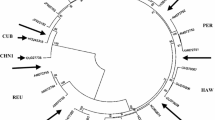
TNAU sorghum varieties grown for fodder, mostly of Sencholam types showing typical mosaic-like symptoms of chlorotic leaves, yellow and green chlorotic stripes, and streaks more clearly visible against the sunlight, and asymptomatic apparently healthy leaves were randomly collected from nearly 35 sorghum fields viz. four in Paramathi (Namakkal), 18 in Coimbatore, two in Sevur (Tiruppur) and one each in Kidaram (Trichy District) and Thiruvannamalai District. All the collected samples were from sorghum fields adjacent to sugarcane crops in Tamil Nadu state during the year 2020-21 (Fig. 1). Young leaf samples were collected mostly from the vegetative stage of the crop in all the places and were stored at -80oC until further processing. Along with sorghum, sugarcane cultivars such as Co 06022, Co 0212, and Co 86032 grown in the adjoining fields were also collected to confirm virus(es) infection.
RNA extraction and RT-PCR
All the collected leaf samples from different places were subjected into RNA extraction, first ground well using liquid nitrogen, re-suspended in 1mL TRI Reagent (Sigma, USA), followed the manufacturer’s protocol and the pellet was dissolved in a final volume of 30 µL RNase-free water and stored at -80oC. In order to remove the DNA contamination in the extracted total RNA, it was treated with DNase I (1U/ µL) (Thermo Fischer scientific, USA) with 10x reaction buffer, MgCl2 and RNase free water to make up the final volume 10 µL, kept at 37oC for 2 hr incubation in water bath. Further, 1 µL of 50mM EDTA was added in the reaction for the DNase enzyme inactivation and continued incubation at 60-65oC for 10 min. By using the Nanodrop™ 2000 Spectrophotometer (Thermo Scientific, USA) the concentration, and purity of RNA were assessed and the integrity were visualized on 1% EtBr-stained agarose gel. For the SCMV and SCSMV diagnosis, one µg total RNA were reverse transcribed using oligo (dT) primer and complementary DNA synthesis was performed using cDNA synthesis kit (Genei®, Bangalore), for ScYLV also one µg total RNA was reverse transcribed using 50 pmol ScYLV reverse primer R615 by following the manufacturer’s protocol in RevertAid H Minus First Strand cDNA Synthesis Kit (MBI Fermentas, USA) in a thermal cycler (C1000, Bio-Rad, USA). The synthesized cDNA was stored at -20°C for RT-PCR assay. SCMV and SCSMV diagnostic primers viz. SCMV-eF 894: 5’ CCCGAAGCTTGCTGGAACAGTCGATGCAGG 3’ SCMV-eR 894: 5’AATCCTCGAGTTAGCCAGCTGTGTGTCTCTCTGTATTC 3’and SCSMV-F 690: 5’GGATCCGGACAAGGAACGCAGCCAC3’; SCSMVR690: 5’AGATCTCGCACGTCGATTTCTGCTGGTG 3’; ScYLVF615: 5’ GGATCCATGAATACGGGCGCTAACCGYYCAC 3’; ScYLVR615: 5’AGATCTGTGTTGGGGRAGCGTCGCYTACC 3’; ScYLV-FP995: 5’GGTTYAAAGTCGAGGTWGCY 3’; ScYLV-RP995: 5’CTGCTAGGCTYSMGTCYCCATTC3’ were designed from the consensus coat protein and movement protein regions based on sequences retrieved from the GenBank using primer blast.
The PCR reaction was performed in a total volume of 25 µl containing 10X Taq buffer with 15 mM MgCl2, 10 mM dNTP mix, 10 µM of each forward and reverse primers, 1.25 units of Taq polymerase (Origin, Kerala, India), 2 µl of cDNA and sterile MilliQ water to make up the final volume. The PCR program was standardized for all three RNA viruses with an initial denaturation at 94oC for 5 min, followed by 34 cycles of denaturation at 94oC for 1 min, annealing at 65oC for 1 min, extension at 72oC for 1 min and final extension at 72oC for 10 min (Thermocycler C1000, Biorad, USA). 1.5% agarose gels stained with ethidium bromide (0.5 µg/ ml) were used for visualization of products in the gel documentation system (Syngene, UK). In case of ScYLV, four different primers starting from overlapping ORF 1 and 2 regions to ORF 3 and ORF 5 covering all the coding regions of the genome were used to confirm the virus presence.
