Abstract
One hundred and seventy-four symptomatic sugarcane samples infected with sugarcane white leaf (SCWL) phytoplasma were collected from sugarcane growing areas in ten provinces across the North, Northeastern and Central plain of Thailand. Genotyping was performed using 16S rRNA and ITS sequence and multilocus sequence typing (MLST) based on six genes including tuf, secY, leuS, secA, AAA1 and groES. The 16S rRNA sequence analysis indicated that the SCWL phytoplasma population showed nucleotide sequence identity of 99.74% to 99.80% to ‘Candidatus Phytoplasma sacchari’. In silico RFLP analysis clustered the strains into 16SrXI-B and 16SrXI-D subgroups. Most strains (77%) were in the 16SrXI-D group, while 16SrXI-B strains were only found in the Northeastern regions. The 16SrXI-D group coexisted with the 16SrXI-B group in Roi Et, Kalasin, and Mookdahan provinces, but it was found alone in Surin province. Multilocal sequence typing showed genetic variations in six genes including tuf, secY, leuS, secA, AAA1, and groES. These variations resulted in the SCWL population being divided into two distinct groups, that are consistent with the 16SrXI subgroups. Additionally, five strains in this population exhibited recombinant DNA in the tuf, secA, leuS, AAA1 and groES genes. This information about the genetic variability of SCWL phytoplasma in Thailand could be used to develop detection methods and monitoring strategies for the disease management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum spp. hybrid) is a global economic crop for sugar and bioenergy production. The major sugarcane-producing countries included Brazil, India, Australia and Thailand (OECD/FAO 2021). Among the major sugarcane diseases, sugarcane white leaf (SCWL) causes significant losses. The disease is distributed in many countries in Asia including Bangladesh, Japan, Taiwan, Thailand (Marcone 2011) and China (Li et al. 2013). In Thailand, this disease has been found in many sugarcane production areas and causes approximately 30-40 million USD in losses annually (Hanboonsong et al. 2017). The symptoms of this disease are found in all developmental stages, starting with white or yellow lines on the leaves, white leaf, stunting, white grassy shoot and eventually plant death. This disease is spread by infected seedcane cuttings and leafhoppers, Matsumuratettix hiroglyphicus and Yamatotettix flavovittatus (Hanboonsong et al. 2017). The disease is associated with the presence of ‘Candidatus Phytoplasma’. These bacteria are pleomorphic, cell wall-less and can infect both plants and insects (IRPCM 2004). They can be divided into different ribosomal subgroups based on the restriction fragment length polymorphism (RFLP) profile of 16S rRNA (Lee et al. 1998) gene sequences. Phytoplasmas within the same ‘Candidatus Phytoplasma' species must have a nucleotide sequence identity of 98.65% or a whole-genome average nucleotide identity (ANI) of 95% (Bertaccini et al. 2022). Sugarcane white leaf phytoplasma is a member of phytoplasma group 16SrXI, which contains phytoplasmas associated with napier grass stunt, flower stunting, sugarcane white leaf, sugarcane grassy shoot, weligama coconut leaf wilt and areca palm yellow leaf (Abeysinghe et al. 2016). This group has many subgroups. The rice yellow dwarf phytoplasma is in subgroup 16SrXI-A (Jung et al. 2003), while the sugarcane white leaf phytoplasma is in subgroups 16SrXI-B and 16SrXI-D (Zhang et al. 2016). The leafhopper phytoplasmas collected from Diplocolenus evansi and Limotettix urnura are in subgroups 16SrXI-C and 16SrXI-G, respectively (Trivellone et al. 2022). The phytoplasma infected creeping thistle is in subgroup 16SrXI-E (Šafářová et al. 2016) and sugarcane grassy shoot phytoplasma is in subgroup 16SrXI-F (Yadav et al. 2017). Previous studies indicated that the provisional classification based only on 16S rDNA was not sufficient for strain differentiation (Al-Abadi et al. 2016). Therefore, the use of several regions/genes of the phytoplasma genome, and multilocus sequence typing (MLST) were explored. Multilocus sequence typing using leuS combined with secA genes and 16S rRNA sequences provided clarity in the differentiation of phytoplasmas in ribosomal groups 16SrXI and 16SrXIV associated with sugarcane white leaf disease, Napier grass stunt, and Bermuda grass white leaf (Abeysinghe et al. 2016). Pusz-Bochenska et al. (2022) reported that 16S rRNA and groEL accurately determined phytoplasma group and subgroups while tuf was able to differentiate the two maize bushy stunt phytoplasma sequences from the canola samples. Recently, multilocus sequence typing up to 10 genes including rp, fusA, secY, tuf, secA, dnaK, rpoB, pyrG, gyrB, and ipt (Kong et al. 2022) and whole genome sequences were reported for sugarcane infecting phytoplasmas (Kirdat et al. 2023; Kirdat et al. 2020).
