Abstract
Symptoms of leaf roll, swollen nodes, flat branch and witches’ broom were observed in five cultivars of sweet cherry from Srinagar, Jammu and Kashmir province, India, during 2019–2021. Phytoplasmas association were confirmed by amplifying 16S rRNA, secA, rp, tuf and secY genes with phytoplasma-specific primers in all symptomatic sweet cherry cultivars in nested PCR assays. Pairwise sequence comparison, phylogeny and virtual RFLP (16S rRNA gene) analyses confirmed the presence of ‘Candidatus Phytoplasma asteris’ and ‘Ca. P. trifolii’ strains in the sweet cherry samples. The incidence of flat branch and witches’ broom symptoms associated with ‘Ca. P. trifolii’ varied from 5.8 to 25% in cultivars Bigarreau Nepoleon (Double), Bigarreau Noir Grossa and CITH-Cherry-9. However, incidence of leaf rolling, swollen nodes and bud proliferation associated with ‘Ca. P. asteris’ was recorded 7.5% in cultivar Stella and 10% in Sunburst, respectively, in the surveyed area. The multigene characterization of sweet cherry phytoplasma strains confirmed the validity of these molecular markers for identification of phytoplasmas enclosed in 16SrI and 16SrVI groups. The presence of phytoplasmas in sweet cherry is the first report from India.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cherry trees occupy an important position among temperate fruit crops all over the world with a high nutritional value and abundant phenolic compounds which contribute greatly to health benefits (Hu et al. 2021). Cherry trees are native to European and Asian regions, and the leading producers are Turkey, the European Union and China. (FAOSTAT 2020). The sweet cherry (Prunus avium L.) is the popularly cultivated commercial type followed by sour cherry (P. cerasus L.), which is used mainly for cooking purpose (Blando and Dave Oomah 2019). Cherry trees require high altitudes and temperate climate to grow, and in India, they are commercially cultivated in Jammu and Kashmir, Uttarakhand and Himachal Pradesh provinces. The sweet cherry production is mainly hindered by several diseases such as cherry leaf spot, bacterial canker, powdery mildew, Coryneum blight, prunus dwarf virus (PDV), prunus ringspot virus (PNRV) and European stone fruit yellows (Sholberg and Kappel 2008; Fiore et al. 2018).
‘Candidatus Phytoplasma’ species are cell wall-less prokaryotes which colonize both plants and insects (IRPCM 2004). The phytoplasmas are mainly classified on the basis of 16S rRNA gene sequence restriction fragment length polymorphism and classified into 35 ribosomal groups, nearly 300 subgroups and 49 ‘Candidatus Phytoplasma’ species (Lee et al. 1998; IRPCM, 2004; Bertaccini 2022; Bertaccini and Lee 2018).
So far, six groups of phytoplasmas (16SrI, 16SrII, 16SrIII, 16SrV, 16SrX and 16SrXII) were reported on cherry from all over the world (Table 1).
In India, phytoplasma strains association have been documented in some stone fruit crops such as peach, plum and apricot, but no report is available so far in sweet cherry cultivars (Rao 2021; Shreenath et al. 2022). In recent years, several phytoplasma-suspected symptoms of witches’ broom, swollen nodes, flat branches and leaf roll were observed on sweet cherry cultivars at ICAR-Central Institute of Temperate Horticulture (ICAR-CITH), Srinagar, Jammu and Kashmir (J and K), India, and was verified for phytoplasmas association using multilocus gene sequence analysis.
Materials and methods
Collection of cherry samples and disease incidence
Survey of cherry fruit orchards was carried out at ICAR-Central Institute of Temperate Horticulture (ICAR-CITH), Srinagar, J and K, from 2019 to 2021 (Fig. 1). An area of 0.5 ha in three cherry orchards was randomly selected for calculating the disease incidence in the surveyed orchards on the basis of symptomatic vs asymptomatic cherry trees. The average disease incidence was calculated with formula: C-T/C × 100, where C is the number of asymptomatic trees and T is the number of symptomatic trees. The phytoplasma-suspected symptomatic leaves and branch samples from ten symptomatic and two asymptomatic cherry plants of five cultivars [Bigarreau Nepoleon (BN), Bigarreau Noir Grossa (BNG), CITH-Cherry-9, Stella and Sunburst] were collected for DNA extraction and PCR analysis.