The positive amplicons from the RT-PCR assay for all the three viruses were sequenced by Sanger method at both ends (Eurofins, Bengaluru, India). The trimmed quality nucleotide sequences (q = > 30) obtained from this study were subjected to Blastn analysis and further pairwise multiple sequence alignment was performed through Bio edit v7.1.9 (Hall 1999) with sugarcane, SrMV and ScYLV sequences representing other countries were retrieved from GenBank. The genetic proximity between sugarcane and sorghum infecting sugarcane viruses were assessed by phylogenetic tree analysis using maximum likelihood Tamura-Nei model with nearest neighbour interchange (NNI) tree options (MEGA X v.10.1.6) with 1000 bootstrap replications (Kumar et al. 2018).
Results
Of the 35 sorghum samples tested for three major sugarcane RNA viruses, SCMV was diagnosed from four symptomatic and two asymptomatic samples from Coimbatore, Paramathi, Thirvannamalai; SCSMV from two asymptomatic samples from Kidaram and Paramathi, and ScYLV from four asymptomatic samples from Paramathi, Kidaram , and Thiruvannamalai. Most of the sorghum samples were collected from nearby sugarcane field cvs Co 86032, Co 0212, and Co 06022, and all the positive samples had shown the expected amplicon sizes of 894, 690, and 615 bp for SCMV, SCSMV and ScYLV, respectively (Fig. 2a-c) and their sequences were submitted to GenBank under the accession numbers of MW580397, MW618113, OK358711, OK358712, OK358713, OK358716 and OK665790 (SCMV); MW654013, OK631531, OK631532, OK631533 and OK 665,794 (ScYLV); OK999998, OK999999, OK665791 and OK665792 (SCSMV). The pairwise multiple sequence alignment of ScYLV sequences from S. bicolor of this study has clearly shown the highest nucleotide similarities of 99 to 100% among themselves and 88.7–95.7% identity with other ScYLV S. bicolor isolates of Florida, USA; 86-86.5% identity with ScYLV- Barley, Tunisia, North Africa. Whereas, it had shown 95.2 to 99.8% identities with other ScYLV- Saccharum sequences from India, China, USA, Brazil, Mauritius, Cuba, Columbia, Reunion and South Africa (Supplementary Table 1). Besides, they had shown 100% nucleotide identity with nearby sugarcane field samples.
The pairwise multiple sequence alignment of SCSMV sequences of this study has clearly shown highest nucleotide similarities of 99.8% among themselves and 96% identity with other SCSMV sequences of sorghum from India; 94.2–97.6% identity with other SCSMV sugarcane from India, Thailand, Indonesia and China, however, they had shown 83–84% identity only with Brazil and SCSMV Thailand isolates (Supplementary Table 2). Besides, it had shown 100% nucleotide identity with nearby sugarcane field samples collected from all the places.
The pairwise multiple sequence alignment of SCMV sequences of this study has shown 99.1–99.8% identity among themselves and 98.3% identity with SCMV Saccharum India; 93.9 to 98.3% with other SCMV isolates of Saccharum, Zea mays, S. bicolor, Setaria viridis and S. faberi reported from China, Thailand, Iran, Vietnam, Australia, USA and shown less similarity with some isolates of SCMV- Canna from China, SCMV-Saccharum and Zea mays from Vietnam (Supplementary Table 3). In phylogenetic analysis, ScYLV- S. bicolor sequences from this study clustered together in a single clade with all ScYLV from Saccharum, sorghum and barley isolates retrieved from GenBank representing other countries (Fig. 3). Similarly, SCSMV and SCMV isolates sequences from this study clustered together in a single clade with other SCMV and SCSMV, S. bicolor, Zea mays and Saccharum isolates retrieved from GenBank representing major sugarcane growing countries such as Brazil, India, China, Australia, Thailand, Vietnam, Iran etc. (Figs. 4 and 5).
