Abstract
A non-white rot fungus, Penicillium simplicissimum (isolate 10), was investigated for its biodegradation activities towards toxic triphenylmethane (TPM) dyes such as Crystal Violet (CV), Methyl Violet (MV), Malachite Green (MG), and Cotton Blue (CB). High decolorization efficiencies of 98.7, 97.5, 97.1, and 96.1% were observed for CV, MV, MG, and CB, respectively, within 2 h of incubation in the dye solutions (50 mg L−1, pH 5.0, 25 ± 2 °C). UV–visible spectral analysis of dyes conducted before and after treatment with P. simplicissimum indicated the occurrence of biodegradation. This was confirmed when enzymatic analyzes revealed induced production of manganese peroxidase (MnP, EC 1.11.1.13; 23.31 U mL−1), tyrosinase (EC 1.14.18.1; 16.18 U mL−1), and triphenylmethane reductase activities (1.15 U mL−1), particularly in biodegrading MG. For MV and CB, increased activities of tyrosinase (20.35 and 18.74 U mL−1, respectively) were detected, whereas no enzyme activities were detected for CV. Dye biodegradation by P. simplicissimum led to reduced toxicity (particularly for MG) based on phytotoxicity and microbial toxicity assays. It was concluded that P. simplicissimum showed potential in biodegrading and detoxifying TPM dyes via MnP, tyrosinase and triphenylmethane reductase activities, resulting in less harmful treated dye solutions.
Graphical abstract

Article Highlights
-
P. simplicissimum biodegraded TPM dyes via enzymatic activities
-
Rapid and efficient decolourization (96.1–98.7%) were achieved within 2 h
-
MnP, tyrosinase, and triphenylmethane reductase biodegrades TPM dyes
-
Dye biodegradation reduced dye toxicity (particularly for MG)
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Triphenylmethane (TPM) dyes such as Crystal Violet (CV), Methyl Violet (MV), Malachite Green (MG), and Cotton Blue (CB) account for one of the largest and most commonly used synthetic colorants for dyeing of textiles, leather, paper, pharmaceuticals, and food (Chaudhry et al. 2014; Jegan et al. 2016). Industrial effluents often contain high concentrations of dyes due to inefficient dyeing and finishing processes. The release of untreated dyestuffs into water bodies can reduce sunlight penetration and consequently, interfere with photosynthetic activities and dissolved oxygen levels (Shedbalkar et al. 2008). TPM dyes are also recalcitrant molecules capable of exerting mutagenic and carcinogenic effects on living organisms (Mani and Bharagava 2016; Vyavahare et al. 2018; Wu et al. 2013). Therefore, effective treatment of wastewater is crucial in preventing environmental pollution.
Wastewater containing TPM dyes are conventionally treated via physico-chemical methods, such as membrane filtration, adsorption, photodegradation, chemical precipitation, and reverse osmosis (Rodrigues et al. 2013; Vergili et al. 2012). However, costly procedures, high chemical demands, and generation of toxic sludge often limit these approaches. An environment-friendly and cheaper alternative is the biological approach, which utilizes plants (phytoremediators) and microorganisms such as fungi, yeast, algae, and bacteria (Casas et al. 2009; Chaturvedi and Verma 2015; Fu et al. 2013; Jegan et al. 2016). Fungi are versatile natural decomposers with degradative capabilities for a diverse range of pollutants, such as synthetic dyes, polycyclic aromatic hydrocarbons, and pesticides (Agrawal et al. 2018; Chen and Ting 2015b; Szewczyk et al. 2018). For dyes, effective decolorization has been reported for fungal species of the genera Phanerochaete, Trametes, Fusarium, Aspergillus and Penicillium (Ali et al. 2016; Casas et al. 2009; Chen and Ting 2015b; Radha et al. 2005). Their removal capabilities are attributed to biosorption and biodegradation mechanisms.
In biosorption, dye molecules are mainly bound onto the cell wall of live and dead cells for sequestration (Chaudhry et al. 2014). This process is aided by the presence of functional groups with excellent binding capacities and the large cell-to-surface ratio of fungal biomass. Biodegradation of dyes, which occurs only in live cells, is attributed to the secretion of extracellular and intracellular ligninolytic enzymes such as laccase, lignin peroxidase, manganese peroxidase, tyrosinase, and triphenylmethane reductase (Barapatre et al. 2017; Jasińska et al. 2012; Shedbalkar et al. 2008). These enzymes have broad substrate specificity and generate free radicals to mediate dye decolorization via reactions like demethylation, hydroxylation, and ring cleavage (Barapatre et al. 2017). Dye decolorization through biodegradation is more desirable than biosorption as complete mineralization may occur to yield less hazardous products (Shedbalkar and Jadhav 2011). For biosorption, dye molecules are entrapped within the biomass matrix in their toxic parent forms, which pose additional waste disposal problems.
