Abstract
Plant diseases are a vital risk to crop yield and early detection of plant diseases remains a complex task for the farmers due to the similar appearance in color, shape, and texture. In this work, authors have proposed an automatic plant disease detection technique using deep ensemble neural networks (DENN). Transfer learning is employed to fine-tune the pre-trained models. Data augmentation techniques include image enhancement, rotation, scaling, and translation are applied to overcome overfitting. This paper presents a detailed taxonomy on the performance of different pre-trained neural networks and presents the performance of a weighted ensemble of those models relevant to plant leaf disease detection. Further, the performance of the proposed work is evaluated on publicly available plant village dataset, which comprises of 38 classes collected from 14 crops. The performance of DENN outperform state-of-the-art pre-trained models such as ResNet 50 & 101, InceptionV3, DenseNet 121 & 201, MobileNetV3, and NasNet. Performance evaluation of the proposed model demonstrates that effective in categorizing various types of plant diseases that comparatively outperform pre-trained models.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In recent days, automated plant disease detection is evolved as one of the vital tasks in precision agriculture. Diseases by fungus, bacteria, and insects affect the yield of the crop and reduce productivity. Classification of plant leaf diseases is a challenging task due to high inter-class similarities and complex pattern variations. The growth of plant infections can be considerably increased due to changes in the climate. Early detection of diseases on leaves of plants is one of the major challenges to ensure good productivity in the agriculture sector. More than 50% of the plant's productivity has been reduced with diseases, so early identification leads to quicker intervention that reduces crop loss. The plant pathologists are feeling difficult to diagnose a plant disease manually, and even this process is time-consuming and not robust. Instead of using traditional shallow machine learning approaches, in this work we have employed pre-trained deep learning models, for the accurate diagnosis of different plant leaf diseases in real scenarios. Researchers have proposed a wide variety of computer vision-based machine learning and deep learning algorithms for accurate classification of plant diseases. In this scenario, CNN models (KC et al. 2019; Klauser 2018) have emerged as powerful in performing image segmentation and classification when the number of data samples is high. The applications of deep learning models are extended to plant disease classification, pest classification, classification of damaged and good fruits, and weed classification. In this paper, authors are intended to improve the performance of plant disease classification using deep ensemble neural network using transfer learning by fine-tuning the hyper-parameters. These deep learning models are optimistic in extracting discriminating features with the use of transfer learning. The proposed model can extract discriminating features from the leaf images and classify the plant leaf diseases with high accuracy.
The main contribution of this work is outlined as follows:
-
Presented a thorough study on research contributions involved in the segmentation and classification of plant leaf diseases.
-
DENN is proposed with the use of transfer learning for pre-trained models to decrease the training convergence time and to tune the vast number of parameters.
-
Performance of the proposed method is evaluated on plant village dataset which comprises a total of 54,305 images belonging to 38 classes. 80% of these images, i.e., 43,456 are used for model building and the remaining 10,849 images are used to evaluate the performance of the proposed method.
-
Performance of the proposed work is evaluated with various hyper-parameters such as activation functions, number of epochs, batch size, learning rate, and L2 regularizer in improving the classification accuracy.
The rest of this paper is organized as follows: “Related work” section presents the thorough survey on literary works published in the domain of plant leaf disease detection. “Plant leaf disease detection using deep ensemble neural network” section illustrates the various deep neural networks and proposed deep ensemble neural networks. “Results and discussions” section presents the performance evaluation of proposed methods with hyperparameters tuning. “Conclusion” section presents the conclusions and future scope of the work.
Related work
Deep learning models are used to optimize the plant disease detection and describe a hyperspectral image benefits on different scales for plant protection and plant disease detection (Thomas et al. 2018). Lee et al. (2020) proposed a technique to examine the most relevant plant disease identification by using a fine-tuned pre-trained model with IPM and Bing test datasets. VGG16 network is used for pest detection and comparisons are made with Fast-RCNN and PestNet methods. Similarly, GoogLeNet BN and InceptionV3 networks are also used to assess the performance in the detection of plant diseases with 34 layers of GoogLeNetBN and 48 layers of InceptionV3 networks. Liu et al. (2019) presented a PestNet to examine the detection of the pest in plant leaves by using the region-based approach, and it mainly focused on feature enhancement and extraction.
