Abstract
Purpose
Human papillomavirus detection is crucial in determining preventive approach in cervical cancer. Different HPV genotypes have been reported worldwide and have shown variations with respect to geographic location and oncogenic capabilities. We aimed to conduct molecular characterization and phylogenetic analysis of human papillomavirus (HPV) in Northern Nigeria, to guide public health intervention.
Methods
Cervical smear samples were collected from 148 consenting subjects attending different Hospitals in Northern Nigeria. It was stained with Papanicolaou method and processed for molecular detection of HPV. Isolates of HPV were sequenced targeting HPVL1 gene. The sequences were analysed for single-nucleotide polymorphism. HPV variant lineages and relatedness was determined by phylogenetic analysis.
Results
The result of the present study showed that 26.4% (34) of the subjects had HPV infection. The result of the cytodiagnosis showed that 29.4% (10/34) had normal cytology, 23.5% (8/34) had ASCUS, and 20.6% (7/34) had LSIL, while those having HSIL and SCC accounts for 20.6% (7/34) and 5.9% (2/34), respectively. The result for sequence analysis shows about fifteen different circulating genotypes in Northern Nigeria. Out of these, HPV 16 has the highest frequency (20.6%), while HPV 11, 70, 18, 31, 33, 35, 52, 56 and 7 has the least frequency (2.9%) each. Total of 21 single-nucleotide polymorphisms were detected, out of which 15 are synonymous, while 6 are non-synonymous mutations. Phylogenetic analysis of HPV genotypes using maximum likelihood method showed that majority of HPV genotypes from Nigeria clustered with two undetermined lineages. Pairwise distance estimation of some sequences from this study and some reference sequence revealed a distance ranging from 0.0082 to 2.0994, indicating that the sequences are majorly closely related.
Conclusion
The present study provided important data about molecular epidemiology, characteristics, phylogeny and polymorphism which will aid in further studies that will advance in-depth understanding of the viral molecular epidemiology and possibly policy on public health intervention.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer is one of the leading causes of mortality among women worldwide, especially in developing countries, and the second most common malignancy among women [1]. Human papillomavirus (HPV) especially type 16 and 18 had been found to be the major causative agent of cervical and some non-cervical cancers [2].
Most HPVs had been found to cause asymptomatic infections of the skin and mucosa, and in some cases cervical cancer and other precancerous lesions, which may in few cases progress to invasive cervical carcinoma [3]. HPVs were classified into high-risk (HR-HPV) and low-risk (LR-HPV) types based on their pathogenesis and association with cervical cancer. Low-risk HPV such as HPV 6 and HPV 11 causes benign lesions and genital warts and have a negligible probability of malignant progression [4], while high-risk HPV such as HPV 16 and HPV 18 causes cervical, anal, head, neck and oral carcinomas [5].
Previous studies have detected different HPV genotypes in clinical specimens, as either single or multiple infections. These HPV genotypes may be the either HR-HPV or LR-HPV, with HPV 16 being the most common [6]. Changes in nucleotide sequences such as deletions, substitution, insertions and single-nucleotide polymorphisms (SNP) have been observed across variants of HPV, with SNP been the most common form of genetic variation, and this can be detected by the ability to distinguish true allelic variation from sequencing error [7]. Such changes if occurred within the epitopes coding region of the virus may result in vaccine failure.
Nigeria is experiencing increased morbidity and mortality rate due to immunizable diseases such as cervical cancer, tuberculosis and measles [8]. In Nigeria, cervical cancer is ranked as the second most prevalent cancer among women aged 15–44, with breast cancer taking the first in the ranking [8]. It has been found that African women with normal cytology had the highest prevalence of HPV accounting for about 22.1%. It was also reported that African women have higher tendency of developing invasive cancer due to the least likelihood of been screened. Furthermore, age at first marriage, poor hygienic conditions and promiscuity contribute high prevalence rate of HPV in some part of Africa. Sub-Saharan Africa has the highest incidence of cervical cancer with high mortality rate affecting women aged 18–45 [8].
A research conducted in Gombe north-eastern part of Nigeria revealed 48.1% prevalence of HPV infection among women that came for cervical cancer screening [9]. Furthermore, a study carried out in Kano, Nigeria, reported HPV prevalence of 76% [10]. Another study carried out in Abuja, Nigeria, reported 10.8% prevalence of high-risk HPV among HIV-negative women [11]. Though none of these authors described detailed molecular characterization of HPV in Northern Nigeria, the high prevalence rate reported may be alarming and warrant further molecular epidemiological studies in this part of the world to further elucidate the virus phylogeny, polymorphism that might possibly influence policy for public health intervention.