Phylogenetictree constructed by the maximum likelihood method using Tamura-Nei model with nearestneighbor-joining interchange tree options (MEGA X v.10.1.6) with 1000 bootstrapreplications showing the closest genetic relationship of ScYLV in S. bicolorfrom this study with other ScYLV from Saccharum, sorghum and barleyisolates from India and other countries. The bootstrap value expressed aspercentage of 1000 replications and branch lengths areproportional to the number of substitutions.
Phylogenetic tree constructed by the maximum likelihood method using Tamura-Nei model with-nearest neighbor-joining interchange tree options (MEGA X v.10.1.6) with 1000 bootstrap replications showing the closest genetic relationship of SCMV in S. bicolor from this study with other SCMV from Saccharum sp, Zea mays, and Setaria sp from India, China, Thailand, Vietnam and the USA. The bootstrap value expressed as a percentage of 1000 replications and branchlengths are proportional to the number of substitutions.
Phylogenetic tree constructed by the maximum likelihood method using Tamura-Nei model with nearest neighbor-joining interchange tree options (MEGA X v.10.1.6) with 1000 bootstrapreplications showing the closest genetic relationship of SCSMV in S. bicolor from this study with other SCSMV from Saccharum sp from India,China, and Thailand. The bootstrap value is expressed as a percentage of 1000 replications and branch lengths are proportional to the number of institutions.
Discussion
Plant viruses and insect vectors might undergo several physiological changes for successful transmission from one host to another. The epidemiology of sugarcane viral disease relies on critical combinations of the viruses, vectors, host plant susceptibility to both viruses, vectors, and environmental conditions (Viswanathan 2021). The present study confirms the natural infection of sugarcane viruses viz. SCMV, SCSMV and ScYLV in grain sorghum, India through RT-PCR assays and by sequences of the highly conserved various coding regions of the genome such as coat protein regions of all the three viruses tested. ScYLV sequences of S. bicolor from this study were 99 to 100% similar among themselves and shared 88.7–95.7% identity with other ScYLV S. bicolor isolates of Florida, USA; 86-86.5% identity with ScYLV- Barley, Tunisia, North Africa. Whereas, it had shown 95.2 to 99.8% identities with other ScYLV- Saccharum sequences from India, China, USA, Brazil, Mauritius, Cuba, Columbia, Reunion and South Africa. Similarly, SCMV and SCSMV sequences from this study has shown 99.1–99.8% similarity among themselves and 98.3% similarity with Saccharum India isolates and 93.9 to 98.3% with other isolates of Saccharum, Zea mays, Sorghum bicolor, Setaria viridis and S. faberi reported from China, Thailand, Iran, Vietnam, Australia, and USA. Besides, sugarcane viruses of sorghum host had shown 100% nucleotide identity with adjacent sugarcane field samples collected from all the places. The results clearly revealed that the sorghum infecting viruses were as same as diagnosed in the nearby sugarcane cultivars and probably they have quickly adapted to the new host environment without any changes in their coat protein and movement protein regions. The results of all three sugarcane RNA viruses from symptomatic and asymptomatic sorghum samples collected from nearby sugarcane fields clearly indicated possible transfer of these viruses by insect species. Mixed infection of all the three viruses were also diagnosed from asymptomatic samples collected from Paramathi (Namakkal District), and Kidaram (Trichy District).
The close similarity between sugarcane and sorghum could be the reason for the efficient transfer of these viruses by the insect species, mostly phloem sap feeders on these plants. In India, M. sacchari was reported as vector for ScYLV (Chinnaraja and Viswanathan 2015) however, role of other reported vectors such as Ceratovacuna lanigera, Rhopalosiphum maidis and R. rufiabdominalis in sugarcane and sorghum fields are to be investigated in detail. In our study on population dynamics of M. sacchari in sugarcane for four consecutive years during 2015 to 2019 under the tropical conditions at ICAR-SBI, Coimbatore, and its Research Centre Agali, Kerala on around 120 varieties including Co canes and Co allied material showed that grand growth period supports higher aphid population irrespective of the varieties and the infestation was higher in old leaves in the diseased plants as compared to the respective healthy ones. Under field conditions, virus-free sugarcane plants become infected with the disease within one or two growing seasons mainly through secondary sources of aphids from nearby fields. The polynomial regression analysis revealed that the number of aphids per plant at their highest density significantly influenced the severity of YLD (Viswanathan et al. 2022).