The performance evaluation of plant disease classification (Too et al. 2019) is done by using different deep learning architectures such as VGG16, Inception V4 101, ResNet 152, and DenseNet 121. The DenseNet 121 shows the highest accuracy than the other deep neural network methods. Fuentes et al. (2017) proposed a robust method to detect and classify the pest diseases that occurred in tomato plants. Zhang et al. (2018) proposed a model to classify the leaf diseases by using deep convolutional neural networks as pre-trained models. The CNN pre-trained models such as AlexNet, GoogLeNet, and ResNet models are used to detect the tomato leaf diseases. The ResNet method is used for identifying the leaf diseases of tomato plants and obtain the best accuracy result with stochastic gradient descent (SGD). Sladojevic et al. (2016) suggested a novel method to identify 13 diseases that occurred in plants and distinguish healthy plant leaves from their surroundings. Ferentinos (2018), Sharif et al. (2018), Arnal Barbedo (2019) projected a machine learning method to explore the lesions and spots of the plants without considering the entire leaf. They introduced a hybridization approach to detect citrus plant diseases by using CNN methods. The lesion spot detection is done using segmentation from the citrus fruits and classified with multiple support vector machine (M-SVM). The color, geometric, and textures features are extracted from the citrus plant and then those are fused into a codebook. Barbedo (2018) made an investigation of deep learning factors influence in recognition of diseases in different plants. Thenmozhi and Srinivasulu Reddy (2019) proposed a model to evaluate the crop pest classification by using standard pre-trained models such as AlexNet, ResNet, VGGNet, and GoogLeNet. Pre-trained models achieved higher accuracy in the agriculture sector for crop protection. Knoll et al. (2018) demonstrated a method that efficiently improves organic farming with the help of deep learning models.
The CNN classifiers are used to identify the diseases in carrots and weeds. Iqbal et al. (2018) conducted a review on citrus plants with different techniques of CNN models. In the pre-processing phase, images are acquisited using sensors and transferred to the next phase. In the next phase, an image has been divided into different segments called segmentation used to detect the region of interest (RoI) from an image. In the third phase, features such as texture, color, shape, SIFT, SURF, and HOG are extracted from RoI. Finally, classification methods are used to access the performance. Karlekar and Seal (2020) proposed a SoyNet to extract the plant leaf disease detection from the complex soybean images. Deep learning models have been used for the recognition of segmented soybean plant diseases. Amara et al. (2017) developed a deep learning model to classify banana leaves. The effectiveness of this model demonstrates that the preliminary results with LeNet CNN architecture to classify the dataset. Türkoğlu and Hanbay (2019) made a performance analysis with deep learning models with traditional methods, the comparison made with nine different and powerful architectures of deep neural networks. The recommended architectures are depending on the extraction of deep features and the features have less computational complexity compared with other methods of transfer learning. Saleem et al. (2019) makes a comprehensive explanation about the different deep learning methods to visualize the various diseases and classification of diseases in plants and evaluating the methods with several performance metrics are used. Rangarajan et al. (2018) proposed a method to classify the tomato crop diseases with the help of standard deep learning algorithms such as VGG16 and AlexNet. Mohanty et al. (2016) described CNN models to identify and analyze the plant disease by utilizing image-based crop disease detection. Cruz et al. (2017) proposed a vision-based system that demonstrates transfer learning and trained on thousands of leaf images. They presented a novel algorithm to show the qualitative, quantitative, and automatic detection of diseases in plants. Saleem and Potgieter (2020) presented a comparative study of various optimizers and Saleem et al. (2020) proposed a deep learning meta-architecture including the SSD (single short multibox detector), F-RCNN (faster RCNN), and RFCN (region-based fully convolutional networks) models to detect the plant diseases. Guo et al. (2020) proposed a model by integrating RPN (region proposal network) algorithm, CV (Chan-Vese) algorithm, and TL (transfer learning) algorithms to effectively recognize the plant diseases in complex environment. Jasim (2020) proposed a robust methodology to detect and classify the plant and leaf diseases with accurate and fast based on computer facilities and deep learning techniques. Harte (2020) demonstrates how CNN models are used to empower small-holder farmers against plant leaf diseases out of healthy leaf tissue. Reddy (2019) proposed a framework to distinguish healthy leaves and diseased leaves by utilizing the preprocessing techniques of image processing. Boulent et al. (2019) developed a web application by using CNN models to automatically be identifying the crop diseases. Author provides procedure and guidelines to maximize the potential of CNNs deployed in real-world applications.