This study evaluates the use of Penicillium simplicissimum as a biological agent to adsorb and biodegrade toxic TPM dyes. This is a departure from the standard approaches of using white rot fungi, to realize the potential of non-white rot fungi as dye degraders. Little is known about Penicillium simplicissimum in regards to dye removal (Bergsten-Torralba et al. 2009; Chen and Ting 2015b). The scarce literatures available on this specific species, were studies by Bergsten-Torralba et al. (2009) on the decolorization potential of P. simplicissimum INCQS 40,211 towards azo dyes Reactive Blue 21, Reactive Red 198, and Reactive Blue 214. The other study is by Chen and Ting (2015b) who have documented the initial decolorization potential of P. simplicissimum (isolate 10) attributed to lignin peroxidase and NADH–DCIP reductase towards TPM dyes (CV, MV, MG, and CB). Hence, this study is expected to further illustrate the biodegradation activities of P. simplicissimum in TPM dyes, using other key enzymes such as manganese peroxidase, tyrosinase, and triphenylmethane reductase. The impact and efficacy of the enzymatic biodegradation is further evaluated via phytotoxicity and microbial toxicity assays. This will bridge the knowledge gap on the role of non-white fungi as dye degraders, the range of enzymes involved and their efficacy in reducing toxicity of TPM dyes. This study also determines the feasibility of using P. simplicissimum as a bioagent to reduce toxicity level of wastewaters and to ensure that the treated wastewaters are environmentally safe for discharge or re-use.
Materials and Methods
Culture Establishment
Penicillium simplicissimum (isolate 10) was isolated by Ting et al. (2011) from the metal-rich wastewater of Atomic Absorption Spectrometer (AAS) located at Monash University Malaysia. The isolate was maintained on Potato Dextrose Agar (PDA, Merck) at 25 ± 2 °C and sub-cultured periodically. Mycelial plugs from 7-day old cultures (5 mm diameter) were used to inoculate 100 mL of Potato Dextrose Broth (PDB, Merck) to yield fungal biomass after 5 days of standing incubation at 25 ± 2 °C. The biomass was washed with sterile distilled water, homogenized and filtered using Miracloth (Calbiochem) according to the procedure described by Chen and Ting (2015b) to obtain “live cells”.
Dye Decolorization Tests
TPM dyes Crystal Violet (CV) and Malachite Green (MG) were purchased from Merck, while Methyl Violet (MV) and Cotton Blue (CB) were from Sigma Aldrich. The respective dye powders were dissolved in sterile distilled water to achieve an initial dye concentration of 50 mg L−1 [concentration commonly observed in industrial effluents (Lima et al. 2017)]. The solutions were then adjusted to pH 5 ± 0.2 using diluted HCl and NaOH (concentration in M or N).
The dye removal experiment was initiated by inoculating 100 mL of each dye solution (50 mg L−1) with 4.0 ± 0.1 g (for CV, MV and MG solutions) and 8.0 ± 0.1 g (for CB solution) wet weight of the biomass (homogenized mycelia). The amount of biomass used here were the optimum biomass determined for the removal of these dyes as established in the earlier study (Chen and Ting 2015b). All mixtures were incubated with agitation at 150 rpm (25 ± 2°C). Similar incubation conditions were used for non-inoculated dye solutions (negative controls). Aliquots of the samples were collected at every 2-h interval for the first 8 h, then at 24 h. The samples were centrifuged at 3500 rpm for 15 min. The cell-free supernatants were then evaluated for the color of dye left via absorbance analysis. The Tecan Infinite M200 plate reader was used to measure the absorbance of MV, CV, CB and MG solutions at their respective λmax at 584, 590, 599, and 617 nm (Chen and Ting 2015b). The decolorization efficiency was calculated as follow (El-Batal et al. 2015):
where Ao initial absorbance of dye (untreated); Af final absorbance of dye (treated).
Ultraviolet–Visible (UV–Vis) Spectral Analysis
UV-Vis spectral analysis was performed to investigate the occurrence of dye biodegradation by P. simplicissimum. The absorbance of cell-free supernatants collected from the dye decolorization tests were analyzed at wavelengths ranging from 300 to 800 nm. Absorption spectra were then plotted and compared for spectra peaks generated from dye solutions before and after treatment with P. simplicissimum.
Enzymatic Assays as Indicators of Dye Biodegradation
The key enzymes responsible for dye-degrading activities, such as manganese peroxidase (EC 1.11.1.13), triphenylmethane reductase, and tyrosinase (EC 1.14.18.1), were assayed to establish biodegradation activities of the TPM dyes. The activity of manganese peroxidase was assayed by monitoring the oxidation of MnSO4 at 238 nm (molar extinction coefficient, ε238 = 6500 M−1 cm−1) in a reaction mixture containing 1mM MnSO4, 50 mM sodium malonate buffer (pH 4.5), and 0.1 mM H2O2 (Asgher et al. 2016). Tyrosinase activity was detected at 420 nm by monitoring the formation of catechol quinone in a reaction mixture (2.0 mL) consisting of 0.01 M pyrocatechol in 50 mM potassium phosphate buffer (pH 7.4) (Bora et al. 2004). Triphenylmethane reductase activity was detected at the respective maximum wavelengths of the TPM dyes (CV at 590 nm, MV at 584 nm, MG at 617 nm, CB at 599 nm). The reaction mixture (2.0 mL) contained 20 µM dye, 20 mM sodium phosphate buffer (pH 7.0), and 0.1 mM β-nicotinamide-adenine dinucleotide, reduced (NADH) (Jang et al. 2005). Reaction mixtures without enzyme solutions (supernatant) served as reference blanks. One enzyme unit was defined as the amount of enzyme required catalyzing the conversion of 1 micromole of substrate per min at 25°C. Enzyme activity was calculated using the following formula (Abd El Monssef et al. 2016):
where \(\Delta A/{\mathrm{m}\mathrm{i}\mathrm{n}}_{\mathrm{s}}\) is change in absorbance per min for sample; \(\Delta A/{\mathrm{m}\mathrm{i}\mathrm{n}}_{\mathrm{b}}\) is change in absorbance per min for blank; V is total volume of the reaction mixture (ml); \(\varepsilon\) is molar extinction coefficient (mM−1 cm−1); d is pathlength (cm); and v is volume of enzyme solution (mL).
