Abstract
Pythium oligandrum is an important potential biological control agent of plant pathogens. The objective of this study was to evaluate the genetic diversity and population structure of 45 isolates from various provinces in Iran. Genetic diversity was characterized based on inter-simple sequence repeat (ISSR) markers. Cluster analysis of ISSR data categorized the isolates into three main clusters. A high level of variability was revealed within P. oligandrum isolates, however, no significant diversity among geographical regions was observed and isolates from different regions were grouped into the same cluster. Genetic diversity was moderate within populations (i.e. Bushehr, Fars and Kermanshah) with high average gene flow and low genetic distances between populations. The observed and effective numbers of alleles were higher in the Fars population than in other studied populations. This is the first report on the genetic diversity of P. oligandrum using ISSR markers. This study paves the way for the selection of isolates to be assessed for their antagonistic efficacy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pythium oligandrum Dreschler is a cosmopolitan oomycete and is one of the five mycoparasite species assigned to clade D of the Pythium genus (Levesque and De Cock 2004). It is considered as non-pathogenic (Kilpatrick 1968; Al-Hamdani and Cooke 1983; Martin and Hancock 1986) and has received considerable attention as a potential biocontrol agent. It is effective in control of damping-off and root diseases caused by several soil-borne fungal phytopathogens (McQuilken et al. 1990; Al-Rawahi and Hancock 1998; Hase et al. 2008; Horner et al. 2012; Yacoub et al. 2016). In protection of plants it acts both directly - through mycoparasitism, antibiosis, competition for nutrients and space, and indirectly - by inducing plant resistance (Gerbore et al. 2014).
Pythium oligandrum was isolated from the soil and rhizosphere of various plants in Iran, including amaryllis (Tehran Province), aptenia (Fars Province), barley (Fars Province), beetroot (Hamadan, Khorasan, Khuzestan, West Azerbaijan Provinces), cucumber, eggplant (Razavi Khorasan Province), lantana (Tehran Province), pine (Mazandaran Province), safflower (Kermanshah Province) and turfgrass (Fars and Tehran Provinces) (Bolboli and Mostowfizadeh-Ghalamfarsa 2015; Mostowfizadeh-Ghalamfarsa 2016). Such a wide geographical distribution would suggest the high variation. However, no information is available on genetic relationship among P. oligandrum isolates from different crops.
DNA molecular markers provide valuable tools to study fungal genetic diversity. Inter simple sequence repeat (ISSR)-PCR has been developed, tested and used in genetic studies of various organisms (Poyraz 2016), including fungal intraspecific genetic variation (Lindblom and Ekman 2005). This technique includes an amplifying DNA sequences between simple sequence repeats (SSRs) with anchored or non-anchored SSR homologous primers (Zietkiewicz et al. 1994). This method does not require genome sequences and generates specific and reproducible patterns due to the highly stringent conditions of the reaction (Bornet and Branchard 2001).
Additionally, the ISSR analysis has a low cost. This method may reveal higher polymorphism compared to the random amplified polymorphic DNA (RAPD) analysis due to the repeated regions in the genome (Es-selman et al. 1999). ISSR has been used to identify repeated motifs in the genome of Pythium group F isolates by Vasseur et al. (2005).
The objective of the present research is to find out the genetic diversity among P. oligandrum isolates from different crops cultivated in various regions of Iran. Pythium oligandrum genetic variation, the relationship among populations, geographical and nutritional preferences may determine the host preferences which can be useful in potential practical biological control.
Materials and methods
Sampling and Pythium isolation
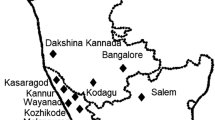
Soil samples were collected around roots of alfalfa (1 location in Kerman Province - southern Iran), barley (1 location in Fars Province - south-western Iran), chickpea (10 locations in Kermanshah, western Iran), eggplant (2 locations in Bushehr Province - south-western Iran), grapevine (1 location in Fars Province), lemon (1 location in Fars Province), lentil (8 locations Kermanshah Province - western Iran), maize (1 location), pine (1 location in Fars Province), pistachio (2 locations in Yazd Province - central Iran), potato (2 locations in Fars Province), rice (1 location in Golestan - north-eastern Iran), soybean (1 location in Golestan), tomato (8 locations in Bushehr, Fars, Ilam - western Iran), turfgrass (2 locations in West Azerbaijan - north-western Iran), walnut (1 location in Chaharmahal and Bakhtiari - central Iran), wheat (2 locations in Chaharmahal and Bakhtiari, and Golestan north-eastern Iran) in December 2013–August 2017 (Fig. 1, Table 1). Pythium spp. were isolated by baiting with 5-mm sour orange (Citrus aurantium L.) leaf disks at 20 °C for 24 h (Banihashemi et al. 1992) and plating on PARP-CMA (PARP: pimaricin 0.01 g, ampicillin 0.25 g, rifampin 0.01 g, pentachloronitrobenzene 0.1 g, CMA: cornmeal 20 g, peptone 20 g, glucose 20 g, agar 15 g, distilled water 1 L) (Jeffers and Martin 1986). After 36 h the single colonies were prepared with hyphal tip method on 1.5% water agar. Cultures were subcultured regularly on CMA and incubated at 25 °C in the dark.