Methods
Study Population
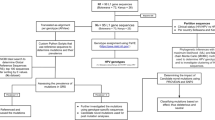
Samples were collected from sexually active women aged 15 years and above attending the gynaecology clinic of Muhammad Abdullahi Wase Specialist hospital Kano, Aminu Kano Teaching Hospital and Federal Teaching Hospital Gombe, all in Nigeria. The study was conducted between August 2018 and July 2019 in the stated hospitals and the states. The subjects of the present study comprised of patients initially diagnosed with either atypical squamous cells of undetermined significance (ASCUS), low-grade squamous intraepithelial lesion (LSIL) or high-grade squamous intraepithelial lesion (HSIL), who were at the clinics for follow-up visit and those patients coming for screening at the first time. A total of 148 women participated in this study, and nineteen out of the 148 samples were excluded because they were found not to contain cells of the squamocolumnar junction.
Sample Collection
Subjects were instructed to lie in a position for pelvic examination. A clean sterile speculum was inserted through the vaginal opening to allow visualization of the cervix. (Subjects were not menstruating at the time of sample collection.) Excess cervical mucus was wiped with a clean swab. A cytobrush was inserted into the endocervix, the brush was rotated in a circular fashion, and cellular smear from both ectocervix and endocervix was collected (squamocolumnar junction).
The cytobrush was gently removed, and the bristle end was detached into the vial containing 10mls of 95% alcohol. The vial was stored at room temperature until use.
Cytology
The vials containing the cervical samples were centrifuged at 3000 rpm, the supernatant was dispensed into a clean tube and a loop full of the sediment was placed on a clean grease-free glass slide. A smear was made and immediately placed in 95% alcohol for 15 min, further fixation. Slides were stained with Harris haematoxylin for 5 min, rinsed in water, differentiated in 1% acid alcohol, blued in Scott’s Tap Water for a minute, rinsed in 95% alcohol, stained with orange G6 for two minutes, rinsed in 95% alcohol, counterstained with Eosin Azure 50 for 2 min, rinsed in 3-changes of 95% alcohol and finally rinsed in absolute alcohol. The slides were cleared in xylene and mounted in DPX according to the method of Papanicolaou [12]. The slides were examined microscopically and graded based on the Bethesda system [16] (Fig. 1).
DNA Extraction
DNA extraction was done using Geneaid (VR100) Viral Nucleic Acid Extraction kit II (Geneaid biotech Ltd., Taiwan). Following manufacturers’ instruction, as stated briefly here, about 200 µl of sample was transferred to a 1.5-ml microcentrifuge tube, and 400 µl of VB lysis buffer was added to the sample, vortexed and incubated at room temperature for 10 min. About 450 µl of AD buffer (with ethanol added) was added to the sample lysate and shaken vigorously to mix. A VB column was then placed in a 2-ml collection tube; then, 600 µl of the lysate mixture was transferred to the VB column and centrifuged at 16,000 × g for 1 min. The flow-through was discarded and the VB column placed back into the 2-ml collection tube. The remaining mixture was transferred to the VB column and centrifuged at 14–16,000 × g for 1 min, and a 2-ml collection tube containing the flow-through was discarded. The VB column was then transferred to a new 2-ml collection tube. Approximately 400 µl of W1 buffer was added to the VB column and centrifuged at 16,000 × g for 30 s. The flow-through was discarded and the VB column was placed back into the 2-ml collection tube, 600 µl of wash buffer was added to the VB column, centrifuged at 16,000 × g for 30 s and the flow-through was discarded. The VB column was placed back into the 2-ml collection tube and centrifuged at 16,000 × g for 3 min to dry the column matrix. The dried VB column was placed in a clean 1.5-ml microcentrifuge tube, followed by the addition of 50 µl of RNase-free water to the centre of the VB column matrix and was allowed to stand for at least 3 min to ensure the RNase-free water was absorbed by the matrix. The column was centrifuged at 16,000 × g for 1 min and the purified nucleic acid was eluted. Extracted DNA was quantified using NanoDrop 2000 (Fisher Scientific, USA).