In Thailand, the genetic diversity of SCWL phytoplasma was studied based on RFLP of 1.35 bp of 16S rRNA covered the 3’ end of 16S rRNA, spacer region between the 16S rRNA and the tRNA (Ile) gene with enzyme HpaII and DNA sequencing which found sugarcane white leaf (SCWL) and sugarcane grassy shoot (SCGS) diseases associated with genetically different phytoplasma (Wongkaew et al. 1997). Later, multilocus sequence typing with leuS and secA indicated that SCWL and SCGS phytoplasmas were likely to be the same phytoplasma (Abeysinghe et al. 2016). This study aims to investigate the genetic diversity of sugarcane white leaf phytoplasmas in Thailand based on the sequences of 16S rRNA and ITS, and six housekeeping genes including tuf (encoding the elongation factor EF-Tu), secA (encoding translocation ATPase subunit), leuS (encoding leucine -tRNA ligase), AAA1 (encoding ATP-dependent Zn protease), gyrB (encoding DNA gyrase subunit B) and groES (encoding heat shock protein type 60).
Material and methods
Sample collection and genomic DNA extraction
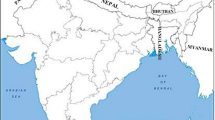
The sugarcane leaf and cane with white leaf or grassy shoot white leaf symptoms were collected from sugarcane fields during the years 2020 -2021. One hundred and seventy-four sugarcane samples were collected from 113 sugarcane fields in ten provinces of Thailand in northern (Kamphaeng Phet, Utai Thanee), central plain (Kanchanaburi, Phetchaburi and Prachuap Khiri Khan) and northeastern (Udon Thani, Kalasin, Mukdahan, Roi Et and Surin) Thailand (Fig. 1 and Table 1). Two hundred milligrams of mid-vein sugarcane leaves were cut into small pieces (0.2 cm by 0.5 cm), ground in liquid nitrogen and extracted with the DNeasy Plant Mini Kit (Qiagen, USA) following the company's protocol. The quality and quantity of DNA were determined using agarose gel electrophoresis and spectrophotometry, respectively. DNA was diluted to 100 ng/μL with water and stored at -20 °C until use.
Sugarcane white leaf collection map and 16SrXI-B and 16SrXI-D group distribution map, subgroup 16SrXI-B is represented by the green triangle, subgroup 16SrXI-D is represented by red dot. (modified from https://aseanup.com/wp-content/uploads/2018/05/thailand-administrative-divisions-2005.jpg)
DNA amplification
Seven phytoplasma genes (tuf, secA, leuS, AAA1, gyrB, groES, and 16S rRNA) were amplified using the primer sets reported in Table 2. The gene specific primers were manually designed based on consensus regions of each gene from SCGS phytoplasma (GenBank accession numbers NZ_VWXM01000000 and VWXM01000002). These regions were also aligned with ‘Ca. P. oryzae’ strain NGS-S10 (GenBank accession number NZ_JHUK01000000), strain Mbita1 (GenBank accession number NZ_LTBM01000000), and ‘Ca. P. cynodontis’ strain LW01 (GenBank accession number NZ_VWOH01000000). Only the sequences that were specific to SCGS phytoplasma sequences were selected and used for manual primer design. The selected sequences were analyzed using the Oligo Calc program (http://biotools.nubic.northwestern.edu/OligoCalc.html) and further validated for their specificity using the Blastn program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=GeoBlast&PAGE_TYPE=BlastSearch).