Plant leaf disease detection using deep ensemble neural network
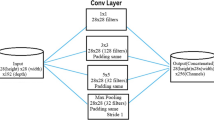
In this work, the authors propose a robust and effective model to detect Bapat et al. (2020) plant leaf diseases. To minimize false positive and false negative with limited computation resources, a deep ensemble neural network is proposed with transfer learning. The architecture of the proposed methodology is given in Fig. 1.
Firstly, preprocessing is applied to improve the quality of the image and quantitatively concern on contrast and brightness of the image. The preprocessing methods include image enhancement, color space transformation, resizing, and noise removal. In this work, firstly performance evaluation of uni-modal pre-trained neural networks such as ResNet, Dense Net, InceptionV3, and NasNet Mobile version is done by resizing the leaf image to 224*224*3.
DenseNet
In CNN Networks, DenseNets are used to simplify the connectivity pattern among the layers. In this net, every layer directly connected with other layers to solve the limitation of maximum information flow. DenseNets have a smaller number of feature maps when compared with traditional CNN. DenseNet exploits the potentials of the layers through feature reuse. DenseNet resolves the issues that occurred by the gradients. Each layer of the DenseNet model has straightforward access with the grades from the input images along with a loss function of gradients. A traditional feed-forward network is used in the DenseNet, and the output of each layer is associated with the preceding layer by performing a composite operation. These operations include pooling layers, batch normalization, and an activation function.
The equation for this would be
In the ResNet, this equation is extended by including the skip connection, reformulating as:
DenseNets are concatenating the input layers and output layers but do not sum the outgoing feature maps with the incoming feature maps. Consequently, the equation reshapes again into:
DenseNets model can be segmented into small blocks called Dense Block. The layers of the Dense Blocks are known as transition layers and take care of down-sampling while applying batch normalization. In this model, there is a change in the number of filters, but the dimensions of the feature maps remain constant. The dimensions of each channel will be increased due to the concatenation of the feature map of every layer. In every time, Hn is used to produce k feature maps then the generalized nth layer is:
k is the growth rate of this hyperparameter. At each layer, the growth rate of hyperparameter k is regulated about the level of information is added to the network. DenseNets are, the input volumes and output of two operations are concatenated, the action of adding information to the shared information of the network and concatenating the k feature maps of information.
NasNet mobile
In convolution neural network, the AutoML and neural architecture search (NAS) are the new kings. NasNet convolutional neural network is trained with millions of images that are collected from the dataset called ImageNet. The network learned with a powerful feature to recognize the images, size of the image we are taken to the input is 224 * 224. The algorithm of the NasNet model searches for the finest CNN architecture and architecture of NasNet is shown in Fig. 2.
NasNet is divided into two variants NasNetMobile and NasNetLarge. NasNetMobile network is a lighter version when compared to NasNetLarge. It utilizes the search strategy to scan for the best convolutional layers or cells on relatively small image datasets. The convolution cells are used to get better performance classification and smaller computational budgets. With these convolutional cells, normal and reduction cells are built to extend the utilization of NasNet for images of any size. The normal cells and reduction cells are combined in various manners, and there was a group of NasNet structures that accomplished the most accessible CNN design with low computation. In NasNet, though the overall architecture is predefined as below, the cells or building blocks that are searched by the strengthening learning search method, i.e., the number of initial convolutional filters is as free parameters, and N is the number of repetitions used for scaling. In the computation with the normal cell, the size of the feature-map will not be changed when compared to the convolutional cells. But reduction cell reduces the height and width of the feature map with the factor of two. The controller recurrent neural network (RNN) searches only the structures of the cells.