Phytotoxicity and Microbial Toxicity Assays
The toxicities of TPM dyes before and after treatment with P. simplicissimum were evaluated by monitoring their effects on the growth of Vigna radiata and the bacteria Bacillus cereus, Staphylococcus aureus, Pseudomonas aeruginosa, and Escherichia coli. The seeds of V. radiata were surface sterilized with 10% v/v hydrogen peroxide for 10 min and washed generously with sterile distilled water (Ahmad et al. 2008). A total of 10 seeds were then placed on a Petri dish layered with two pieces of sterile Whatman No. 1 filter paper at the bottom of the Petri dish (Pandey et al. 2016). The seeds were immersed with either 7 mL of sterile distilled water (control), fungal-treated dye solutions, or untreated dye solutions on a daily basis (from dye decolorization tests; sterile filtered with 0.45 µm membrane filter). The length of roots and shoots were recorded after 7 days (25 ± 2°C). The germination rate was also calculated as in Eq. 2 (Vyavahare et al. 2018):
Microbial toxicity assays were conducted using the agar diffusion method (Chaturvedi and Verma 2015). Sterile filter paper disks containing 20 µL of fungal-treated TPM dye solutions were loaded on nutrient agar containing bacterial lawn. After incubation for 24 h at 37°C, the clear zone of inhibition around the disks were measured (diameter in mm). Both assays were also performed on untreated TPM dyes, sterile distilled water (negative control), and 10 µg/µL antibiotic Tetracycline (positive control).
Statistical Analysis
All experiments were carried out in triplicates. The data were analyzed by One-Way Analysis of Variance (ANOVA) using the Statistical Package for the Social Sciences (SPSS) software (version 20.0). Means were compared using the Tukey’s Honestly Significant Difference test (HSD, P < 0.05), or independent t-test analysis (P < 0.05) where relevant.
Results
Dye Decolorization Activities of P. simplicissimum
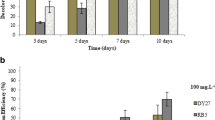
P. simplicissimum decolorized the TPM dyes (CV, MV, MG, and CB) with high decolorization efficiencies (between 93.0 and 95.8%) within 1 day of incubation under optimum conditions (as established in the previous study by Chen and Ting (2015b)) (Fig. 1). Fungal cells decolorized MV, MG, and CV at similar efficiencies of 95.8, 95.80, and 94.4% respectively, while 93.0% of CB were removed (Fig. 1). The removal of TPM dyes by P. simplicissimum in this study occurred rapidly, with relatively high decolorization efficiency observed even within the first 1 h (91.2% for MV, 86.4% for MG, 72.7 for CV, and 65.4% for CB). Maximum decolorization efficiencies were achieved within 2 h for CV (98.7%), CB (97.5%), and MG (97.1%), and 8 h for MV (97.7%) (Fig. 1). Nevertheless, 2 h was sufficient to decolorize MV at 96.1%.
Decolorization efficiency of live cells of P. simplicissimum for 50 mg L−1 of TPM dyes (
 ) CV (
) CV (
 ) MV (
) MV (
 ) MG, and (
) MG, and (
 ) CB over 24 h at 25 ± 2 °C. Means with the same letters within dye–group are not significantly different at Honestly Significant Difference (HSD(0.05)). Bars represent standard error of mean (± SEM)
) CB over 24 h at 25 ± 2 °C. Means with the same letters within dye–group are not significantly different at Honestly Significant Difference (HSD(0.05)). Bars represent standard error of mean (± SEM)
UV–Vis Spectral Analysis
The major absorption peak present at 580 nm for untreated CV and 580–590 nm for untreated MV disappeared completely after application with P. simplicissimum (Fig. 2a, b). Similar observations were recorded for the absorption peaks of CB at 600 and 310–320 nm (Fig. 2c). For MG, the major peak at 610–620 nm decreased in intensity such that it nearly disappeared after fungal treatment (Fig. 2d). On the contrary, complete disappearance of minor peaks at 310–320 and 420–430 nm were observed. UV–Vis spectral analysis was thus able to establish that biodegradation occurred as a mechanism of dye removal by P. simplicissimum. This was evident from the type of changes to the absorption spectra before and after treatment with fungal cells after 1 day. According to Ayed et al. (2008), a proportional decrease in peak intensity was indicative of the occurrence of biosorption, whereas complete disappearance of major absorption peak or emergence of a new peak suggested biodegradation as chromophoric groups were broken-down. Nevertheless, the involvement of biosorption in dye decolorization, especially for CV, MV and MG, was implied with fungal cells retaining the bright colors of the dyes, as described by Casas et al. (2009). This corresponded with the high removal rates of P. simplicissimum (65.4–91.2%) for TPM dyes within the first hour (Fig. 1). The return of the original greenish-white color of fungal cells after CB treatment suggested that decolorization occurred mainly by biodegradation.