Komor (2011) reported that the monocotyledon grass species wheat, rice and corn did not acquire the ScYLV during their growth phase even though their fields were next to YL infected sugarcane fields, similarly, wheat, rice, corn, barley and oat grown in pots together with pots of infected sugarcane were not infected with the disease-causing agent. After one decade, natural infections of ScYLV were reported in barley, Erianthus sp, Columbus grass and grain sorghum based on RT-PCR and nested RT-PCR analysis due to cross infections (Bouallegue et al. 2014; ElSayed et al. 2018; Tang Lihua et al. 2019). In USA, aphid has been reported as a perennial pest to sorghum and causing severe damage to most of the sorghum growing regions (Uchimiya and Knoll 2019). Apart from ScYLV, M. sacchari was earlier reported to be transmitting other viruses such as millet red leaf virus (MRLV, Luteovirus), SrMV (Potyvirus), and SCMV (Potyvirus) (Yang 1986). Recently, natural infection of ScYLV was reported in Sorghum almum (Columbus grass) in sugarcane ecosystem in USA. S. almum is a weed species related to sugarcane and maize grown along the canals and near sugarcane fields, now identified as a secondary host of ScYLV and M. sacchari in virus transmission studies (Delgado et al. 2016). Similarly, M. sacchari is reported to infest other grass hosts such as Hordeum vulgare (barley), Avena sativa (oats), Sorghum bicolor (sorghum), Zea mays var. saccharata (sweet corn), and Triticum aestivum (wheat) by Singh et al. 2004.
Recent reports on molecular characterization of natural infection of ScYLV from S. bicolor revealed its 99% identity to a sugarcane isolate from Florida that was sequenced 20 years ago (Moonan et al. 2000) and it was suspected that the same virus strain might have evolved to infect a new host crop (Boukari et al. 2020). However, a recombinant new genotype of ScYLV was also reported in S. bicolor (ElSayed et al. 2018). High prevalence of SCMV was reported in asymptomatic S. bicolor based on RT-PCR and HTS analysis (Boukari et al. 2020), similarly, sugarcane infected by ScYLV in Florida was reported as generally asymptomatic and no disease symptoms associated with this virus in S. almum and S. bicolor. Earlier, natural infections of maize dwarf mosaic virus (MDMV), SCMV (Gordon and Thottappily 2003) and SCSMV (Srinivas et al. 2010) in sorghum were reported. In this study, we diagnosed SCMV, SCSMV and ScYLV from both symptomatic and asymptomatic samples and the results are in accordance with the earlier reports from other countries. However, natural occurrence of ScYLV in S. bicolor is the first report from this study.
During the year 2013, massive outbreaks of M. sacchari occurred on sorghum which altogether changed the aphid population structure in USA led to invasion of a new super clone H1 haplotype in sorghum, and within two years, the new haplotype had spread to ≥ 98% sorghum growing regions (Armstrong et al. 2015; Bowling et al. 2016; Nibouche et al. 2018). Recently, existence of haplotypes H1-H6 was reported in USA, of which H1 type mainly infests Sorghum species and sugarcane; H3 mainly infests sugarcane, sorghum and Johnsongrass (S. halepense) and H6 infests Johnsongrass and sugarcane (Nibouche et al. 2018). Earlier it was reported that spread of YLD by insect vector depends on cultivar susceptibility and prevailing climatic conditions (Daugrois et al. 2011).