Residual networks (ResNet)
ResNet is one of the popular deep learning architectures with variants in number of layers as 50, 101 and 152 (Too et al. 2019). The learning of ResNet is done through residual representation of a framework. The prior layer output is taken as an input to the very next layer without performing any kind of modifications. ResNet has characteristics such as localization and recognition; divide the large image into small segments to recognize the diseased part in the plant leaf. ResNet comprises of two kinds of blocks called Convolutional and Identity. A collection of residual models can form a building block to construct the Resnet. These residual entities are organized as pooling, convolution, and batch normalization layers (Fig. 3).
Each unit can be expressed as
where \(x_{n}\) is the input, \(x_{n + 1}\) is the output of nth unit, and f is the residual function of the nth unit.
h(xn) is the identity mapping of ReLu function. In ResNet, the vanishing gradient problem does not encounter due to backpropagation. The residual representation has the shortcut networks which are equivalent to the simple convolutional layers. After some weight layers, the skip connections are used to add the input values to the output after some weight layers (Figs. 4 and 5).
Fifty parameterized layers are used to deploy the ResNet-50 model with recurrent connections as residual blocks with transfer learning. Similarly, ResNet-101 consists of 101 parameterized layers. These residual blocks are meant to expand the depth of the layer and reduce the size of the output. Ultimately, a personalized Softmax layer can be defined for the identification of plant diseases with L2 regularizer. The ResNet-50 includes a fully connected layers, which is created prior to the Softmax output layer, a 7 × 7 average pooling layers through stride7, residual building blocks are 16, a 7 × 7 convolution layer and a 3 × 3 max pooling layer with stride 2.
Proposed deep ensemble neural network
In recent literature, the ensemble approach is evolved as one of the efficient and robust approaches and known as a meta-algorithm. It could be used for various objectives that are reducing the variance of a model, improve predictions, and optimizes bias. The ensemble method is used to create a new model from the information gathered from the various individual predictive models. The ensemble model overtakes the uni-models with its soothing nature and architecture of the proposed work is given in Fig. 6.
In this work, the ensemble model is designed using various pre-trained CNN models such as Resnet, Inception V3, NasNet Mobile and Densenet. Given algorithm presents a detailed explanation of the proposed method. Let D = {ResNet 50, Resnet 101V2, Inception V3, Dense201, Densenet121, Mobile Net V3, and NasNet Mobile} is the set of pre-trained models. Each model in the ensemble method is fine-tuned with the diseased and healthy images of 13 different types of plants in the dataset (Mi, Ni), where M consisting the number of images, each of size, 224 × 224, and N is the corresponding labels of images, N = {n/n ∊ {Black_rot, Leaf_blight, Bacterial_spot, Early_blight, Healthy, etc.}}. Mini batches are formed by dividing the training set with the size of n, which enhances the accuracy of deep learning models by decreasing the empirical loss:
Overfitting and feature selection are carried through L2 regularizer also known as ridge regression. Ridge regression adds a squared magnitude of coefficient as penalty term to the above loss function. If \(\lambda\) is high, then it adds too much weight and leads to underfitting. In this work, it was set to 0.01.
Here, the CNN models are defined as h(m,w) that predicts the class n with input m, and d(·) is the defined entropy loss function. The Nesterov-accelerated adaptive moment estimation is used to update the learning parameters;
The ensemble method is used to combine the predictions of various methods and produce a cohesive result. The ensemble method combines the predictive scores of separate models and ensures better accuracy. The proposed ensemble result is given as:

Results and discussions
In this section, performing the evaluation of various pre-trained models of CNN and proposed deep ensemble neural network (DENN) is carried on plant village dataset on a high-end GPU machine.
About dataset
Kaggle is the largest data science community with tools and resources such as datasets. In this work, the performance of the recommended model is estimated on a dataset collected from plant village. This dataset comprises RGB images of both infected and healthy leaves collected uniformly from 14 different types of crops including Tomato, Blueberry, Cherry, Corn, Apple, Strawberry, Orange, Peach, etc. The plant village dataset comprises of 38 various crop diseases put together with a total of 43,456 plant leaf samples for model building and other 10,849 images for estimating the performance of the proposed model. The 38 different classes of crops and the number of train and test samples are given in Table 1.