Enzyme Activities as Indicators of Biodegradation
Enzyme assays revealed that the activities of manganese peroxidase, triphenylmethane reductase, and tyrosinase were generally induced in cultures supplemented with dyes as compared to control (without dye) (Table 1). The range of manganese peroxidase, triphenylmethane reductase, and tyrosinase were 4.68–23.31, 1.15–3.85, and 16.18–20.35 U mL−1, respectively compared to 0.019, 0.003, and 0.003 U mL−1 from control (Table 1). All three enzymes showed enhanced activities in response to MG, whereas only tyrosinase activities were higher when inoculated into MV and CB (Table 1). This suggested that dye degradation by P. simplicissimum involved oxidative and reductive reactions catalyzed by enzymes (Jang et al. 2005; Radha et al. 2005). On the contrary, the levels of all three enzymes were not significantly enhanced when exposed to CV. In CV, levels of manganese peroxidase, triphenylmethane reductase, and tyrosinase were 14.37, 2.06, and 9.65 U mL−1, compared to 0.019, 0.003, and 0.003 U mL−1 from control (Table 1). The large standard error for some of the enzyme activities in Table 1 could be due to variation between biomass in their expression of enzymes in response to TPM dyes. A possible solution would be to increase the number of fungal biomass tested.
Phytotoxicity Tests
Both phytotoxicity and microbial toxicity assays revealed that biodegradation of toxic TPM dyes by P. simplicissimum may yield degradation products with similar, higher or lower toxicities. The toxicity of untreated solutions of 50 mg L−1 CV, MV, and MG towards Vigna radiata was evident with shorter roots observed after dye exposure (Fig. 3). In most cases, shoot growth was less affected than root growth, suggesting that roots might have higher sensitivity towards dye toxicity compared to shoots. Among the four TPM dyes tested, CB (at 50 mg L−1) was considered as less toxic than others as untreated CB solutions resulted in seedlings with root length similar to those in control (without dye application), and the shoots were slightly longer (Fig. 3). The dyes responded differently to biodegradation by P. simplicissimum. MG was most effectively degraded by P. simplicissimum and the resulting treated dye was reduced in toxicity, evident with longer root lengths of Vigna radiata seedlings watered with the treated dye (Fig. 3). For treated CV and MV, toxicity of the degradation products was similar as for the parent dye compounds (untreated dyes) since root elongation of Vigna radiata was still inhibited (Fig. 3). The most significant observation was that treated CB was even more toxic than untreated CB, as the growth of both roots and shoots of Vigna radiata was inhibited (Fig. 3). Despite the reduction in shoot or root length, the germination rate of V. radiata applied with treated or untreated dyes solutions were not affected (100% germination), except for treated MG solutions (97% germination). It is concluded that the possible by-products from the biodegradation activities may have yielded components that are possibly detrimental to growth but not to the germination of seeds.
Microbial Toxicity Assays
Untreated CV, MV, and MG inhibited the growth of Bacillus cereus and Staphylococcus aureus as formation of large inhibition zones were observed (8.17–10.25 and 16.08–21.25 mm, respectively) (Table 2; Fig. 4). The application of treated solutions of the same three dyes did not inhibit growth. The results suggested possible detoxification of CV, MV, and MG. The validity of the microbial toxicity assay was confirmed with non-inhibited growth in response to sterile distilled water (negative control) and occurrence of inhibition zone for broad-spectrum antibiotic tetracycline (positive control; 22.08–31.33 mm) (Table 2). Nevertheless, the phytotoxicity test showed toxicity to the tested seeds, with only lowered toxicity for treated MG. This suggested that microbial toxicity assays might be less sensitive than phytotoxicity tests. On the contrary, the growth of test pathogens Pseudomonas aeruginosa and Escherichia coli was not inhibited by both untreated and treated TPM dyes (Fig. 4).
Discussion
In this study, TPM dyes CV, MV, MG and CB were effectively decolorized by cells of P. simplicissimum. Successful decolorization of these TPM dyes are similar to other species of Penicillium such as P. ochrochloron (Shedbalkar et al. 2008; Shedbalkar and Jadhav 2011), and P. pinophilum (Jasińska et al. 2012), revealing the nature of Penicillium species as dye degraders. P. simplicissimum is now part of the fungal species cluster capable of degrading dye molecules, which include Aspergillus niger (Ali et al. 2016), Mucor mucedo (Moturi and Singara Charya 2009), Coriolopsis sp. (Chen and Ting 2015a), Phanerochaete chrysosporium (Radha et al. 2005), and Trametes versicolor (Casas et al. 2009). This clearly indicated that non-white rot fungi also have the potential to decolorize recalcitrant dyes just as white rot fungi (i.e., Coriolopsis sp., Phanerochaete sp., Trametes sp.). The rapid decolorization rate of TPM dyes by P. simplicissimum is similar to the decolorization capacities by other Penicillium species. According to Shedbalkar et al. (2008) and Shedbalkar and Jadhav (2011), P. ochrochloron decolorized 93% of 50 mg L−1 CB and MG within 2.5 and 14 h, respectively. P. pinophilum was discovered to remove 87.1% of 10 mg L−1 MG within 48 h, whereas P. janthinellum decolorized 150 mg L−1 CV at 56.9% after 24 h of incubation (Jasińska et al. 2012; Wang et al. 2015). Moturi and Singara Charya (2009) reported that Mucor mucedo removed 0.02% CV (78%) and MG (65%) within 15 days. Aspergillus niger took 10 days to decolorize 80.9% of 10 mg L−1 CV, while complete decolorization of 20 mg L−1 MV by Aspergillus sp. occurred within 1 day (Ali et al. 2016; Kumar et al. 2011). Coriolopsis sp. removed 97, 94, 91, and 52% of MV, CV, CB, and MG, respectively, within 7, 7, 1, and 9 days (Chen and Ting 2015a). The different dye removal rates in these studies compared to present study may be attributed to the type of fungal species used, chemical structures of the dyes, and different experiment parameters (e.g., pH, temperature, agitation speed, nutrient availability, and concentration of fungal cells and dyes) (Jasińska et al. 2012). In this study, homogenization of mycelia during the biomass preparation stage could have enhanced the decolorization efficiency of P. simplicissimum by increasing the surface area for dye adsorption (Jin et al. 2015).