Detection of HPV
HPV was detected by PCR assay using MY09/11 (5' CGT CCM ARR GGA WAC TGA TC 3') and (5' GCM CAG GGW CAT AAY AAT GG 3') and GP5 + /6 + primer (5’ TTT GTT ACT GTG GTA GAT ACT AC 3’) and (5’ GAA AAA TAA ACT GTA AAT CAT ATT C 3’) L1 general consensus primers [13]. Each PCR mix contained 2.6 µl of primer pair, 10 µl of dNTPs, 25 µl of polymerase chain reaction (PCR) buffer, 10 µl of nuclease free water, 1 µl of KOD FX Neo (Toyobo Life Science department, Inc, Japan) and 1.4 µl of the sample. For MY09/11primer, the dsDNA was denatured at 94 °C for 2 min, followed by 35 cycles of 94 °C for 45 s, 55 °C for 45 s and 72 °C for 45 s, and then 1 cycle of 72 °C for 5 min. For GP5 + /6 + primer, the dsDNA was denatured at 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 45 °C for 60 s and 72 °C for 90 s, and then 1 cycle of 16 °C for 5 min.
PCR products were visualized using 1% agarose gel electrophoresis stained with ethidium bromide.
HPV L1 Gene Sequencing
Amplicons of positive samples were subjected to gene sequencing at 1st BASE laboratories, Malaysia, by next-generation sequencing method using Applied Biosystems 3730xl sequencer (Applied Biosystems, Massachusetts USA). The generated sequence data were analysed using oligo 7 and bioEdit. The sequences were subsequently analysed by NCBI Blast (blast.ncbi.nlm.nih.gov/Blast.cgi) to determine the identity of the HPV genotypes.
Phylogenetic Analysis
The L1 nucleotide sequences were used in constructing a phylogenetic tree using maximum likelihood and neighbour joining methods [14]. The nucleotide sequences were aligned by applying the ClustalW Multiple Alignment. Tamura–Nei model was used as the recommended model of evolution. Finally, maximum likelihood (ML) tree-building algorithm was used to construct a single tree using MEGA X software [15]. Reference sequences were obtained from the GenBank and used to construct distinct phylogenetic branches (Table 1).
Single-Nucleotide Polymorphism Analysis
Sequences were aligned and compared using NCBI BLAST. Sequences with highest maximum score, query coverage, E value and percentage identity were selected. Alignment was observed, and positions with nucleotide variability between the query sequence and subject sequence were identified. The reading frames for the sequences were determined by identifying the Start codon (AUG). The sequences were read from the start codon and the codon(s) at which a single-nucleotide change occurred were identified. This was further investigated to determine whether a change in a single nucleotide in each codon may change the amino acid that is coded for. The observed polymorphisms were then checked for similarity to reported polymorphisms across different molecular databases and published articles.
Statistical Analysis
Frequencies and percentages were used in presenting the data obtained. The distribution of HPV genotypes according to lesion grades and also PCR result in relation to age was compared by two-tailed Fisher exact test using SPSS version 20 (SPSS, Inc., Chicago, USA).
Results
Cytological Analysis
In this study, 129 women whose age ranges from 15 to 82 years with a mean age of 39 years participated in this study, majority (62%) of which falls within the age of 20 to 40 years, and the least (3.2%) falls within the age range of 71–90 years. The subjects were grouped into five categories based on whether they are having epithelial cell abnormality or not; the result showed that 65(50.3%) had normal cytology, 14(10.9%) had ASCUS, 32(24.8%) had LSIL, 15(11.6%) had HSIL and 3(2.4%) had SCC.
Molecular Detection and Distribution of HPV in Relation to the Lesion Grades
In general, 26.4% (34/129) of the study subjects were positive for HPV infection, out of which 29.4% (10/34) were found to have normal cytology. The result further showed that 23.5% (8/34) of the total HPV positive subjects have ASCUS, and 20.6% (7/34) have LSIL, while those having HSIL and SCC were 20.6% (7/34) and 5.9% (2/34), respectively. It was observed that 8 out 15 subjects with HSIL and 1 out of 3 subjects with SSC were negative for HPV. This might be due to the fact that some of our samples are retrospective samples, it is possible that the virus DNA degraded, or there is presence of PCR inhibitors in the sample.
Furthermore, the result of the analysis of whether an individual is infected with a single HPV genotype or more showed that 76.5% (26/34) of all infected subject have single HPV genotype infection, while multiple infections accounts for 23.5% (8/34) of the positive samples (Table 2). The finding of this study indicates that HPV 16 was the most common genotype (20.6%) in the study area, this is followed by HPV 51 (8.8%), HPV 6, 66 and 81 (5.9%) and the least types are HPV 11, 70, 18, 31, 33, 35, 52, 56 and 7 each accounting for 2.9% (Table 2).