The PCR reaction was composed of the final concentration of the following: 1X AllTaq PCR buffer, 0.2 mM each of dNTPs, 0.2 µM of each primer (forward and reverse), 1.25 U of AllTaq DNA polymerase (Qiagen, USA), 100 ng of DNA template in a total volume 10 µl. The PCR cycling conditions were as follows: 93 °C for 3 min; 40 cycles of 93 °C for 15 sec, Tm °C for 30 sec, 68 °C for 2 min and a final extension of 68 °C for 5 min. Gene specific primer sequences and their annealing temperatures (Tm) were shown in Table 2. The amplification was visualized by agarose gel electrophoresis. The target gene PCR products were purified and directly sequenced. The PCR products that were larger than 1,000 bp were sequenced in both directions while those under 1,000 bp were sequenced with reverse primers twice without cloning using the Sanger sequencing method by 1st Base Malaysia - Apical Scientific Sdn Bhd (Selangor, Malaysia).
Multilocus sequence genotyping
The chromatogram of the target DNA fragment was manually analyzed for quality control. DNA sequences were trimmed and aligned using Clustal W algorithm (Thompson et al. 1994). The nucleotide sequences were concatenated and then translated into amino acid sequences. The DNA and amino acid sequences of the concatenated six genes were clustered using the neighbor-joining method (Saitou and Nei 1987). The pairwise distance and percentage of identity were calculated using the Tamura-Nei model (Tamura and Nei 1993). The analyses were performed using the MEGA 11 program (Tamura et al. 2021).
Virtual RFLP analysis of 16S rRNA gene sequence
The sequences of 16S rRNA of nine representative strains were virtually digested with 17 restriction enzymes and compared to the three members of ‘Ca. P. sacchari’ using the iPhyClassifier program available at https://plantpathology.ba.ars.usda.gov/cgi-bin/resource/iphyclassifier.cgi (Zhao et al. 2009).
Population structure analysis
The DNA sequences of six genes from 174 strains were aligned using Clustal W and variable positions in the sequences were identified. These variable sequences were converted into binary data using KK3-MDH9-2 as the reference strain. Nucleotides identical to the reference strain were represented as 0, while those that differed were represented as 1. Recombination strains were identified by population structure analysis using the Structure 2.3.4 software (Pritchard et al. 2000). The population structure was done by using admixture model applying the Markov Chain Monte Carlo (MCMC) estimation process which repeated at 100,000 generations while the burnin period was set to 50,000 iterations. We tested from K = 1 to 10 with five independent replicate runs per test. Pophelper (Francis 2017) was used to estimate K, which was calculated by the Evanno method.
Results
16S rRNA genotyping
The 1,520 bp long partial sequences of 16S rRNA and ITS nucleotides from 174 strains in this study (GenBank accession numbers OQ851109 – OQ851282) were analysed. They had a high identity to two strains of ‘Ca. P. sacchari’; SCWL1 phytoplasma strain (GenBank accession number CP115156.1 position 3311-4829) and the sugarcane grassy shoot phytoplasma (SCGS) (GenBank accession number VWXM01000011.1 position 14955-16473) at 99.74% to 99.93% and 99.80% to 100%, respectively and were classified as ‘Ca. P. sacchari’. The population showed four nucleotide variations in the 16S rRNA gene (at position 152, 812, 1137 and 1426) (Table 3).
In silico RFLP analysis of 16S rRNA sequences from the population was compared with ‘Ca. P. sacchari’ sequences (CP115156.1, VWXM01000011.1 and X76432) using 17 restriction enzymes. The results showed two distinct RFLP patterns, differing only in the HaeIII pattern (Supplement Table 1). The RFLP pattern of the first group was identical to the RFLP patterns of the three ‘Ca. P. sacchari’ sequences, with a similarity coefficient of 1.0, while the RFLP pattern of the second group had a similarity coefficient of 0.97 (Supplement Table 2). The HaeIII RFLP pattern of the SCWL from the first and the second groups were identical to RFLP patterns of 16SrXI-B and 16SrXI-D, respectively (Fig. 2), indicating that they belonged to their respective subgroups. The differences in HaeIII digestion pattern corresponded to the nucleotide variation (A/G) at position 152 in SCWL phytoplasma population (Table 3).