The decolorization of TPM dyes by P. simplicissimum presumably involved biosorption and biodegradation. Biosorption was deduced based on the high decolorization efficiencies observed at the start of the test (within the first 1 h) (Fig. 1) and dye-colored biomass at the end (especially for CV, MV and MG). The initial rapid removal could be attributed to the availability of sorption sites on the biomass surface for binding with dye molecules (Banerjee et al. 2017). According to Bouras et al. (2017), biosorption of dyes on the cell surface of fungi involved functional groups such as carboxyl, sulphonate, amide and amine groups that were determined via Fourier Transform InfraRed (FTIR) analysis on Penicillium glabrum biomass exposed to Congo Red. The profiling of key enzymes in P. simplicissimum is not known, thus this study revealed the key enzymes responsible for dye biodegradation.
It was discovered that the removal of TPM dyes (except for CV) by P. simplicissimum involved the enzymes tyrosinase, manganese peroxidase and triphenylmethane reductase (Table 1). According to Yang et al. (2016), degradation of MG by crude manganese peroxidase of Irpex lacteus involved N-demethylation and oxidative cleavage of C–C double bond. The degradative mechanisms of tyrosinase for TPM dyes remains to be elucidated. Nevertheless, tyrosinase of Brevibacterium sp. was found to cleave the azo and sulfonate bonds of Reactive Red 198 (azo dye) (Franciscon et al. 2012). Triphenylmethane reductase of Citrobacter sp. is known to catalyze the degradation of TPM dyes via NADH-dependent reduction (Jang et al. 2005). MG reductase of P. pinophilum mediated the decolorization of MG to yield leucomalachite green, N-demethyl-leucomalachite green and N-demethyl-malachite green (Jasińska et al. 2012). The lower specificity of triphenylmethane reductase for CV than for MG observed in this study has been attributed to the higher number of dimethylamino group present in the CV structure (Kim et al. 2008). Nevertheless, CV still has a relatively high decolorization, and this may be attributed to other enzymes produced by P. simplicissimum such as NADH–DCIP reductase (Chen and Ting 2015b). This enzyme has also been observed to render similar biodegradation activities towards MV, MG, and CB (Chen and Ting 2015b). Chen and Ting (2015b) also discovered the contributions of lignin peroxidase in MV and CB removal. Hence, all these results suggested that effective decolorization of TPM dyes by P. simplicissimum may require the contributions of several enzymes. The decolorization of CV by Trichoderma asperellum was mediated by laccase, while CB removal by P. ochrochloron involved lignin peroxidase, tyrosinase, and aminopyrine N-demethylase (Shanmugam et al. 2017; Shedbalkar et al. 2008). Mucor mucedo decolorized CV and MG with the aid of lignin peroxidase and triphenylmethane reductase (Moturi and Singara Charya 2009). Future studies on P. simplicissimum could investigate the decolorization efficiency of purified enzymes for TPM dyes and the influence of heavy metals on dye removal as industrial effluents often contain both hazardous substances.
Phytotoxicity tests revealed that the roots of Vigna radiata had stunted growth when exposed to untreated TPM dyes (particularly CV, MV and MG), while the shoots were unaffected. Similar observations have been reported for V. radiata, Brassica chinensis Tsen & Lee, and Arabidopsis thaliana in other TPM dye studies (Chaturvedi and Verma 2015; Fu et al. 2013; Matpang et al. 2017). According to Jayanthy et al. (2014), toxic untreated dyes may impede the growth of V. radiata roots and leaves by reducing protein and carbohydrate contents. For CB, the non-toxic nature of 50 mg L−1 untreated dye towards V. radiata in this study could be attributed to more prominent toxicity occurring at higher concentration (700 mg L−1), as reported for Triticum aestivum and Ervum lens Linn (Shedbalkar et al. 2008). Of the four TPM dyes treated with P. simplicissimum, only the degradation products of MG showed lower toxicities than the parent compound (Fig. 3). The similar or higher toxicities of treated CV, MV, and CB than for untreated dyes could be due to partially degraded products or the formation of toxic metabolites during the biodegradation process. Similar observations were recorded for the degradation of MG by Aspergillus flavus (Barapatre et al. 2017), and Procion Red MX-5B (azo dye) decolorization by A. terreus (Almeida and Corso 2014). This suggests that a longer incubation period with fungal biomass may be required to reduce dye toxicity.