Geographical locations of Pythium oligandrum isolates studied (Original map is from https://d-maps.com/carte.php?num_car=5494&lang=fr)
Identification of P. oligandrum isolates
Pythium oligandrum was identified on morphological characters (oogonia, sporangia, antheridia) and molecular data of the ITS rDNA (with primers P.OLIG.F1 (5’-CTGTGCTTCGTCGCAAGACT-3′ and P.OLIG.R.04 (5′- CTTTAAAAAGACAGCGCGAGA-3′) and specific genes encoding elicitin-like proteins (with primers OilgandrinF (5’-ATGTTCACCAAGACCTTGG-3′ and OligandrinR (5′- TTAAGCGGAGCCAACCACGG-3′) (Godfrey et al. 2003; Masunaka et al. 2010). Amplifications were performed in a 20-μL reaction volume (Vasseur et al. 2005), with an initial denaturation for 2 min at 95 °C, followed by 30 cycles of 40 s denaturation at 94 °C, 50 s of primer annealing at 60 °C for P.OLIG.F1/P.OLIG.R.04 and at 58 °C for OligandrinF/OligandrinR, 60 s extension at 72 °C, and a final extension at 72 °C for 10 min. The presence of PCR products was confirmed by gel electrophoresis.
Genetic diversity analysis with inter simple sequence repeat (ISSR)-PCR
The ISSR markers were amplified using seven primers (Table 2). The PCR amplification was carried out in 20-μL reaction mixture (Weiland et al. 2015) with an initial denaturation for 2 min at 95 °C, followed by 35 cycles of 1 min denaturation at 94 °C, 50 s of primer annealing at the specific temperature, 60 s extension at 72 °C, and a final extension at 72 °C for 10 min. Effectiveness of amplification was checked by electrophoresis on a 1.5% agarose gel.
Amplification products were scored for the presence (1) or absence (0) of bands, and a binary matrix was used to estimate the genetic similarities between pairs. The unweighted pair group method with arithmetic mean (UPGMA) clustering method was used to generate a dendrogram for P. oligandrum by computing the GS values with Jaccard coefficient in NTSYSpc 2.2 program (Rohlf 2000). The genetic diversity was analyzed in populations of 5,10 and 18 isolates, from Bushehr, Fars and Kermanshah Provinces, respectively. Genetic variation for each locus and population was analyzed with Nei’s gene diversity index (H), Shannon index (I), observed (Na) and effective (Ne) number of alleles, number of polymorphic loci (PL), percentage of polymorphic loci (PPL) and also Nei’s genetic distance (Yeh et al. 1999). Total genetic diversity (Ht), mean diversity within each population (Hs), Nei’s coefficient of genetic diversity (Gst) and gene flow (Nm) were calculated according to Nei (1986). Principal coordinate analysis (PCA) and analysis of molecular variance (AMOVA) was performed using GenAlEx 6.41 (Peakall and Smouse 2006).
Results
Collection of P. oligandrum
Forty-five P. oligandrum isolates were collected from soil around roots of alfalfa, barley, chickpea, eggplant, grapevine, lemon, lentil, maize, pine, pistachio, potato, rice, soybean, tomato, turfgrass, walnut and wheat in eight provinces in Iran (Fig. 1, Table 1). The isolates were characterized by ornamented oogonia with acute spines, contiguous sporangia with (sub-) globose elements connected by hypha and often absence of antheridia (Van der Plaäts-Niterink 1981). Identification was confirmed by PCR with specific primers.