Transmission of SCMV and SrMV by several aphid vectors viz. Dactynotus ambrosiae, Hysteroneura setariae, Longiunguis sacchari, Rhopalosiphum maidis, and Toxoptera graminum in a non-persistent manner were reported. Ants have also been reported for indirect transmission if they interact actively with aphids in diseased sugarcane fields. Although SCSMV transmitting insect vectors have not yet been identified, mites (Aceria sp) were reported as vectors for the closely related virus species Triticum mosaic virus (TriMV) and wheat streak mosaic virus (WSMV) transmission (Lockhart and Autrey 2000; Singh et al. 2005). Since sorghum and sugarcane has very close genetic proximity, the resistance level in sorghum reported as a critical factor for M. sacchari outbreaks when sugarcane and sorghum coexist in the field or in crop succession. In this study, we have also detected ScYLV from the aphid samples collected from the sorghum fields in Paramathi (Namakkal) (Viswanathan, unpublished) and with these findings, S. bicolor has been conclusively found as another natural host to ScYLV in India in the sugarcane ecosystem. Since M. sacchari has been reported to affect many monocots, investigating its role in sugarcane virus transmission to sorghum and other grass hosts is important in India to understand the viral disease epidemiology.
Future work focusing on sequencing of complete genomes of SCSMV, SCMV and ScYLV from infected sorghum will provide more information regarding new virus strains association with sorghum plants. Plant growth and yield reduction due to these sugarcane viruses, presence of sugarcane aphid and any haplotype emergence in Indian M. sacchari population in sorghum as like in USA need to be investigated as virus and vector threaten both sugarcane and sorghum crops. Similarly, regular screening of the viral hosts in sugarcane ecosystem by metagenomics will allow us to monitor the spread of sugarcane viruses to new host ranges and new vectors which will eventually benefit us to develop crop advisory practices for the management.
References
Armstrong JS, Rooney WL, Peterson GC, Villenueva RT, Brewer MJ, Sekula-Ortiz D (2015) Sugarcane aphid (Hemiptera: Aphididae): host range and sorghum resistance including cross-resistance from greenbug sources. J Econ Entomol 108:576–582. https://doi.org/10.1093/jee/tou065. PMID: 26470168
Bagyalakshmi K, Viswanathan R, Ravichandran V (2019) Impact of the viruses associated with mosaic and yellow leaf disease on varietal degeneration in sugarcane. Phytoparasitica 47:591–604. https://doi.org/10.1007/s12600-019-00747-w
Bouallegue M, Mezghani-Khemakhem M, Makni H, Makni M (2014) First report of sugarcane yellow leaf virus infecting barley in Tunisia. Plant Dis 98:1016
Boukari W, Wei C, Tang L, Hincapie M, Naranjo M, Nuessly G, Beuzelin J, Sood S, Rott P (2020) Lack of transmission of sugarcane yellow leaf virus in Florida from Columbus grass and sugarcane to sugarcane with aphids or mites. PLoS ONE 15(3):e0230066. https://doi.org/10.1371/journal.pone.0230066
Bowling RD, Brewer MJ, Kerns DL, Gordy J, Seiter N, Elliott NE, Buntin GD, Way MO, Royer TA, Biles S, Maxson E (2016) Sugarcane aphid (Hemiptera: Aphididae): a New Pest on Sorghum in North America. J Integr Pest Manag 7(1):12. https://doi.org/10.1093/jipm/pmw011
Braidwood L, Muller SY, Baulcombe D (2019) Extensive recombination challenges the utility of sugarcane mosaic virus phylogeny and strain typing. Sci Rep 9:20067
Chatenet M, Mazarin C, Girard JC, Fernandez E, Gargani D, Rao GP, Royer M, Lockhart B, Rott P (2005) Detection of sugarcane streak mosaic virus in sugarcane from several asian countries. Sugar Cane Int 23(4):12–15
Chen J, Chen JP (2002) Sugarcane mosaic disease in Zhejiang Province was caused by Sorghum mosaic virus and sugarcane mosaic virus. Chin J Virol 18(4):362–366
Chinnaraja C, Viswanathan R (2015) Quantification of sugarcane yellow leaf virus in sugarcane following transmission through aphid vector, Melanaphis sacchari. Virus Dis 26:237–242