In this study, the microorganisms used in toxicity assays were deduced to be tolerant towards TPM dyes as degraded dye products that were toxic towards V. radiata (such as CV and MV) (Fig. 3) did not inhibit microbial growth (Table 2). Bacteria, especially of the Gram negative variety, have been known to tolerate dye toxicity with the aid of the outer membrane. This layer of the cell serves as a permeability barrier to impair entry of dye molecules into the cells, as well as act synergistically with efflux pumps to actively transport these molecules out of the cytoplasm (Stancu and Grifoll 2011). The growth of test pathogens Pseudomonas aeruginosa and Escherichia coli was not inhibited by both untreated and treated TPM dyes (Fig. 4), which suggested possible dye tolerance in these strains (Kalyani et al. 2012).
Conclusion
This study demonstrated the rapid decolorization of 50 mg L−1 TPM dyes Crystal Violet, Methyl Violet, Malachite Green, and Cotton Blue by cells of P. simplicissimum. Biosorption of dyes occurred at the initial stage of removal as high decolorization efficiencies were recorded within 2 h. The absence of major absorption peaks of the dyes upon treatment with P. simplicissimum indicated the involvement of biodegradation in dye removal as well. Extracellular enzymes such as manganese peroxidase, tyrosinase, and triphenylmethane reductase, secreted by P. simplicissimum, mediated this process. This reveals that P. simplicissimum also produces key enzymes that allow for degradation of dye molecules. The lower toxicity of dye degradation products (especially for treated Malachite Green) towards plant seeds and microorganisms suggested detoxification capabilities of P. simplicissimum. Hence, the fungal isolate is a potential candidate for the bioremediation of TPM dyes in wastewater.
References
Abd El Monssef RA, Hassan EA, Ramadan EM (2016) Production of laccase enzyme for their potential application to decolorize fungal pigments on aging paper and parchment. Ann Agric Sci 61:145–154. https://doi.org/10.1016/j.aoas.2015.11.007
Agrawal N, Verma P, Shahi SK (2018) Degradation of polycyclic aromatic hydrocarbons (phenanthrene and pyrene) by the ligninolytic fungi Ganoderma lucidum isolated from the hardwood stump. Bioresour Bioprocess 5:11. https://doi.org/10.1186/s40643-018-0197-5
Ahmad MSA, Hussain M, Ijaz S, Alvi AK (2008) Photosynthetic performance of two mung bean (Vigna radiata) cultivars under lead and copper stress. Int J Agri Biol 10:167–172
Ali HM, Shehata SF, Ramadan KMA (2016) Microbial decolorization and degradation of crystal violet dye by Aspergillus niger. Int J Environ Sci Technol 13:2917–2926. https://doi.org/10.1007/s13762-016-1117-x
Almeida EJ, Corso CR (2014) Comparative study of toxicity of azo dye Procion Red MX-5B following biosorption and biodegradation treatments with the fungi Aspergillus niger and Aspergillus terreus. Chemosphere 112:317–322. https://doi.org/10.1016/j.chemosphere.2014.04.060
Asgher M, Ramzan M, Bilal M (2016) Purification and characterization of manganese peroxidases from native and mutant Trametes versicolor IBL-04. Chin J Catal 37:561–570. https://doi.org/10.1016/S1872-2067(15)61044-0
Ayed L, Chaieb K, Cheref A, Bakhrouf A (2008) Biodegradation of triphenylmethane dye Malachite Green by Sphingomonas paucimobilis. World J Microbiol Biotechnol 25:705–711. https://doi.org/10.1007/s11274-008-9941-x
Banerjee S, Dubey S, Gautam RK, Chattopadhyaya MC, Sharma YC (2017) Adsorption characteristics of alumina nanoparticles for the removal of hazardous dye, Orange G from aqueous solutions. Arab J Chem. https://doi.org/10.1016/j.arabjc.2016.12.016
Barapatre A, Aadil KR, Jha H (2017) Biodegradation of Malachite Green by the Ligninolytic Fungus Aspergillus flavus. Clean (Weinh) 45:1600045. https://doi.org/10.1002/clen.201600045
Bergsten-Torralba LR, Nishikawa MM, Baptista DF, Magalhães DP, da Silva M (2009) Decolorization of different textile dyes by Penicillium simplicissimum and toxicity evaluation after fungal treatment. Braz J Microbiol 40:808–817. https://doi.org/10.1590/S1517-838220090004000011
Bora P, Holschuh H, da Silva Vasconcelos M (2004) Characterization of polyphenol oxidase of soursop (Annona muricata L.) fruit and a comparative study of its inhibition in enzyme extract and in pulp. Cienc Tecnol Aliment 4:267–273. https://doi.org/10.1080/11358120409487770
Bouras HD, Yeddou AR, Bouras N, Hellel D, Holtz MD, Sabaou N, Chergui A, Nadjemi B (2017) Biosorption of Congo red dye by Aspergillus carbonarius M333 and Penicillium glabrum Pg1: Kinetics, equilibrium and thermodynamic studies. J Taiwan Inst Chem Eng 80:915–923. https://doi.org/10.1016/j.jtice.2017.08.002