Cluster analysis
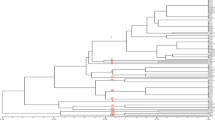
A total of 102 DNA fragments were amplified using ISSR primers. Three clusters were obtained with UPGMA (Fig. 2). The cluster 1 consisted of 11 isolates from lemon, lentil, pistachio, rice, soybean, tomato and wheat soils in Chaharmahal & Bakhtiari, Fars, Golestan and Kermanshah. The major cluster 2 consisted of 32 isolates from alfalfa, chickpea, grapevine, grass, lentil, maize, pine, potato, tomato and walnut soils in Chaharmahal & Bakhtiari, Fars, Kerman, Kermanshah, Uromia, Yazd. The cluster 3 included only two isolates from barley and tomato soils in Fars Province. All eighteen isolates from Kermanshah Province were in clusters 1 and 2 and all 10 isolates from Fars Province were in clusters 2 and 3. Genetic similarity between isolates from soils of chickpea and lentil was 100% and 96% respectively in Kermanshah Province. Genetic similarity between isolates from soil of eggplant in Bushehr Province, and soils of soybean and rice in Golestan Province was 90%.
Genetic diversity of populations
Nei’s gene diversity index (H), Shannon’s index (I), observed (Na) and effective (Ne) number of alleles, number of polymorphic loci (PL) and percentage of polymorphism (PPL) indicate the highest and the lowest polymorphs of P. oligandrum from Fars and Bushehr, respectively (Table 3). Nei’s pairwise genetic distances between the populations ranged from 0.037 to 0.077 (Table 4). The highest genetic distance was obtained between isolates from Bushehr and Fars, and the lowest genetic distance was obtained between isolates from Bushehr and Kermanshah. Nei’s gene diversity statistics, including total genetic diversity (Ht) and mean diversity within each population (Hs) indices were 0.31 and 0.27, respectively. Nei’s coefficient of genetic diversity (Gst) was 0.11, indicating that genetic variance among the populations was 11%. Gene flow (Nm) showing the average number of migrants among the population was 3.96. The PCA scatter plot based on the ISSR dataset showed vague borders between the three populations (Fig. 3). No clear separation between the different populations was highlighted. The first and the second principal coordinates accounted for 21.81% and 33.21% of variation, respectively. Variation among and within populations from three different provinces was 7% and 93%, respectively (Table 5).
Discussion
The genetic structure and diversity of P. oligandrum population from Iran was evaluated using ISSR markers. A moderate level of genetic diversity was found within three P. oligandrum populations. The level of genetic diversity of our P. oligandrum was: (i) similar to diversity in Pythium aphanidermatum (Edson) Fitzp. in Pennsylvania (USA) (Lee et al. 2010) and (ii) higher than diversity of P. aphanidermatum in Oman (Al-Sa'di et al. 2012).
Pythium oligandrum is a homothallic organism, and self-fertilization in sexual reproduction (Huzar-Novakowiski and Dorrance 2018) should reduce the genetic variation. It happened in homothallic Globisporangium irregulare (Buisman) Uzuhashi, Tojo & Kakish. (syn. Pythium irregulare Buisman) and Globisporangium ultimum (Trow) Uzuhashi, Tojo & Kakish. (syn. Pythium ultimum Trow) from forest nursery soils (Weiland et al. 2015). However, different oomycetes behave differently. Genetic diversity among populations of various oomycetes seems to be variable, depending on the level of inbreeding and out-crossing, location and climate. The dominating out-crossing in sexual reproduction of Globisporangium sylvaticum (W.A. Campb. & F.F. Hendrix) Uzuhashi, Tojo & Kakish. (syn. Pythium sylvaticum W.A. Campb. & F.F. Hendrix) and P. aphanidermatum contributed to the high diversity of the former (Weiland et al. 2015) but not of the latter (Al-Sa'di et al. 2012).
High and low genetic variability respectively within and among populations emphasize that the most of the variation exists within populations. Similar results were reported for another oomycete, e.g. Phytophthora capsici Leonian (Li et al. 2012). Many individuals within such populations are likely to be genetically different.
Grouping of the isolates in three major clusters was not associated with geographical or nutritional preferences. It partly agrees with Zhao et al. (2007), Al-Sa'di et al. (2008), Amini et al. (2014) and Bentes et al. (2018) who reported that there is not significant positive correlation between genetic variation and geographical or nutritional preferences of species, including P. aphanidermatum. We showed that the external factors, such as host, location and climate do not affect the genetic structure of P. oligandrum.
Gene flow is one of the evolutionary forces that may have a significant impact on the genetic diversity of a population (Nourollahi et al. 2011). It indicates the frequent migration and high genetic exchange (Weiland et al. 2015). The high level of gene flow among our populations indicates the lack of clear-cut boundaries. This is shown also by principal coordinate analysis (PCA), which showed no definite separation between individuals from different populations. The high level of gene flow has also been observed in G. irregulare and G. sylvaticum in forest nursery soils (Weiland et al. 2015). In the absence of gene flow, genetic drift causes different allele frequencies at neutral loci, leading to differentiation in isolated populations (Keller et al. 1997). Genetic diversity of population can be caused by migration or transfer (with soil or plants) of microorganisms’ genotypes. It is limited, but possible (Lee et al. 2010; Weiland et al. 2015; Huzar-Novakowiski and Dorrance 2018).