Performance evaluation metrics
The experimentation is conducted using Keras framework using TensorFlow-GPU NVIDIA-based workstation. To calculate the performance of the proposed method, authors used metrics such as accuracy, precision, F-score, recall, and loss. The calculated accuracy can be defined in terms of positive and negative classes:
where TP (True Positives) can be defined as the number of instances classified correctly, FP (False Positives) can be defined as the number of misclassified instances. TN (True Negatives) can be defined as the number of instances that are classified correctly from the rest of the classes, and FN (False Negatives) is the number of instances misclassified from the rest of the classes under observation.
Recall/sensitivity is defined as the ratio of TP and total actual positive
Precision is defined as the ratio of TP and Total predicted as positive
F1-score is the weighted harmonic mean of precision and recall. It is used to measure the balance between precision and recall values.
All these metrics yields score amongst 0 and 1, here 1 defined as the best score and 0 for the worst.
Hyperparameter tuning
The set of parameters that can affect the learning of the model is referred to as hyperparameters. These parameters include the number of layers, number of epochs, activation functions, learning rate, etc. The following tables illustrate the configuration of hyper-parameters used in these pre-trained models. During experimentation, after several attempts, authors fixed the learning rate, momentum, and regularizer factor. The performance of the proposed plant leaf disease detection is done using different pre-trained models such as NasNet, DenseNet, InceptionV3, and ResNet50. Every model has been evaluated with various optimizers for 30 epochs (Table 2).
Performance evaluation of various pre-trained networks
The pre-trained model ResNet101 has101 layers, with trainable parameters as 42,606,758 and 97,664 as non-trainable parameters. ResNet 101 has achieved high accuracy of 99% with Adamax and lower accuracy of 76% with rmsprop. Inception V3 has a depth of 48 layers, having 13,701,926 as trainable and 8,178,720 as non-trainable parameters. Inception V3 has achieved 97% of accuracy with Adam and higher accuracy of 98% with all other optimizers.
DenseNet 201 has a depth of 201 with 6,992,806 as trainable parameters and 83,648 as non-trainable parameters. With ADAMAX, ADAGRAD and NADAM as an optimizer, DenseNet 201 got an accuracy of 99% and given 96.9% with Adam. DenseNet121 has a depth of 121 layers, with a number of trainable parameters as 6,992,806 and a number of non-trainable parameters as 83,648. DenseNet 121 has achieved an accuracy of 98% with SGD and AdaGrad, and poor performance was given with NADAM. MobileNet V3 has 3,245,926 trainable parameters and 21,888 non-trainable parameters. This MobileNet version 3 with SGD has achieved an accuracy of 96%. With ADAM and AdaMax, MobileNet V3 has given a maximum of 99% accuracy. It has consistent performance with all the optimizers and bit low accuracy was given with SGD.
NasNetMobile version has 4,273,144 trainable parameters and 36,738 non-trainable parameters. During experimentation, the performance of NasNet outperformed other pre-trained models with majority of optimizers. NasNet has yielded 99% of accuracy with Adam, Adamax, Nadam, and RmsProp. 96% is the lowest performance presented with Adagrad.
ResNet 50 has a depth of 50 layers and it has a total of 23,665,574 parameters. This ResNet 50model has 23,612,454 trainable and 53,120 non-trainable parameters. As like NasNet Mobile, ResNet 50 also shown better performance with all activation functions. But the trainable parameters are quite high when compared with NasNet Mobile. ResNet 50 have yielded an accuracy of 99% with SGD, ADAM, ADAGRAD, and ADAMAX. It has presented the poor performance with RMSPROP. Figures 7 and 8 present the loss and accuracy curves of pre-trained models with various optimizers.
Accuracy comparison of pre-trained models with various optimizers is given in Fig. 9.
Further, the performance of the deep ensemble neural network is evaluated on plant village (Fig. 10) dataset by combining various pre-trained models. In this section, to check the performance of DENN, two individual low-performing models are combined with one good model. Firstly, DenseNet201 and ResNet101 are combined with Mobile NasNet, and then performance is evaluated on plant village dataset, and the proposed DENN has achieved an accuracy of 99.99%. Later, InceptionV3 and ResNet101 are combined with Mobile Net V3 and achieved an accuracy of 99.99%. In all the other aspects, the proposed ensemble model has achieved an accuracy of 100%. From the results, the proposed DENN has outperformed all the pre-trained models (Table 3).