Casas N, Parella T, Vicent T, Caminal G, Sarra M (2009) Metabolites from the biodegradation of triphenylmethane dyes by Trametes versicolor or laccase. Chemosphere 75:1344–1349. https://doi.org/10.1016/j.chemosphere.2009.02.029
Chaturvedi V, Verma P (2015) Biodegradation of malachite green by a novel copper-tolerant Ochrobactrum pseudogrignonense strain GGUPV1 isolated from copper mine waste water. Bioresour Bioprocess 2:1. https://doi.org/10.1186/s40643-015-0070-8
Chaudhry MT, Zohaib M, Rauf N, Tahir SS, Parvez S (2014) Biosorption characteristics of Aspergillus fumigatus for the decolorization of triphenylmethane dye acid violet 49. Appl Microbiol Biotechnol 98:3133–3141. https://doi.org/10.1007/s00253-013-5306-y
Chen SH, Ting ASY (2015a) Biodecolorization and biodegradation potential of recalcitrant triphenylmethane dyes by Coriolopsis sp. isolated from compost. J Environ Manage 150:274–280. https://doi.org/10.1016/j.jenvman.2014.09.014
Chen SH, Ting ASY (2015b) Biosorption and biodegradation potential of triphenylmethane dyes by newly discovered Penicillium simplicissimum isolated from indoor wastewater sample. Int Biodeterior Biodegrad 103:1–7. https://doi.org/10.1016/j.ibiod.2015.04.004
El-Batal AI, ElKenawy NM, Yassin AS, Amin MA (2015) Laccase production by Pleurotus ostreatus and its application in synthesis of gold nanoparticles. Biotechnol Rep (Amst) 5:31–39. https://doi.org/10.1016/j.btre.2014.11.001
Franciscon E, Grossman MJ, Paschoal JAR, Reyes FGR, Durrant LR (2012) Decolorization and biodegradation of reactive sulfonated azo dyes by a newly isolated Brevibacterium sp. strain VN-15. SpringerPlus 1:37–37. https://doi.org/10.1186/2193-1801-1-37
Fu XY, Zhao W, Xiong AS, Tian YS, Zhu B, Peng RH, Yao QH (2013) Phytoremediation of triphenylmethane dyes by overexpressing a Citrobacter sp. triphenylmethane reductase in transgenic Arabidopsis. Appl Microbiol Biotechnol 97:1799–1806. https://doi.org/10.1007/s00253-012-4106-0
Jang MS, Lee YM, Kim CH, Lee JH, Kang DW, Kim SJ, Lee YC (2005) Triphenylmethane reductase from Citrobacter sp. strain KCTC 18061P: purification, characterization, gene cloning, and overexpression of a functional protein in Escherichia coli. Appl Environ Microbiol 71:7955–7960. https://doi.org/10.1128/AEM.71.12.7955-7960.2005
Jasińska A, Różalska S, Bernat P, Paraszkiewicz K, Długoński J (2012) Malachite green decolorization by non-basidiomycete filamentous fungi of Penicillium pinophilum and Myrothecium roridum. Int Biodeterior Biodegrad 73:33–40. https://doi.org/10.1016/j.ibiod.2012.06.025
Jayanthy V, Geetha R, Rajendran R, Prabhavathi P, Karthik Sundaram S, Dinesh Kumar S, Santhanam P (2014) Phytoremediation of dye contaminated soil by Leucaena leucocephala (subabul) seed and growth assessment of Vigna radiata in the remediated soil. Saudi J Biol Sci 21:324–333. https://doi.org/10.1016/j.sjbs.2013.12.001
Jegan J, Vijayaraghavan J, Bhagavathi Pushpa T, Sardhar Basha SJ (2016) Application of seaweeds for the removal of cationic dye from aqueous solution. Desalin Water Treat 57:25812–25821. https://doi.org/10.1080/19443994.2016.1151835
Jin S, Zhang G, Zhang P, Fan S, Li F (2015) High-pressure homogenization pretreatment of four different lignocellulosic biomass for enhancing enzymatic digestibility. Bioresour Technol 181:270–274. https://doi.org/10.1016/j.biortech.2015.01.069
Kalyani DC, Telke AA, Surwase SN, Jadhav SB, Lee JK, Jadhav JP (2012) Effectual decolorization and detoxification of triphenylmethane dye malachite green (MG) by Pseudomonas aeruginosa NCIM 2074 and its enzyme system. Clean Technol Environ Policy 14:989–1001. https://doi.org/10.1007/s10098-012-0473-6
Kim MH, Kim Y, Park HJ, Lee JS, Kwak SN, Jung WH, Lee SG, Kim D, Lee YC, Oh TK (2008) Structural insight into bioremediation of triphenylmethane dyes by Citrobacter sp. triphenylmethane reductase. J Biol Chem 283:31981–31990. https://doi.org/10.1074/jbc.M804092200
Kumar CG, Mongolla P, Basha A, Joseph J, Sarma VU, Kamal A (2011) Decolorization and biotransformation of triphenylmethane dye, methyl violet, by Aspergillus sp. isolated from Ladakh, India. J Microbiol Biotechnol 21:267–273. https://doi.org/10.4014/jmb.1011.11010
Lima DR, Klein L, Dotto GL (2017) Application of ultrasound modified corn straw as adsorbent for malachite green removal from synthetic and real effluents. Environ Sci Pollut Res Int 24:21484–21495. https://doi.org/10.1007/s11356-017-9802-y