Extraction of nucleic acids and PCR assays
Total DNA was extracted from the leaf midrib or phloem tissues of young growing symptomatic shoot branch of the cherry tree and the positive phytoplasma controls (brinjal little leaf, ‘Ca. P. trifolii,’ GenBank accession number KX689234, and sesame phyllody, ‘Ca. P. asteris,’ GenBank accession number KC920747) maintained in periwinkle using the CTAB protocol (Ahrens and Seemüller 1992). The extracted DNA was subjected to nested PCR assay with universal phytoplasma-specific primers, P1/P7 (Deng and Hiruki 1991; Schneider et al. 1995) followed by 3For/3Rev (Manimekalai et al. 2010) and/or R16F2n/R2 primer pairs (Gundersen and Lee 1996). The PCR assays were carried out in a final reaction volume of 25 μl containing of nuclease-free water (Sisco Research Laboratories Pvt. Ltd., India), OnePCR™2X PCR Master Mix (GeneDireX, Taiwan), for/rev primer 10 pmol/μl (0.2 μM) and the DNA template (= 100 ng). PCRs were performed in a thermal cycler (Mastercycler, Eppendorf, Hamburg, Germany). Reaction mix without DNA was used as negative control.
For finer differentiation of phytoplasmas, ribosomal protein (rps3, rps19 and rpl22) and secA, secY and tuf genes were analyzed. The rpF1/rpR1 followed by rp (I) F1A/rp (I) R1A primers specific to 16SrI group were used to amplify rp gene (Lim and Sears 1992; Lee et al. 2003). Similarly, primer pairs rpF1C/rp (I) R1A followed by rp (VI) F2/rp (VI) R2 specific to 16SrVI group were used to amplify rp gene (Martini et al. 2007). The secA gene amplification was carried out in semi-nested PCR assay with primer pairs SecAfor1/SecArev3 as earlier described (Hodgetts et al. 2008). The amplification of secY gene was performed in semi-nested PCR assay with primer pairs SecYF1 (VI)/SecYR1(VI) followed by SecYF2 (VI)/SecYR1(VI) using PCR cycling parameters reported by Lee et al. (2010). Further, fTuf1/rTuf1 followed by fTufAy/rTufAy primer pairs were used to amplify tuf gene in cherry samples (Schneider et al. 1997). The amplified products of PCR assays were diluted 1:20 with nuclease-free water and then utilized as template in the nested PCR assays. Six microliters of nested PCR products was electrophoresed in a 1.0% (w/v) agarose gel, stained with GoodView™ nucleic acid stain and observed under Gel Doc (Azure Biosystems, USA). The amplified 16S rRNA, secA, rp, secY and tuf gene fragments were purified using the WizardR SV Gel and PCR Cleanup System (Promega, Madison, USA).
Sequence analysis
The purified PCR products of 16Sr RNA and multigenes were further ligated to TA plasmid vector using pGEM®T Easy Vector Kit (Promega, Madison, USA) and cloned in competent Escherichia coli (DH5-α) cells. The cloned products were outsourced for sequencing using M13F/M13R universal primer pair in both directions at Eurofins Genomics India Pvt. Ltd., Bengaluru, Karnataka, India. Pairwise BLAST analysis of sequence was performed using the similar sequences from NCBI database. The sequence data of each gene were edited and assembled using Qiagen CLC Sequence viewer 11 (https://digitalinsights.qiagen.com). Multiple alignments were performed using ClustalW software (Thompson et al. 1994), and the sequences generated in the study were submitted in the NCBI database.