Virtual RFLP patterns generated from 16S rRNA genes from the representative strains in this study (LKKPP16, KK3KRI383, KK3MDH12, KK3MDH102, KK3MDH92, KK3KLS23, KK3KLS192, KK3SR22, KK3SR53) and reported strains (AB052874, FM208260, FM208259, KT270944, AB052874, FM208260, FM208259, KT270944, AB646271, KC295286 and KM280678) comparing with the HaeIII RFLP patterns of the 16SrXI subgroups. MW, ΦX174 DNA HaeIII marker
16SrXI-B subgroup comprised 40 SCWL strains (23% of the population) from Kalasin, Mukdahan, Roi Et and Surin Provinces. Strains from Surin Province were only in the 16SrXI-B subgroup while Kalasin, Mukdahan and Roi Et provinces contained strains from both subgroups (Fig. 1). Analysis of previously published SCWL sequences showed that SCWL strains previously collected from Thailand (GenBank accession numbers FM208260 and AB052874), India (GenBank accession numbers DQ459438 and DQ459439), Japan (GenBank accession number JX862179), China (GenBank accession numbers KR020690 and KR020691), SCWL (GenBank accession number X76432) and ‘Ca. P. sacchari’ strain SCGS and SCWL1 (GenBank accession number VWXM01000011 and CP115156) also clustered into the 16SrXI-B subgroup (Fig. 3). The members of this subgroup had a sequence identity of 99.00% to 100%.
Phylogenetic tree reconstructed with the MEGA 11 program with the neighbour-joining method and the evolutionary distances with the Tamura-Nei method using the 16S rRNA gene sequence (1,241 bp) of strains from this study (name starts with KK3 and LK, red color) with those of other 16SrXI group phytoplasmas and selected strains of sugarcane white leaf phytoplasma collected in Asian countries published in Genbank. 16S rRNA sequence from Bacillus subtilis was used as an outgroup. Numbers on branches indicate bootstrap values based on 1,000 replicates (values less than 50 are not shown)
16SrXI-D subgroup consisted of 134 SCWL strains (77 % of the population) from all collected sites except those from Surin Province. Additional analysis of previously reported sequences showed that SCWL strains from Thailand (GenBank accession number FM208259), Vietnam (GenBank accession numbers KC295286 and KM280678), Myanmar (GenBank accession number AB646271), Laos (GenBank accession numbers KT270944) and China (GenBank accession numbers KR020685, KR020686, KR020687, KR020688) had a sequence identity of 99.50% to 100% (Fig. 3).
Multilocus sequence genotyping (MLST)
The partial sequences of six phytoplasma genes (3,672 bp in length) including tuf, secA, leuS, gryB, AAA1 and groES in SCWL phytoplasma population were analysed. Their had nucleotide variations of 1.66% (11/664), 1.54% (5/325), 1.04% (7/675), 0.87% (8/929), 1.04% (9/869) and 1.91% (4/210), respectively. At the amino acid level, the variations were 4, 1, 3, 3 and 2 amino acids in tuf, leuS, gyrB, AAA1 and groES, respectively. The amino acid change was not observed in the secA gene. The alterations of amino acid side chain properties were observed in tuf, leuS, gyrB and groES such as negative charge side chains to polar uncharged side chains (D to N in tuf and N to D in leuS) and positive charged side chains to hydrophobic side chains (K to I in gyrB) (Table 3).
Concatenated partial gene sequences of tuf, secA, leuS, gyrB, AAA1 and groES of 174 SCWL were compared to selected phytoplasma sequences and grouped into two populations represented by each color in Supplement Figure 1. The members of each population were as in the 16S rRNA subgroups (16SrXI-B and 16SrXI-D). The nucleotide and amino acid sequences of representative strains from each population were aligned with the sequences of reported SCWL phytoplasma strains and other phytoplasmas (‘Ca. P. cynodontis’, ‘Ca. P. oryzae’ and ‘Ca. P. luffae’). Subgroup 16SrXI-B phytoplasma clustered with two strains of ‘Ca. P. sacchari’: SCWL1 (GenBank accession number CP115156) and SCGS (GenBank accession number VWXM01000000) and had a nucleotide identity of 99.39% to 100% and an amino acid identity of 99.18% to 100%, while subgroup 16SrXI-D phytoplasma had a nucleotide identity of 98.77% to 100% and an amino acid identity of 98.77% to 100% (Fig. 4A and B).