Mani S, Bharagava RN (2016) Exposure to Crystal Violet, its toxic, genotoxic and carcinogenic effects on environment and its degradation and detoxification for environmental safety. In: Voogt WP (ed) Reviews of environmental contamination and toxicology. Springer, Switzerland, pp 71–104
Matpang P, Sriuttha M, Piwpuan N (2017) Effects of malachite green on growth and tissue accumulation in pak choy (Brassica chinensis Tsen & Lee). ANRES 51:96–102. https://doi.org/10.1016/j.anres.2016.10.008
Moturi B, Singara Charya MA (2009) Decolourisation of Crystal Violet and Malachite Green by fungi. Sci World J 4:28–33
Pandey AK, Sarada DVL, Kumar A (2016) Microbial decolorization and degradation of Reactive Red 198 azo dye by a newly isolated alkaligenes species. Proc Natl Acad Sci India B Biol Sci 86:805–815. https://doi.org/10.1007/s40011-015-0497-x
Radha KV, Regupathi I, Arunagiri A, Murugesan T (2005) Decolorization studies of synthetic dyes using Phanerochaete chrysosporium and their kinetics. Process Biochem 40:3337–3345. https://doi.org/10.1016/j.procbio.2005.03.033
Rodrigues CS, Madeira LM, Boaventura RA (2013) Treatment of textile dye wastewaters using ferrous sulphate in a chemical coagulation/flocculation process. Environ Technol 34:719–729. https://doi.org/10.1080/09593330.2012.715679
Shanmugam S, Ulaganathan P, Sivasubramanian S, Esakkimuthu S, Krishnaswamy S, Subramaniam S (2017) Trichoderma asperellum laccase mediated crystal violet degradation–Optimization of experimental conditions and characterization. J Environ Chem Eng 5:222–231. https://doi.org/10.1016/j.jece.2016.11.044
Shedbalkar U, Jadhav JP (2011) Detoxification of malachite green and textile industrial effluent by Penicillium ochrochloron. Biotechnol Bioprocess Eng 16:196–204. https://doi.org/10.1007/s12257-010-0069-0
Shedbalkar U, Dhanve R, Jadhav J (2008) Biodegradation of triphenylmethane dye cotton blue by Penicillium ochrochloron MTCC 517. J Hazard Mater 157:472–479. https://doi.org/10.1016/j.jhazmat.2008.01.023
Stancu MM, Grifoll M (2011) Multidrug resistance in hydrocarbon-tolerant Gram-positive and Gram-negative bacteria. J Gen Appl Microbiol 57:1–18
Szewczyk R, Kusmierska A, Bernat P (2018) Ametryn removal by Metarhizium brunneum: Biodegradation pathway proposal and metabolic background revealed. Chemosphere 190:174–183. https://doi.org/10.1016/j.chemosphere.2017.10.011
Ting ASY, Lim SJ, Tan WS (2011) Diversity and metal tolerance of filamentous fungi from analytical wastewater in laboratory. International Congress of the Malaysian Society for Microbiology. Penang, Malaysia, pp 110–113
Vergili I, Kaya Y, Sen U, Gönder ZB, Aydiner C (2012) Techno-economic analysis of textile dye bath wastewater treatment by integrated membrane processes under the zero liquid discharge approach. Resour Conserv Recycl 58:25–35. https://doi.org/10.1016/j.resconrec.2011.10.005
Vyavahare GD, Gurav RG, Jadhav PP, Patil RR, Aware CB, Jadhav JP (2018) Response surface methodology optimization for sorption of malachite green dye on sugarcane bagasse biochar and evaluating the residual dye for phyto and cytogenotoxicity. Chemosphere 194:306–315. https://doi.org/10.1016/j.chemosphere.2017.11.180
Wang M-X, Zhang Q-L, Yao S-J (2015) A novel biosorbent formed of marine-derived Penicillium janthinellum mycelial pellets for removing dyes from dye-containing wastewater. Chem Eng J 259:837–844. https://doi.org/10.1016/j.cej.2014.08.003
Wu Y, Xiao X, Xu C, Cao D, Du D (2013) Decolorization and detoxification of a sulfonated triphenylmethane dye aniline blue by Shewanella oneidensis MR-1 under anaerobic conditions. Appl Microbiol Biotechnol 97:7439–7446. https://doi.org/10.1007/s00253-012-4476-3
Yang X, Zheng J, Lu Y, Jia R (2016) Degradation and detoxification of the triphenylmethane dye malachite green catalyzed by crude manganese peroxidase from Irpex lacteus F17. Environ Sci Pollut Res 23:9585–9597. https://doi.org/10.1007/s11356-016-6164-9
Acknowledgements
This work was supported by the Malaysian Ministry of Higher Education [FRGS/2/2013/STWN01/MUSM/02/2]; and Monash University Malaysia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chen, S.H., Cheow, Y.L., Ng, S.L. et al. Biodegradation of Triphenylmethane Dyes by Non-white Rot Fungus Penicillium simplicissimum: Enzymatic and Toxicity Studies. Int J Environ Res 13, 273–282 (2019). https://doi.org/10.1007/s41742-019-00171-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41742-019-00171-2





 ) before and (
) before and (
 ) after 24 h of treatment with P. simplicissimum at 25 ± 2 °C
) after 24 h of treatment with P. simplicissimum at 25 ± 2 °C