Finally, the performance of the proposed model is compared with other literature models on plant village dataset. In Szegedy et al. (2016), authors applied Inception V3 to detect pest diseases and achieved an accuracy of 98.33%. In Simonyan and Zisserman (2015), authors demonstrated the performance of VGG 16 on plant village dataset and achieved 99% accuracy. Further, Mohanty et al. (2016) applied GoogLeNet and Too et al. (2019) applied DenseNet 121 achieved an accuracy of 99.35% and 99.75%, respectively. From the experimentation, our proposed DENN model with transfer learning outperforms the pre-trained models such as DenseNet, ResNet, Inception and NasNet on plant village dataset (Fig. 11).
Conclusion
Plant diseases affect the growth of their respective crops; therefore, their early detection is very important. Crop yield and quality are drastically affected by various kinds of diseases, fungus, and insects. In this work, pre-trained models are employed for vision-based plant disease classification including Inception V3, ResNet 50 & 101, DenseNet 121 & 201, NasNet Mobile. In this work, the authors proposed a robust and efficient deep ensemble neural network with various pre-trained models for plant leaf disease classification. Data augmentation techniques and transfer learning are employed to fine-tune the parameters in the network. Further, the performance of the proposed work is evaluated on publicly available plant village dataset, which comprises of 38 classes collected from 14 crops. From the experimental results, it is evident that the proposed ensemble technique with a minimum number of computations has achieved high classification accuracy when compared with pre-trained models.
References
Amara J, Bouaziz B, Algergawy A (2017) A deep learning-based approach for banana leaf diseases classification. In: Lecture notes in informatics (LNI), Proceedings - series of the gesellschaft fur informatik (GI). 266:79–88
Arnal Barbedo JG (2019) Plant disease identification from individual lesions and spots using deep learning. Biosys Eng 180:96–107. https://doi.org/10.1016/j.biosystemseng.2019.02.002
Bapat A, Sabut S, Vizhi K (2020) Plant leaf disease detection using deep learning. Int. J. Adv. Sci. Technol. 29(6):3599–3605
Barbedo JGA (2018) Factors influencing the use of deep learning for plant disease recognition. Biosys Eng 172:84–91. https://doi.org/10.1016/j.biosystemseng.2018.05.013
Boulent J, Foucher S, Théau J, St-Charles PL (2019) Convolutional neural networks for the automatic identification of plant diseases. Front Plant Sci. 10:941. https://doi.org/10.3389/fpls.2019.00941
Cruz AC, Luvisi A, De Bellis L, Ampatzidis Y (2017) Vision-based plant disease detection system using transfer and deep learning. In: 2017 ASABE Annual international meeting, 1–9. https://doi.org/10.13031/aim.201700241
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318. https://doi.org/10.1016/j.compag.2018.01.009
Fuentes A, Yoon S, Kim SC, Park DS (2017) A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors (Switzerland). https://doi.org/10.3390/s17092022
Guo Y, Zhang J, Yin C, Hu X, Zou Y, Xue Z, Wang W (2020) Plant disease identification based on deep learning algorithm in smart farming. Discrete Dynamics in Nature and Society, 2020.
Harte E (2020) Plant disease detection using CNN. September. https://doi.org/10.13140/RG.2.2.36485.99048
Iqbal Z, Khan MA, Sharif M, Shah JH, ur Rehman MH, Javed K (2018) An automated detection and classification of citrus plant diseases using image processing techniques: a review. Comput Electron Agric 153:12–32. https://doi.org/10.1016/j.compag.2018.07.032
Jasim MA, AL-Tuwaijari JM (2020) Plant leaf diseases detection and classification using image processing and deep learning techniques. In 2020 International Conference on Computer Science and Software Engineering (CSASE) (pp. 259–265). IEEE.