Phylogenetic tree of concatenated partial gene sequences with a total length of 3,765 bp (A) and 1,254 AA (B) of tuf, secA, leuS, gyrB, AAA1 and groES from selected SCWL phytoplasma strains in this study (name starts with KK3 and LK) and sequences from selected phytoplasma genomes in GenBank database using the MEGA 11 program with the neighbour-joining method and Tamura-Nei evolutionary distances. Numbers on branches indicate bootstrap values based on 1,000 replicates (values less than 50 are not shown)
Population structure analysis
The population structure analysis of SCWL 174 strains based on nucleotide variation of six genes was analysed. The 44 nucleotides were used as genetic markers. At K=2, inferred using the Evanno method, which identified the most probable number of subpopulations, has shown two subpopulations as shown by different colors; 39 strains in dark blue (subpopulation I), 135 strains in light blue (subpopulation II) (Fig. 5), which consistent result with multilocus sequence type analysis (MLST) result (Supplement Figure 1). The mixed color in each column represented the recombination of sequence patterns in each subpopulation. In subpopulation I, three strains including KK3-SR 5-1, KK3-KLS 19-2 and KK3-SR 8-1 strains had the recombination in groES, secA and tuf, respectively. In subpopulation II, KK3-RE 15-3 strain had the recombination only in leuS while KK3-SR 5-3 strain exhibited recombination in secA, leuS, and AAA1. In addition, the KK3-KLS8-2 and KK3-KLS42-1 were found point mutation in leuS and AAA1, respectively, and CSB09-31-UD3-3 strain was found point mutation in leuS and AAA1 (Fig. 5 and Supplemental Figure 1). Notably, changes in the amino acid sequence of secA, leuS, and AAA1 were only observed in the KK3-SR 5-3 strain. Consequently, the KK3-SR 5-3 strain, which was classified into the 16SrXI-B subgroup based on virtual RFLP analysis (Figure 2), was clustered in another clade based on multilocus sequence analysis (Supplemental Figure 1).
A box plot visualizing the population structure of 174 SCWL strains based on 44 nucleotide variations within concatenated partial sequences of the tuf, secA, leuS, gyrB, AAA1, and groES genes generated by using Structure 2.3.4 software. The admixture model was employed with the Markov Chain Monte Carlo (MCMC) estimation process, repeated for 100,000 generations with a burn-in period of 50,000 iterations. The Evanno method inferred K=2 as the most probable number of subpopulation. Recombinant populations were represented by a mixture of colors within each column
Discussion
The 16S rRNA sequences of SCWL phytoplasma from Thailand were reported previously, including Udon thani (UD) strain (GenBank accession number AB052874) (Jung et al. 2003) and six were directly submitted (GenBank accession number FM208255 to FM208260) in 2008. Previously reported strains were found in both subgroups, as indicated by the RFLP analysis. The results of the present survey indicated that both subgroups were found with 16SrXI-D as the major subgroup covering 134/174 (77%) strains. The 16SrXI-D group has also been detected in China (Zhang et al. 2023), Myanmar (GenBank accession number AB646271), Laos (GenBank accession number KT270944) and Vietnam (GenBank accession numbers KC295286 and KM280678). However, the 16SrXI-B subgroup was found to be prevalent in China (Zhang et al. 2023) and Sri Lanka (Dayasena et al. 2021).