Sequencing
A total of 9 sequences of HPV 16, 31, 81, 66 and 56 were deposited in GenBank with accession numbers MN075932 to MN075940.
Phylogenetic Analysis
Phylogenetic analysis of HPV genotypes using maximum likelihood method showed that majority of HPV genotypes from this study clustered with two undetermined lineages (Fig. 2). Some genotypes were found to cluster with European (KU298917.1, KU298917.1 and EF177178) and African (EF177176.1 and (EF177177.1) lineages. Also, HR-HPV isolates from this study were found to cluster with some LR-HPV (HPV 70 and HPV 11).
SNPs
A total of 21 single-nucleotide polymorphisms (SNP) were found among four study subjects. It was observed that 6/21 of the SNP were non-synonymous mutations which result in the change in the amino acid sequence. The remaining 15 polymorphisms were synonymous mutations (Table 3).
Discussion
Human papillomavirus is a silent killer that causes cervical and other non-genital cancers through long-term exposure and progressive process that takes 10-15 years to develop into a full-blown invasive carcinoma. Several approaches are employed in curtailing the menace of HPV, one of which is the use of L1 prophylactic HPV vaccines that elicit the production of neutralizing antibodies against the virus. In the present study, we analysed cervical smear samples of 148 women collected, of which 129 were analysed for HPV infection using PCR. HPV DNA was found in 34/129 of the samples giving a prevalence of 26.4%. With regard to lesion grades, this study found a HPV prevalence of 7.8% in subjects with normal cytology and 18.6% in those with squamous intra-epithelial lesion, confirming the fact that squamous intra-epithelial lesion is caused by HPV. The present study found 15 HPV genotypes including 5 LR types (HPV 6, 11, 40, 70, 81), 8 HR types (HPV 16, 18, 31, 33, 35, 51, 52 and 56) and two HPV other types (HPV 7 and 66). The present study also found that 66.7% of HPV 16 isolates belong to the same lineage, with the remaining 33.3% belonging to African (C) lineage. However, some isolates from this study were found to belong to the European lineage. Phylogenetic tree drawn from sequences obtained from this study and reference sequences from Jing [17] using maximum likelihood showed that two HPV 16 and three HPV 56 genotypes clustered with lineage A (European), and a genotype each of HPV 6 and HPV 66 clustered with lineage B (African-1). The remaining sequences were found to cluster with two undetermined lineages. But by using the neighbour joining method, the tree showed almost similar phylogeny, with three HPV 56 genotypes clustering with lineage A, while HPV 6, 66 and two HPV 16 genotypes clustered with lineage B. This method also showed same phylogeny for the remaining isolates as seen using the maximum likelihood method. Pairwise distance estimation between some sequences from this study and some references sequences showed variable homology between the DNA sequences. Pairwise distance estimation between sequences from this study showed a distance ranging from 0.0082 to 2.0994, indicating close relationship between the sequences. This study has detected a total of 21 single-nucleotide polymorphisms using different reference sequences from NCBI with accession numbers (U89349.1, U89348.1, X74483.1, U31794.1 and M12732.1) which to the best of our knowledge has never been previously reported. Of these, 6 are non-synonymous mutations, while the remaining 15 were synonymous. The non-synonymous mutations were found to cause a change in the secondary structure of the proteins involved (T2233I, Q315R, Y2207L, S2237F and C2240Y).
The prevalence in this study is similar to 21.6% found in a study by Fadahunsi [18] in Ile Ife, and higher than 10% found by Gage [19] in Ondo state. However, a study by Manga [9] in Gombe State (one of the study areas of this study) revealed a prevalence of 48.1%, which is almost twice that found in this study. This may be due to the nested PCR method employed by Manga and colleagues with GP5 + /6 + and PGMY 09/11 consensus primers resulting in higher detection rate of the viral DNA. A study by Bedoya-Pilozo [6] in the coastal city of Ecuador revealed a prevalence of 68%, a finding much higher than that found in this present study. This may be due to the fresh biopsies collected in addition to the cervical smear samples, giving more chances of amplifying the viral DNA. Another factor that may lead to this is the ethnic composition of the Ecuadorian population and also the high rate of migration from border countries of Peru and Columbia.