Karlekar A, Seal A (2020) SoyNet: soybean leaf diseases classification. Comput Electron Agric. https://doi.org/10.1016/j.compag.2020.105342
KC K, Yin Z, Wu M, Wu Z (2019) Depthwise separable convolution architectures for plant disease classification. Comput Electron Agric 165:104948. https://doi.org/10.1016/j.compag.2019.104948
Klauser D (2018) Challenges in monitoring and managing plant diseases in developing countries. J Plant Dis Prot 125(3):235–237. https://doi.org/10.1007/s41348-018-0145-9
Knoll FJ, Czymmek V, Poczihoski S, Holtorf T, Hussmann S (2018) Improving efficiency of organic farming by using a deep learning classification approach. Comput Electron Agric 153:347–356. https://doi.org/10.1016/j.compag.2018.08.032
Lee SH, Goëau H, Bonnet P, Joly A (2020) New perspectives on plant disease characterization based on deep learning. Comput Electron Agric 170:105220. https://doi.org/10.1016/j.compag.2020.105220
Liu L, Wang R, Xie C, Yang P, Wang F, Sudirman S, Liu W (2019) PestNet: an end-to-end deep learning approach for large-scale multi-class pest detection and classification. IEEE Access 7:45301–45312. https://doi.org/10.1109/ACCESS.2019.2909522
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1–10. https://doi.org/10.3389/fpls.2016.01419
Rangarajan AK, Purushothaman R, Ramesh A (2018) Tomato crop disease classification using pre-trained deep learning algorithm. Procedia Comput Sci 133:1040–1047. https://doi.org/10.1016/j.procs.2018.07.070
Reddy JN (2019) Analysis of classification algorithms for plant leaf disease detection. In: 2019 IEEE international conference on electrical, computer and communication technologies (ICECCT), 1–6
Saleem MH, Potgieter J, Arif KM (2020) Plant disease classification: a comparative evaluation of convolutional neural networks and deep learning optimizers. Plants 9(10):1319
Saleem MH, Potgieter J, Arif KM (2019) Plant disease detection and classification by deep learning. Plants 8(11):32–34. https://doi.org/10.3390/plants8110468
Saleem MH, Khanchi S, Potgieter J, Arif KM (2020) Plant Disease Classification: A Comparative Evaluation of Convolutional Neural Networks and DeepLearning Optimizers. Plants 9(10):1319. https://doi.org/10.3390/plants9111451
Sharif M, Khan MA, Iqbal Z, Azam MF, Lali MIU, Javed MY (2018) Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection. Comput Electron Agric 150:220–234. https://doi.org/10.1016/j.compag.2018.04.023
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: 3rd International conference on learning representations, ICLR 2015 - conference track proceedings, pp 1–14
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci. https://doi.org/10.1155/2016/3289801
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition, 2016-Decem, pp 2818–2826. https://doi.org/10.1109/CVPR.2016.308
Thenmozhi K, Srinivasulu Reddy U (2019) Crop pest classification based on deep convolutional neural network and transfer learning. Comput Electron Agric 164:104906. https://doi.org/10.1016/j.compag.2019.104906
Thomas S, Kuska MT, Bohnenkamp D, Brugger A, Alisaac E, Wahabzada M, Behmann J, Mahlein AK (2018) Benefits of hyperspectral imaging for plant disease detection and plant protection: a technical perspective. J Plant Dis Prot 125(1):5–20. https://doi.org/10.1007/s41348-017-0124-6
Too EC, Yujian L, Njuki S, Yingchun L (2019) A comparative study of fine-tuning deep learning models for plant disease identification. Comput Electron Agric 161:272–279. https://doi.org/10.1016/j.compag.2018.03.032
Türkoğlu M, Hanbay D (2019) Plant disease and pest detection using deep learning-based features. Turk J Electr Eng Comput Sci 27(3):1636–1651. https://doi.org/10.3906/elk-1809-181
Zhang K, Wu Q, Liu A, Meng X (2018) Can deep learning identify tomato leaf disease? Adv Multimed. https://doi.org/10.1155/2018/6710865
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vallabhajosyula, S., Sistla, V. & Kolli, V.K.K. Transfer learning-based deep ensemble neural network for plant leaf disease detection. J Plant Dis Prot 129, 545–558 (2022). https://doi.org/10.1007/s41348-021-00465-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-021-00465-8