The prevalence of the 16SrXI-D subgroup in Thailand may be linked to the sugarcane variety. Diverse phytoplasma strains have been reported to be associated with different plant host genotypes such as in chrysanthemum (Taloh et al. 2020) and rose genotypes in India (Rihne et al. 2021). In this study ‘Khon Kaen 3’ or ‘KK3’ hybrid variety (clones 85-2-352 x K84-200) was developed between 1995 and 2007 and approved as a sugarcane variety in 2008. Although this variety was originally developed for cultivation in the Northeast region (Ponragdee et al. 2011), it was distributed and grown in 35% of sugarcane planted areas in Thailand in 2011 (Office of the Cane and Sugar Board 2011). The adoption of “KK3” steadily increased to 53% in 2014, 86% in 2018 and 90% in 2020 (Field and Renewable Energy Crops Research Institute 2022), establishing it as the predominant variety cultivated in Thailand for at least a decade. Although phytoplasma can be transmitted by insect vectors and infected cane setts, the latter is prevalent for long distance spread (Wongkaew 2012; Zhang et al. 2020). Treatments such as hot water treatment were implemented to reduce the spread of SCWL disease (Hanboonsong et al. 2021; Sushil et al. 2022). These treatments, however, were proven to be infective as a high concentration of SCWL phytoplasma was still detected in sugarcane cuttings after treatments (Kaewmanee and Hanboonsong 2011).
In this study, recombinant DNA between the two subgroups was found in five strains. Interspecies or intraspecies recombination is common in bacteria populations. Recombination has been reported in ‘Ca. P. pyri’, which had recombinant of some genes with ‘Ca. P. prunorum’ (Danet et al. 2011) and in Xanthomonas euvesicatoria pv. euvesicatoria and pv. perforans strains (Jibrin et al. 2018).
In conclusion, the genetic diversity of SCWL phytoplasma population in Thailand differentiates the Thai strains into two subgroups: 16SrXI-B and 16SrXI-D. The 16SrXI-D group was the predominant population may be due to changes in sugarcane varieties. Infected sugarcane setts are the main factor in the distribution and movement of phytoplasma and they need to be attentioned on for disease control. In addition, multilocus sequence typing (MLST) and bioinformatic analysis provided additional information about genetic recombination, as seen in tuf, secA, leuS, AAA1 and groES. Extensive genetic diversity studies of SCWL phytoplasma in Thailand using multilocus sequence typing will help to develop more focused detection methods for monitoring and consequently disease management.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Abeysinghe S, Abeysinghe PD, Kanatiwela-de Silva C, Udagama P, Warawichanee K, Aljafar N, Kawicha P, Dickinson M (2016) Refinement of the taxonomic structure of 16SrXI and 16SrXIV phytoplasmas of gramineous plants using multilocus sequence typing. Plant Disease 100:2001–2010
Al-Abadi SY, Al-Sadi AM, Dickinson M, Al-Hammadi MS, Al-Shariqi R, Al-Yahyai RA, Kazerooni EA, Bertaccini A (2016) Population genetic analysis reveals a low level of genetic diversity of ‘Candidatus Phytoplasma aurantifolia’causing witches’ broom disease in lime. SpringerPlus 5:1–11
Bertaccini A, Arocha-Rosete Y, Contaldo N, Duduk B, Fiore N, Montano HG, Kube M, Kuo C-H, Martini M, Oshima K (2022) Revision of the ‘Candidatus Phytoplasma’species description guidelines. International Journal of Systematic and Evolutionary Microbiology 72:005353
Danet JL, Balakishiyeva G, Cimerman A, Sauvion N, Marie-Jeanne V, Labonne G, Lavinˇa A, Batlle A, Krizanac I, Škorić D, Ermacora P, Serçe ÇU, Çağlayan K, Jarausch W, Foissac X (2011) Multilocus sequence analysis reveals the genetic diversity of European fruit tree phytoplasmas and supports the existence of inter-species recombination. Microbiology 157:438–450
Dayasena Y, Panda P, Thushari A, Rao G (2021) Geographical distribution and identification of phytoplasma strain associated with sugarcane white leaf disease in Sri Lanka. Sugar Tech 23:1351–1358