Comstock JC, Miller JD, Schnell RJ (2001) Incidence of sugarcane yellow leaf virus in clones maintained in the world collection of sugarcane and related grasses at the United States National Repository in Miami, Florida. Sugar Tech 3:128–133
Daugrois JH, Edon-Jock C, Bonoto S, Vaillant J, Rott P (2011) Spread of sugarcane yellow leaf virus in initially disease-free sugarcane is linked to rainfall and host resistance in the humid tropical environment of Guadeloupe. Eur J Plant Pathol 129:71–80
Daugrois JH, Roumagnac P, Kouakou Y, Oura OJDT, Pita JS (2020) First report of sugarcane streak mosaic virus in sugarcane (Saccharum spp.) in Côte d’Ivoire. New Dis Rept 41:22. https://doi.org/10.5197/j.2044-0588.2020.041.022
Delgado HE, Kaye C, Hincapie M, Boukari W, Wei C, Fernandez J, Mollo DS, Comstock J, Rott P (2015) First report of sugarcane yellow leaf virus infecting Columbus grass (Sorghum almum) in Florida. Plant Dis 100:1027
Delgado HVE, Kaye C, Hincapie M, Boukari W, Wei C, Fernandez JV, Mollov D, Comstock JC, Rott P (2016) First report of sugarcane yellow leaf virus infecting Columbus grass (Sorghum almum) in Florida. https://doi.org/10.1094/PDIS-10-15-1158-PDN. Disease Notes
Elsayed AI, Boulila M, Odero DC, Komor E (2018) Phylogenetic and recombination analysis of sorghum isolates of sugarcane yellow leaf virus. Plant Pathol 67:221–232
Feng XY, Shen LB, Zhao TT, Xiong GR, Wang JG, Yang BP, Wang WZ, Feng CL, Zhang SZ (2018) Molecular identification of virus diseases in sugarcane in Yunnan. J South Agric 49:2198–2203
Grisham MP, Rott P, Bailey RA, Comstock JC, Croft BJ, Eds (2011) CIRAD Publication Services: Montepellier, France, 249–254
Gordon DT, Thottappily G (2003) Maize and Sorghum. In: Loebenstein G, Thottappily G (eds) Virus and virus-like diseases of major crops in developing countries. Kluwer Academic Publishers, Netherlands, pp 295–335
Grisham MP, Pan YB (2007) A genetic shift in the virus strains that cause mosaic in Louisiana sugarcane. Plant Dis 91:453–458
Ha C, Revill P, Harding RM, Vu M, Dale JL (2008) Identification and sequence analysis of potyviruses infecting crops in Vietnam. Arch Virol 153:45–60
Holkar SK, Parameswari B, Kumar A, Nithya K, Shingote PR, Chhabra ML, Kumar S, Praveen Kumar P, Viswanathan R, Jain RK, Pathak AD (2020) Present status and future management strategies for sugarcane yellow leaf virus: a major constraint to the global sugarcane production. Plant Pathol J 36(6):536–557
Komor E (2011) Susceptibility of sugarcane, plantation weeds and grain cereals to infection by sugarcane yellow leaf virus and selection by sugarcane breeding in Hawaii. Eur J Plant Pathol 129:379–388
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Liang SS, Alabi OJ, Damaj MB, Fu WL, Gao SJ (2016) Genomic variability and molecular evolution of Asian isolates of sugarcane streak mosaic virus. Arch Virol 161:1493–1503
Lockhart BEL, Autrey LJC (2000) Mild mosaic. In: A Guide to Sugarcane Diseases; Rott, P., Bailey, R.A., Comstock, J.C., Croft, B.J., Eds.; La Librairie du Cirad: Montpellier, France, 2000: 245–248
Lu G, Wang Z, Xu F, Pan YB, Grisham MP, Xu L (2021) Sugarcane mosaic disease: characteristics, identification and control. Microorganisms 9: 1984. https://doi.org/10.3390/microorganisms9091984
Moonan F, Molina J, Mirkov TE (2000) Sugarcane yellow leaf virus: an emerging virus that has evolved by recombination between luteoviral and poleroviral ancestors. Virology 269:156–171
Nibouche S, Costet L, Holt JR, Jacobson A, Pekarcik A, Sadeyen J et al (2018) Invasion of sorghum in the Americas by a new sugarcane aphid (Melanaphis sacchari) superclone. PLoS ONE 2018; 13(4): e0196124 https://doi.org/10.1371/journal.pone.0196124
Nithya K, VishnuVardhan J, Balasaravanan S, Vishalakshi D, Kaverinathan K, Viswanathan R (2021) First report of Maize yellow mosaic virus (MaYMV) infecting sugarcane in India and its molecular characterization. Austras Plant Pathol 50:633–638. https://doi.org/10.1007/s13313-021-00809-w
Parameswari B,Bagyalakshmi K, Viswanathan R, Chinnaraja C (2013) Molecular characterization of Indian Sugarcane streak mosaic virus isolate Virus Genes 46: 186-189.