Conclusion
Study show the diversity and evolutionary potential of antagonistic P. oligandrum. The collection of P. oligandrum isolates representing the range of its genetic diversity in Iran could help to screen isolates for biocontrol agent.
References
Al-Hamdani AM, Cooke RC (1983) Effects of the mycoparasite Pythium oligandrum on cellulolysis and sclerotium production by Rhizoctonia solani. Trans Br Mycol Soc 81:619–621
Al-Rawahi AK, Hancock JG (1998) Parasitism and biological control of Verticillium dahliae by Pythium oligandrum. Plant Dis 82:1100–1106
Al-Sa'di AM, Drenth A, Deadman ML, Aitken EAB (2008) Genetic diversity, aggressiveness and metalaxyl sensitivity of Pythium aphanidermatum populations infecting cucumber in Oman. Plant Pathol 57:45–56
Al-Sa'di AM, Al-Ghaithi AG, Al-Balushi ZM, Al-Jabri AH (2012) Analysis of diversity in Pythium aphanidermatum populations from a single greenhouse reveals phenotypic and genotypic changes over 2006 to 2011. Plant Dis 96:852–858
Amini M, Safaie N, Saify Nabiabad H (2014) Assessment of genetic diversity in Pythium aphanidermatum isolates using ISSR and rep-PCR methods. Iran J Genet Plant Breed 3:41−52
Banihashemi Z, MacDonald JD, Stite J (1992) Combine baiting and ELISA to detect and quantify Phytophthora spp. in container media. Phytopathology 82:1101
Bentes JLDS, Sousa FMG, Lopes MTG, Valente MSF, Almeida FV, Demosthenes LR (2018) Genetic variability of Corynespora cassiicola isolates from Amazonas, Brazil. Arq Inst Biol 85
Bolboli Z, Mostowfizadeh-Ghalamfarsa R (2015) Phylogenetic relationships and taxonomic characteristics of Pythium spp. isolates in cereal fields of Fars Province. Iran J Plant Pathol 51:471–492
Bornet B, Branchard M (2001) Nonanchored inter sequence repeat (ISSR) markers: reproducible and specific tools for genome fingerprinting. Plant Mol Biol Report 19:09–215
Dinolfo MI, Stenglein SA, Moreno MV, Nicholson P, Jennings P, Salerno GL (2010) ISSR markers detect high genetic variation among Fusarium poae isolates from Argentina and England. Eur J Plant Pathol 127:483–491
Es-selman EJ, Jianqiang L, Crawford DJ, Windus JL, Wolfe AD (1999) Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Mol Ecol 8:443–451
Geleta M, Bryngelsson T (2009) Inter simple sequence repeat (ISSR) based analysis of genetic diversity of Lobelia rhynchopetalum (Campanulaceae). Hereditas 146:122−130
Gerbore J, Benhamou N, Vallance J, Le Floch G, Grizard D, Regnault-Roger C, Rey P (2014) Biological control of plant pathogens: advantages and limitations seen through the case study of Pythium oligandrum. Environ Sci Pollut Res Int 21:4847–4860
Godfrey SA, Monds RD, Marshall JW (2003) Identification of Pythium oligandrum using species-specific ITS rDNA PCR oligonucleotides. Mycol Res 107:790–796
Gupta M, Chyi YS, Romero-Severson J, Owen JL (1994) Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theor Appl Genet 89:998−1006
Hase S, Takahashi S, Takenaka S, Nakaho K, Arie T, Seo S, Ohashi Y, Takahashi H (2008) Involvement of jasmonic acid signalling in bacterial wilt disease resistance induced by biocontrol agent Pythium oligandrum in tomato. Plant Pathol 57:870–876
Horner NR, Grenville-Briggs LJ, Van West P (2012) The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation. Fungal Biol 116:24–41
Huzar-Novakowiski J, Dorrance AE (2018) Genetic diversity and population structure of Pythium irregulare from soybean and corn production fields in Ohio. Plant Dis 102:1989–2000
Jeffers SN, Martin SB (1986) Comparison of two media selective for Phytophthora and Pythium species. Plant Dis 70:1038–1043
Keller SM, McDermott JM, Pettway RE, Wolfe MS, McDonald BA (1997) Gene flow and sexual reproduction in the wheat glume blotch pathogen Phaeosphaeria nodorum (anamorph Stagonospora nodorum). Phytopathology 87:353–358
Kilpatrick RA (1968) Seedling reaction of barley, oats and wheat to Pythium species. Plant Dis Rep 52:209–212