Field and Renewable Energy Crops Research Institute (2022) Sugarcane Breeding Manual. Field and Renewable Energy Crops Research Institute. Department of Agriculture, Thailand
Francis RM (2017) Pophelper: an R package and web app to analyse and visualize population structure. Molecular Ecology Resources 17:27–32
Hanboonsong Y, Wangkeeree J, Kobori Y (2017) Integrated management of the vectors of sugarcane white leaf disease in Thailand: an update. International Sugar Journal 119:220–223
Hanboonsong Y, Sakuanrungsirikul S, Saengsai W, Kumhong S, Pituk S, Hamarn T, Kobori Y (2021) Healthy seedcane propagation and distribution manual against sugarcane white leaf disease. Office of the Cane and Sugar Board, Ministry of Industry, Thailand. Available at: https://old.ocsb.go.th/upload/journal/fileupload/12686-3399.pdf. Accessed 5 Dec 2023
IRPCM (2004) ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. International Journal of Systematic and Evolutionary Microbiology 54:1243–1255
Jibrin MO, Potnis N, Timilsina S, Minsavage GV, Vallad GE, Roberts PD, Jones JB, Goss EM (2018) Genomic inference of recombination-mediated evolution in Xanthomonas euvesicatoria and X. perforans. Applied and Environmental Microbiology 84:e00136-00118
Jung H-Y, Sawayanagi T, Wongkaew P, Kakizawa S, Nishigawa H, Wei W, Oshima K, Miyata S-i, Ugaki M, Hibi T (2003) ‘Candidatus Phytoplasma oryzae’, a novel phytoplasma taxon associated with rice yellow dwarf disease. International Journal of Systematic and Evolutionary Microbiology 53:1925–1929
Kaewmanee C, Hanboonsong Y (2011) Evaluation of the efficiency of various treatments used for sugarcane white leaf phytoplasma control. Bulletin of Insectology 64:S197–S198
Kirdat K, Tiwarekar B, Thorat V, Narawade N, Dhotre D, Sathe S, Shouche Y, Yadav A (2020) Draft genome sequences of two phytoplasma strains associated with sugarcane grassy shoot (SCGS) and bermuda grass white leaf (BGWL) diseases. Molecular Plant-Microbe Interactions 33:715–717
Kirdat K, Tiwarekar B, Sathe S, Yadav A (2023) Complete genome sequences of phytoplasma strains in group 16SrII associated with Parthenium phyllody in India. Phytopathogenic Mollicutes 13:23–24
Kong D-Z, Lin C-L, Yu S-S, Tian G-Z, Ma H-B, Wang S-J (2022) Molecular diversity and evolutionary relatedness of Paulownia witches’-broom phytoplasma in different geographical distributions in China. Biology 11:1611
Lee I-M, Dawn EG-R, Davis RE, Bartoszyk IM (1998) Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. International Journal of Systematic Bacteriology 48:1153–1169
Li W-F, Wang X-Y, Huang Y-K, Shen K, Shan H-L, Luo Z-M, Yin J, Jia Y-M, Bai Z-G, Zhang R-Y (2013) First report of sugarcane white leaf phytoplasma in Yunnan province, China. Canadian Journal of Plant Pathology 35:407–410
Marcone C (2011) Sugarcane white leaf phytoplasma (white leaf of sugarcane). CABI compendium, CABI Digital Library. Available at: https://www.cabi.org/isc/datasheet/49804. Accessed 18 Aug 2022
OECD/FAO (2021) OECD-FAO Agricultural Outlook 2021–2030. OECD Publishing, Paris
Office of the Cane and Sugar Board (2011) Sugarcane planted area in producing year 2010/2011. Office of the Cane and Sugar Board. Ministry of Industry, Thailand
Ponragdee W, Sansayawichi T, Sarawat P, Moulanon T, Kapetch P, Leabwon U (2011) Khon Kaen 3 a sugarcane cultivar for the northeast. Thai Agricultural Research Journal 29:283–301
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pusz-Bochenska K, Perez-Lopez E, Wist TJ, Bennypaul H, Sanderson D, Green M, Dumonceaux TJ (2022) Multilocus sequence typing of diverse phytoplasmas using hybridization probe-based sequence capture provides high resolution strain differentiation. Frontiers in Microbiology 13:959562
Rihne T, Namita Singh KP, Singh M, Talukdar A, Rao G (2021) Multilocus gene typing, mixed infection of phytoplasma strains associated with rose genotypes and confirmation of their natural reservoir sources. Tropical Plant Pathology 46:596–607
Šafářová D, Zemánek T, Válová P, Navrátil M (2016) ‘Candidatus Phytoplasma cirsii’, a novel taxon from creeping thistle [Cirsium arvense (L.) Scop.]. International Journal of Systematic and Evolutionary Microbiology 66:1745–1753