Parameswari B, Bagyalakshmi K, Chinnaraja C, Viswanathan R (2014) Molecular characterization of indian sugarcane streak mosaic virus isolates reveals recombination and negative selection in the P1 gene. Gene 552:199–203
Perera MF, Filippone MP, Noguera AS, Cuenya MI, Castagnaro AP (2012) An overview of the sugarcane mosaic disease in South America. In: Viswanathan R, Sundar AR (eds) Functional Plant Science and Biotechnology 6. Global Science Books, Japan, pp 98–107
Putra LK, Kristini A, Achadian EM, Damayanti TA (2014) Sugarcane streak mosaic virus in Indonesia: distribution, characterisation, yield losses and management approaches. Sugar Tech 16:392–399
Scagliusi SM, Lockhart BEL (2000) Transmission, characterization, and serology of a luteovirus associated with yellow leaf syndrome of sugarcane. Phytopathology 90:120–124
Schenck S, Lehrer AT (2000) Factors affecting the transmission and spread of sugarcane yellow leaf virus. Plant Dis 84:1085–1088. https://doi.org/10.1094/PDIS.2000.84.10.1085
Srinivas KP, Subba Reddy V, Ramesh B, Lava Kumar P, Sreenivasulu P (2010) Identification of a virus naturally infecting sorghum in India as Sugarcane streak mosaic virus. Eur J Plant Pathol 127:13–19
Singh BU, Padmaja PG, Seethara N (2004) Biology and management of the sugarcane aphid Melanaphis sacchari (Zehntner) (Homoptera: Aphididae), in sorghum: a review. Crop Protect 23:739–755
Singh M, Singh A, Upadhyaya PP, Rao GP (2005) Transmission studies on an indian isolate of sugarcane mosaic potyvirus. Sugar Tech 7:32–38
Sorho F, Sereme D, Kouame K, Kone N, Kone D, Yao K, Ouattara M, Tapsoba W, Ouattara B, Kone D (2021) First report of sugarcane streak mosaic virus (SCSMV) infecting sugarcane in Cote d’Ivoire. Plant Dis 105:519
Tang Lihua B, Wardatou S, Sushma H, Martha RP (2019) Detection of Sugarcane yellow leaf virus and Sugarcane mosaic virus in arthropods collected from corn, sorghum and sugarcane in Florida. American Society of Sugar Cane Technologists Annual Meeting (ASSCT 2019) (https://agritrop.cirad.fr/594381/)
Uchimiyam M, Knoll JE (2019) Rapid data analytics to relate sugarcane aphid (Melanaphis sacchari (Zehntner) population and damage on sorghum (Sorghum bicolor (L.) Moench). Sci Rep 9:370. https://doi.org/10.1038/s41598-018-36815-0
Viswanathan R (2016) Varietal degeneration in sugarcane and its management in India. Sugar Tech 18(1):1–7. https://doi.org/10.1007/s12355-015-0369-y
Viswanathan R (2018) Changing scenario of sugarcane diseases in India since introduction of hybrid cane varieties: path travelled for a century. Journal of Sugarcane Research 8(1):1–35
Viswanathan (2021) Impact of yellow leaf disease in sugarcane and its successful disease management to sustain crop production. Indian Phytopathol 74:573–586. https://doi.org/10.1007/s42360-021-00391-7
Viswanathan R, Balamuralikrishnan M, Karuppaiah R (2007) Sugarcane mosaic in India: a combined infection of sugarcane mosaic virus and sugarcane streak mosaic virus. Sugar Cane Intern 25(2):10–18
Viswanathan R, Balamuralikrishnan M, Karuppaiah R (2008) Duplex - reverse transcription - polymerase chain reaction (D-RT-PCR) - a technique for the simultaneous detection of viruses causing sugarcane mosaic. Sugar Tech 10(1):81–86. https://doi.org/10.1007/s12355-008-0014-0