Lee S, Garzón CD, Moorman GW (2010) Genetic structure and distribution of Pythium aphanidermatum populations in Pennsylvania greenhouses based on analysis of AFLP and SSR markers. Mycologia 102:774−784
Levesque CA, De Cock AW (2004) Molecular phylogeny and taxonomy of the genus Pythium. Mycol Res 108:1363–1383
Li P, Cao S, Dai YL, Li XL, Xu DF, Guo M, Pan YM, Gao ZM (2012) Genetic diversity of Phytophthora capsici (Pythiaceae) isolates in Anhui Province of China based on ISSR-PCR markers. Genet Mol Res 11:4285–4296
Lindblom L, Ekman S (2005) Molecular evidence supports the distinction between Xanthoria parietina and X. aureola (Teloschistaceae, lichenized Ascomycota). Mycol Res 109:187–199
Martin FN, Hancock JG (1986) Association of chemical and biological factors in soils suppressive to Pythium ultimum. Phytopathology 76:1221–1231
Masunaka A, Sekiguchi H, Takahashi H, Takenaka S (2010) Distribution and expression of elicitin-like protein genes of the biocontrol agent Pythium oligandrum. J Phytopathol 158:417–426
McQuilken MP, Whipps JM, Cooke RC (1990) Control of damping‐off in cress and sugar‐beet by commercial seed‐coating with Pythium oligandrum. Plant Pathol 39:452−462
Mostowfizadeh-Ghalamfarsa R (2016) Pythium species in Iran. Shiraz University Press, Shiraz
Nei M (1986) Definition and estimation of fixation indices. Evolution 40:643–645
Nourollahi K, Javannikkhah M, Naghavi M, Lichtenzveig J, Okhovat SM, Oliver RP, Ellwood SR (2011) Genetic diversity and population structure of Ascochyta rabiei from the western Iranian Ilam and Kermanshah provinces using MAT and SSR markers. Mycol Prog 10:1–7
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Poyraz I (2016) Comparison of ITS, RAPD and ISSR from DNA-based genetic diversity techniques. C R Biol 339:171–178
Rohlf FJ (2000) NTSYSpc: numerical taxonomy and multivariate system. Version 2.1 Exeter software. Applied Biostatics Inc., New York
Sharma M, Gupta SK, Sharma TR (2005) Characterization of variability in Rhizoctonia solani by using morphological and molecular markers. J Phytopathol 153:449−456
Van der Plaäts-Niterink AJ (1981) Monograph of the genus Pythium. Studies in Mycology. Vol. 21. Netherlands: Centraalbureau voor Schimmelcultures
Vasseur V, Rey P, Bellanger E, Brygoo Y, Tirilly Y (2005) Molecular characterization of Pythium group F isolates by ribosomal-and intermicrosatellite-DNA regions analysis. Eur J Plant Pathol 112:301–310
Weiland JE, Garrido P, Kamvar ZN, Espíndola AS, Marek SM, Grünwald NJ, Garzón CD (2015) Population structure of Pythium irregular, P. ultimum, and P. sylvaticum in forest nursery soils of Oregon and Washington. Phytopathology 105:684–694
Yacoub A, Gerbore J, Magnin N, Chambon P, Dufour MC, Corio-Costet MF, Guyoneaud R, Rey P (2016) Ability of Pythium oligandrum strains to protect Vitis vinifera L., by inducing plant resistance against Phaeomoniella chlamydospora, a pathogen involved in Esca, a grapevine trunk disease. Biol Control 92:7–16
Yeh FC, Yang RC, Boyle T (1999) POPGENE version 1.32: Microsoft window-based freeware for population genetics analysis. University of Alberta, Edmonton
Zhao W, Wang Y, Chen T, Jia G, Wang X, Qi J, Pang Y, Wang S, Huang H, Pan Y, Yang Y (2007) Genetic structure of mulberry from different ecotypes revealed by ISSRs in China: an implications for conservation of local mulberry varieties. Sci Hortic 115:47–55
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgements
This study was funded by the Iran National Science Foundation (INSF, award number 96008191).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Haghi, Z., Mostowfizadeh-Ghalamfarsa, R. & Edel-Hermann, V. Genetic diversity of Pythium oligandrum in Iran. J Plant Pathol 102, 1197–1204 (2020). https://doi.org/10.1007/s42161-020-00613-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-020-00613-3