Phylogenetic trees were constructed by neighbor-joining method using MEGA 7.0 (Kumar et al. 2016) with 1000 bootstrap values. The sequences of Acholeplasma laidlawii (GenBank accession Number NR074448) for 16S rRNA gene, A. oculi (GenBank accession Number LK028559) for secY and rp genes and Bacillus subtilis (GenBank accession Number D10279) for secA and tuf genes, respectively, were used as out-groups to root the phylogeny trees.
Virtual RFLP analysis
About ~ 1.25 kb and ~ 1.3 kb sequences of 16S rDNA fragment (corresponding to the R16F2n/R2 and 3For/3Rev, respectively) of cherry phytoplasma strains were subjected to in silico RFLP comparison analysis using the iPhyClassifier and pDRAW32 program, respectively (http://www.acaclone.com). The computer-generated restriction patterns were compared, and the similarity coefficient value was calculated for the identification of respective phytoplasma subgroup strains (Zhao et al. 2009).
Results
Symptomatology and incidence
Leaf rolling, swollen nodes, bud proliferation and malformation symptoms were recorded in the cherry cultivars Stella and Sunburst with an incidence of about 7.5% to 10% at ICAR-CITH, Srinagar during May–September 2019 – 2021 (Fig. 2a,–c). Witches’ broom and flat branch with swollen node symptoms were observed in cvs Bigarreau Nepoleon (BN), Bigarreau Noir Grossa (BNG), and CITH-Cherry-9 with disease incidence varied from 5.8 to 25% in cherry orchards (Fig. 2d–f; Table 2).
Phytoplasma disease symptoms in sweet cherry cultivars: leaf rolling in cv Stella (a); swollen nodes, bud proliferation and malformation in cv Sunburst (b, c); witches’ broom with flat branch in cv Bigarreau Nepoleon (BN) (d); flat braches in cv BN (1,2); cv Bigarreau Noir Grossa (BNG) (3,4); e flat branch in young cherry plant cv CITH-Cherry-9 at CITH, Srinagar, Jammu and Kashmir
Molecular detection and identification of phytoplasma
PCR assay with the P1/P7 primer pair did not yield any amplification in the gel electrophoresis from DNA templates extracted from any of the symptomatic cherry samples. However, about ~ 1.3 kb amplicons were amplified from the three symptomatic sweet cherry cvs BN, BNG and CITH-Cherry-9, and ~ 1.25 kb amplicons were seen in two cherry cvs Stella and Sunburst in nested PCR assays with the primers 3For/3Rev and R16F2n/R2, respectively (data not shown). Faint bands of ~ 1.2 kb were also amplified with R16F2n/R2 nested primers from sweet cherry cvs BN, BNG and CITH-Cherry-9 (data not shown). The faint amplification products visualized in gels by employing 3Far/3Rev primer pair in two cvs Stella and Sunburst were attempted to clone but failed to get good cloned products (data not shown). Hence, the 3Far/3Rev amplified products of three cherry cvs BN, BNG and CITH-Cherry-9 and R16F2n/R2 amplified products of two cherry cvs Stella and Sunburst were further processed for cloning and sequencing in the study. Further, no amplifications were obtained from any of the asymptomatic cherry samples from five tested cultivars in the nested PCR assays (data not shown).
Amplification of DNAs from symptomatic cherry cultivars was also performed using two sets of rp gene-specific primers. The products of ~ 1000 bp were consistently obtained with symptomatic cherry cvs BN, BNG and CITH-Cherry-9 and the positive control (brinjal little leaf, 16SrVI group) in the nested PCR assays with 16SrVI group-specific rp gene primer pairs. Similarly, with another set of rp gene primer pairs specific to 16SrI group, rp(I)F1A/rp (I)R1A, an amplicon of ~ 1.2 kb was obtained in the symptomatic cherry cvs Stella and Sunburst along with the positive control (sesame phyllody, 16SrI group) in nested PCR assays. An amplified product of ~ 1.7 kb was obtained in three cherry cvs (BN, BNG and CITH-Cherry-9) with SecYF2 (VI)/SecYR1 (VI) primer specific to 16SrVI group in the semi-nested PCR assay.