The findings of the study by Kleter [20] which reported a prevalence of 12% in subjects with normal cytology and 35% in those with squamous intra-epithelial lesion and another study by De Sanjos [21] which reported a HPV prevalence of 22.1% in women with normal cytology are in agreement with the findings from this study. This implies that there is increased rate of HPV infection among sexually active women, and this may be due to lack of awareness about routine gynaecological screening tests that are focused towards diagnosing cervical changes or abnormalities, so as to avoid malignant progression. Other factors that might lead to this are multiple sex partners, poor personal hygiene, diet, access to medical care and possibly failed vaccine efficacy or vaccine resistance.
A study by Badial [22] reported a HR-type prevalence of 57.5%, which is almost close to 61.5% found in this study. A study by Manga [9] reported a finding of 10 HPV types in Gombe Nigeria, with HPV 38, 45, 56, 58 and 82 reported, but not found in this present study. He also reported HPV 18 to be the most common (44.7%), which is in contrast to what was obtained in the present study; HPV 16 was found to be the most common in the present study (20.6%). This indicated the need for frequent review of the most circulating HPV genotype to guide public health intervention, at any given time. There is also an urgent need to intensify research with local isolate of the virus to find virus therapeutics and vaccine variant [23]
Phylogenetic analysis of HPV 16 from Tunisian women by Ghedira [24] revealed that 85.7% of HPV 16 variants belong to the European (A) lineage. Ghedira, however, reported variants of European lineage to be spread all over the world except for sub-Saharan Africa where the African variants are more prevalent.
A limitation of this study is the low number of samples (148) analysed and that no biopsies were collected in addition to the cervical smear samples. Therefore, further studies are recommended to address these limitations and provide in-depth understanding of the molecular characteristic of HPV genotypes circulating in the study area.
Conclusion
The present study provided important basic data about the molecular epidemiology, characteristics, phylogeny and polymorphism which will aid in further studies involving larger number of study subjects to advance in-depth understanding of the viral genomics. Furthermore, it has been established that there is variation in the most common circulating HR-HPV in a particular time and at a given region, warranting the need for frequent updating at interval, which genotype of the virus is more prevalent so as to guide public health interventions. The existence of SNPs among the circulating HR-HPV may indicate that vaccine designed using wild type of such isolates might require review to ascertain if the available vaccine can still be relevant in the control of such variant, thought larger study subjects needs to be recruited to establish this fact.
Data Availability
All data and information regarding this manuscript will be made available on request.
Abbreviations
- HPV:
-
Human papillomavirus
- HR-HPV:
-
High-risk human papillomavirus
- LR-HPV:
-
Low-risk human papillomavirus
- SNP:
-
Single-nucleotide polymorphism
- ASCUS:
-
Atypical squamous cells of undetermined significance
- LSIL:
-
Low-grade squamous intraepithelial lesion
- HSIL:
-
High-grade squamous intraepithelial lesion
- PCR:
-
Polymerase chain reaction
References
Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, Bray F. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. The Lancet Global Health. 2020. https://doi.org/10.1016/S2214-109X(19)30482-6.
Smith JS, Lindsay L, Hoots B, Keys J, Francheschi S, Winer R, Clifford GM. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121:621–32.
Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16(1):1–17.
Bosch FX, de Sanjosé S, Castellsagué X. HPV and genital cancer: the essential epidemiology. Vacc Prev Cerv Cancer. 2008;12:18–22.
Stanley MA. Pathology and epidemiology of HPV infection in females. Gynecol Oncol. 2010;117(2):5–10.
Bedoya-Pilozo CH, Medina Magues LG, Espinoga-Garcia M, Sanches M, Parrales Valdiviezo JV, Molina D, Badano I. Molecular epidemiology and phylogenetic analysis of human papillomavirus infection in women with cervical lesions and cancer from the coastal region of Ecuador. Rev Argent Microbiol. 2017;50(2):136–46.
Marth GT, Korl I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Gish WR. A general approach to single-nucleotide polymorphism discovery. Nat Genet. 1999;23(4):452–6.
Thomas JO, Herrero R, Omigbodun AA, Ojemakinde K, Ajayi IO, Fawole A, Franceschi S. Prevalence of papillomavirus infection in women in Ibadan, Nigeria: a population-based study. Br J Cancer. 2004;90(3):638–45.