Viswanathan R, Ganesh Kumar V, Karuppaiah R, Scindiya M, Chinnaraja C (2013) Development of duplex-immunocapture (duplex-IC) RT-PCR for the detection of sugarcane streak mosaic virus and sugarcane mosaic virus in sugarcane. Sugar Tech 15:399–405.https://doi.org/10.1007/s12355-013-0216-y
Viswanathan R, Chinnaraja C, Malathi P, Gomathi R, Rakkiyappan P, Neelamathi D, Ravichandran V (2014) Impact of sugarcane yellow leaf virus (ScYLV) infection on physiological efficiency and growth parameters in sugarcane in India. Acta Physiol Plant 36:1805–1822
Viswanathan R, Parameswari B, Nithya K (2018) Molecular characterization of sugarcane viruses and their diagnostics. In: Prasad R, Gill SS, Tuteja N (eds) Crop improvement through microbial biotechnology. Elsevier, Amsterdam, pp 175–193
Viswanathan R, Ramasubramanian T, Chinnaraja C, Selvakumar R, Lakshmi Pathy T, Manivannan K, Nithyanantham R (2022) Population dynamics of Melanaphis sacchari (Zehntner), the aphid vector of sugarcane yellow leaf virus under tropical conditions in India. Trop Plant Pathol 47:260–2779
Wang J, Roe B, Macmil S, Yu Q, Murray JE, Tang H, Chen C, Najar F, Wiley G, Bowers J et al (2010) Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genom 11:261
Wang XY, Li WF, Huang YK, Zhang RY, Shan HL, Yin J, Luo ZM (2017) Molecular detection and phylogenetic analysis of viruses causing mosaic symptoms in new sugarcane varieties in China. Eur J Plant Pathol 148:931–940
Wei C, Hincapie M, Larsen N, Nuessly G, Rott P (2016) First report of sugarcane yellow leaf virus infecting grain sorghum (Sorghum bicolor) in the United States. Plant Dis 8:1798
Wu L, Zu X, Wang S, Chen Y (2012) Sugarcane mosaic virus-long history but still a threat to industry. Crop Prot 42:74–78
Yang CR (1986) Transmission of sugarcane mosaic virus by three kinds of aphids. Chin J Entomol 6:43–49
Yang Z, Mirkov T (1997) Sequence and relationships of sugarcane mosaic and sorghum mosaic virus strains and development of RT-PCR based RFLPs for strain discrimination. Phytopathology 87:932–939
Zhou G, Li J, Xu D, Shen W, Deng H (2006) Occurrence of sugarcane yellow leaf virus in south China and its transmission by the sugarcane colonizing aphid, Ceratovacuna lanigera. Scientia Agricultura Sinica 39:2023–2027
Acknowledgements
Financial support received from the ICAR-CRP on the Vaccines and Diagnostics project is greatly acknowledged and authors are thankful to the Director, ICAR-Sugarcane Breeding Institute for providing the necessary facilities to conduct the research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standard
The present research did not involve human participants and/or animals.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Viswanathan, R., Nithya, K., Vishnuvardhan, J. et al. Sorghum (Sorghum bicolor) a new host to sugarcane yellow leaf and mosaic viruses in India. Indian Phytopathology 76, 867–877 (2023). https://doi.org/10.1007/s42360-023-00662-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-023-00662-5