In another set of experiment, ~ 480 bp amplicons were observed only in samples of two symptomatic cherry cvs Stella and Sunburst with secA gene universal phytoplasma-specific primer pair SecAfor2/SecArev3 in semi-nested PCR assays. However, no amplicons were achieved with rest of the three cherry cultivars. With tuf gene, the phytoplasma amplicons of ~ 940 bp were achieved in the nested PCR assay by using fTufAy/rTufAy primers specific to 16SrI group in two symptomatic cherry samples (cvs Stella and Sunburst) and in the positive control (Ca. P. asteris, GenBank accession number KC920747). Further, no amplification was obtained by nested or semi-nested PCR assay from any of the asymptomatic cherry (control) samples with rp, secY, secA and tuf gene-specific primers used in the study (data not shown).
The amplified PCR products were cloned, sequenced and analyzed, and the partial 16S rRNA, tuf, secA, secY and rp gene sequences were submitted in the GenBank (Table 2).
Sequence analysis
BLASTn sequence identity search and pairwise comparison of ~ 1.3 kb amplicons of 16S rRNA gene sequences of sweet cherry phytoplasma strains (cv. BN, GenBank accession numbers OM019094 and OM019095; cv. BNG, GenBank accession numbers OM019096 and OM019097 and cv. CITH-Cherry-9, Acc. Nos. OM019098 and OM019099) shared 99.92–100% sequence identity with the 16SrVI-D subgroup reference strain of periwinkle little leaf (GenBank accession number AF228053) as well as with the earlier identified phytoplasma strains of brinjal little leaf (GenBank accession number KX284698) and Datura stramonium witches’ broom (GenBank accession numbers KY078230) belonging to ‘Ca. P. trifolii.’ Sweet cherry phytoplasma strains (cv. Stella, GenBank accession numbers OP093761 and OP093762) and sweet cherry phytoplasma strains (cv. Sunburst, GenBank accession numbers OP093763 and OP093764) had 99.60–100% 16S rRNA gene sequence identity with the 16SrI-B subgroup reference strain of Oenothera phytoplasma (GenBank accession number M30790) as well as with the earlier identified phytoplasma strains of rapeseed phyllody (GenBank accession number CP055264), sapota flat stem little leaf (GenBank accession number MK271071) and pineapple shoot proliferation (GenBank accession number MK209105) belonging to ‘Ca. P. asteris.’
Sequence comparison of ~ 1700 bp amplicons of secY gene of sweet cherry phytoplasma strains (BN, BNG and CITH-Cherry-9) showed 99.84–100% sequence identity with Cannabis sativa little leaf phytoplasma (GenBank accession number KU297165), Catharanthus roseus phytoplasma (GenBank accession number MW654230) and brinjal little leaf phytoplasma (GenBank accession number KT970077) related to ‘Ca. P. trifolii’ strains.
Sequence comparison of ~ 1000 bp amplicons of partial rp gene (specific to 16SrVI group) of sweet cherry phytoplasma strains (BN, BNG and CITH-Cherry-9) showed 99.58–100% sequence similarity with phytoplasma strains: potato witches’ broom (AY197683) and brinjal little leaf (GenBank accession number EF183489) belonging to ‘Ca. P. trifolii.’ However, pairwise sequence comparison of ~ 1200 bp of partial rp gene (specific to 16SrI group) of sweet cherry phytoplasma strains (cvs Stella and Sunburst) shared 99.56 to 99.65% sequence identity with C. roseus aster yellows (GenBank accession number CP035949) and bougainvillea shoot proliferation (GenBank accession number MN477308, MN477310) all related to ‘Ca. P. asteris.’