Manga MM, Fowotade A, Abdullahi YM, El-nafaty AU, Adamu DB, Pindiga HU, Osoba AO. Epidemiological patterns of cervical human papillomavirus infection among women presenting for cervical cancer screening in North-Eastern Nigeria. Infect Agent Cancer. 2015;10:39.
Auwal IK, Aminu M, Atanda AT, Tukur J, Sarkinfada F. Prevalence and risk factors of high risk human papillomavirus infections among women attending gynaecology clinics in Kano. Northern Nigeria Bajopas. 2014;6:67–71.
Dareng EO, Ma B, Famooto AO, Akarolo-Anthony SN, Offiong RA, Olaniyan O, Adebamowo CA. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol infect. 2016;2016(144):123–37.
Papanicolaou GN. A new procedure for staining vaginal smears. Science. 1942;95(2469):438–9.
Bernard HU, Chan SY, Manos MM, Ong CK, Villa LL, Delius H, Wheeler M. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170(5):1077–85.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26.
Kumar S, Stecher G, Tamura K. MEGAX: Molecular Evolutionary Genetics Analysis version X for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Nayar R, Wilbur DC. The pap test and Bethesda 2014. Acta Cytol. 2015;59(2):121–32.
Jing Y, Wang T, Chen Z, Ding X, Xu J, Mu X, Cao M, Chen H. Phylogeny and Polymorphism in the long control regions E6, E7, and L1 of HPV Type 56 in women from Southwest China. Mol Med Rep. 2008;17:7131–41.
Fadahunsi OO, Omoniyi-Esan GO, Banjo AAF, Esimai OA, Osiagwu D, Clement F, Iyola S. Prevalence of high risk oncogenic human papillomavirus types in cervical smears of women attending well woman clinic in Ile Ife Nigeria. Gynecol Obst. 2013. https://doi.org/10.4172/2161-0932.1000185.
Gage JC, Ajenifuja KO, Wentzensen NA, Adepiti AC, Eklund C, Reilly M, Schiffman M. The age-specific prevalence of human papillomavirus and risk of cytologic abnormalities in rural Nigeria: Implications for screen-and-treat strategies. Int J Cancer. 2012;130(9):2111–7.
Kleter BL, van Doorn J, Schrauwen L, Molijn A, Sastrowijoto S, Terschegget J, Quint M. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection of anogenital human papillomavirus. J Clin Microbiol. 1999;37(8):2508–17.
De Sanjosé S, Diaz M, Castellsagué X, Clifford G, Bruni L, Munoz L, Bosch FX. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: a meta-analysis. Lancet. 2007;7(7):453–9.
Badial RM, Dias MC, Stuqui B, Melli P, Quintana SM, Bonfim C, Rahal P. Detection and genotyping of human papillomavirus (HPV) in HIV-infected women and its relationship with HPV/HIV co-infection. Medicine. 2018;97(14):e9545.
Aliyu IA, Ling KL, Md Hashim NF, Chee HY. Annexin A2 Translocation and Virus Interaction; a Potential Target for Antivirus Drug Discovery. Rev Med Virol. 2019. https://doi.org/10.1002/rmv.2038.
Ghedira R, Mahfoudh W, Hadhri S, Gabbouj S, Bouanene I, Khairi H, Hassen E. Human papillomavirus genotypes and HPV-16 variants distribution among Tunisian women with normal cytology and squamous intraepithelial lesions. Infect Agent Cancer. 2016;11:61.
Acknowledgements
The authors gratefully acknowledge MLS. Sani Aliyu Mohammed, Dr. Yusuf M. Abdullahi, Dr. Aliyu Ibrahim Lawan and Dr. Aliyu Bala Umar for their useful contributions towards this research.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
KI, DU and AIA presented the methods, carryout the experiment, analysed the data and wrote the manuscript, KI, KA, BJ and AIA, conceptualized the work, supervised and approved the manuscript, and KI, YL, BH, AS, DA, NL and AU, performed laboratory work. All authors approved the manuscript as submitted.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Ethics Approval
Ethical approval was obtained from the research and ethics committee of the hospitals from which the study subjects were recruited.
Informed Consent
Samples were collected from consenting individuals.
Consent for Publication
Consent to publish findings was obtained from the study subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Malik, K.I., Aliyu, D.U., Abubakar, B.J. et al. Identification of high-risk human papillomavirus isolates circulating in Nigeria and phylogenetic analysis based on the virus essential protein. Indian J Gynecol Oncolog 19, 87 (2021). https://doi.org/10.1007/s40944-021-00587-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40944-021-00587-9