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4:406–425
Sushil S, Joshi D, Roy S, Rao G, Pathak A (2022) Plant quarantine regulations with reference to sugarcane in India: strengths and challenges. Sugar Tech 24:1319–1329
Taloh A, Raju D, Banyal N, Kumar G, Panda P, Manimekalai R, Marcone C, Rao G (2020) Genetic diversity of phytoplasma strains infecting chrysanthemum varieties in India and their possible natural reservoirs. 3 Biotech 10:1–13
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512–526
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 38:3022–3027
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22:4673–4680
Trivellone V, Cao Y, Dietrich CH (2022) Comparison of traditional and next-generation approaches for uncovering phytoplasma diversity, with discovery of new groups. Subgroups and potential vectors 11:977
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology 173:697–703
Wongkaew P (2012) Sugarcane white leaf disease characterization, diagnosis development, and control strategies. Functinal Plant Science and Biotechnology 6:73–84
Wongkaew P, Hanboonsong Y, Sirithorn P, Choosai C, Boonkrong S, Tinnangwattana T, Kitchareonpanya R, Damak S (1997) Differentiation of phytoplasmas associated with sugarcane and gramineous weed white leaf disease and sugarcane grassy shoot disease by RFLP and sequencing. Theoretical and Applied Genetics 95:660–663
Yadav A, Thorat V, Deokule S, Shouche Y, Prasad DT (2017) New subgroup 16SrXI-F phytoplasma strain associated with sugarcane grassy shoot (SCGS) disease in India. International Journal of Systematic and Evolutionary Microbiology 67:374–378
Zhang R-Y, Li W-F, Huang Y-K, Wang X-Y, Shan H-L, Luo Z-M, Yin J (2016) Group 16SrXI phytoplasma strains, including subgroup 16SrXI-B and a new subgroup, 16SrXI-D, are associated with sugar cane white leaf. International Journal of Systematic and Evolutionary Microbiology 66:487–491
Zhang R-Y, Shan H-L, Huang Y-K, Wang X-Y, Li J, Li W-F, Cang X-Y, Yin J, Luo Z-M (2020) Survey of incidence and nested PCR detection of sugarcane white leaf in different varieties. Plant Disease 104:2665–2668
Zhang R-Y, Wang X-Y, Shan H-L, Li J, Li Y-H, Li W-F, He X-H, Huang Y-K (2023) Multilocus sequence typing reveals two distinct populations of 'Candidatus Phytoplasma sacchari’ in China. Tropical Plant Pathology 48:199–206
Zhao Y, Wei W, Lee I-M, Shao J, Suo X, Davis RE (2009) Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). International Journal of Systematic and Evolutionary Microbiology 59:2582–2593
Acknowledgments
This work was funding supported by National Research Council of Thailand (NRCT), 2020 under the project “Genetic diversity of sugarcane white leaf phytoplasma and development for accurate and rapid detection of sugarcane white leaf phytoplasma” and the laboratory facilities were supported by the Center of Excellence on Agricultural Biotechnology (AG-BIO/PERDO-MHESI) and Center for Agricultural Biotechnology, Kasetsart University.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by Patchara Chaphakdee, Wuttitharuk Boonsonti and Pimpilai Saengmanee. The data analysis, first draft and revised version of the manuscript were written by Jutatape Watcharachaiyakup, Parichart Burns and Wichai kositratana and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Research Notes
Nucleotide sequences of genes obtained from this study were deposited into GenBank database. Their accession numbers are as follows: tuf (OQ805871 - OQ806044), secA (OQ806045 - OQ806218), leuS (OQ806219 - OQ806392), gyrB (OQ806393 - OQ806566), AAA1 (OQ806567 - OQ806740), groES (OQ806741 - OQ806914) and 16S rRNA and ITS (OQ851109 – OQ851282).
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Watcharachaiyakup, J., Burns, P., Chaphakdee, P. et al. Multilocus genotyping of sugarcane white leaf phytoplasma in Thailand. Trop. plant pathol. 49, 346–356 (2024). https://doi.org/10.1007/s40858-024-00635-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-024-00635-7