Sequence comparison of ~ 480 bp amplicons of partial secA gene of sweet cherry phytoplasma strains (cvs Stella and Sunburst) shared 99.40–100% sequence identity with rapeseed phyllody (GenBank accession number CP055264) and sesame phyllody (GenBank accession number JN977032) phytoplasma strains of ‘Ca. P. asteris’ (16SrI group). Similarly, sequence comparison of ~ 940 bp amplicons of partial tuf gene of sweet cherry phytoplasma strains (cvs Stella and Sunburst) shared 97.47% to 97.57% sequence identity with rapeseed phyllody (GenBank accession number CP055264) and aster yellows (GenBank accession number MN526022) strains of ‘Ca. P. asteris.’
Phylogenetic and virtual RFLP analysis
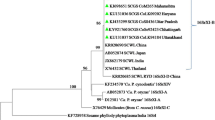
Phylogenetic analysis confirmed that the sequences of the amplified genes (16S rRNA, secA, secY, rp and tuf) of the phytoplasma strains from sweet cherry clustered with sequences of ‘Ca. P. trifolii’ (16SrVI-D subgroup; cvs. BN, BNG and CITH-Cherry-9) and ‘Ca. P. asteris’ (16SrI-B subgroup; cvs Stella and Sunburst) (Figs. 3, 4, 5, 6 and 7). Virtual RFLP analysis of the 16S rDNA of phytoplasma strains from symptomatic sweet cherry cultivars (BN, BNG and CITH-Cherry-9) was performed for assigning the 16Sr subgroup classification. Amplicons of ~ 1.3 kb of 16S rRNA genes of sweet cherry phytoplasmas digested with 17 restriction endonucleases yielded profiles referable to phytoplasma strains belonging to 16SrVI-D subgroup (GenBank accession number AF228053) in virtual RFLP analysis (Fig. 8a–d). Similarly virtual RFLP analysis results of the 16S rDNA gene fragments of cherry cvs Stella and Sunburst phytoplasma strains showed identical restriction profiles with Oenothera phytoplasma reference strain belonging to 16SrI-B with a similarity coefficient of 1.0 (sweet cherry, CSR-1 and CRR-1, Acc. No. OP093761 and OP093763 with reference strain Acc. No. M30790; Fig. 9a–c).
Phylogenetic tree constructed by neighbor-joining method based on 16SrDNA gene sequences of sweet cherry phytoplasma strains with other selected phytoplasma strains from GenBank. Accession numbers are specified in the tree. A. laidlawii was used as an out group. Numbers on branches are bootstrap values obtained for 1000 replicates
Comparison of virtual RFLP patterns generated from in silico digestion of 1.3 kb 16SrDNA sequences of a reference strain (periwinkle little leaf phytoplasma, GenBank accession number AF228053), b sweet cherry (cv Double) phytoplasma strain-1 (GenBank accession number OM019094), c sweet cherry (cv Mishri) phytoplasma strain-1 (GenBank accession number OM019096), d sweet cherry (cv CITH-Cherry-9) phytoplasma strain-1 (GenBank accession number OM019098) digested using 17 different endonucleases (AluI, BamHI, BfaI, BstUI, DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, MboI, MseI, RsaI, SspI and TaqI), indicating that sweet cherry cvs belonged to 16SrVI-D phytoplasma subgroup
Virtual RFLP patterns from in silico digestion of 16SrDNA R16F2n/R2 fragments of phytoplasma strains infecting sweet cherry cvs Stella and Sunburst in India. The patterns are compared for 16SrI-B reference strain (GenBank accession number M30790) (a) with sweet cherry phytoplasma strain (cv Stella, CSR-1, GenBank accession number OP093761) (b) and sweet cherry phytoplasma strain (cv Sunburst, CRR-1, GenBank accession number OP093761 (c) indicating that sweet cherry cvs belonged to 16SrI-B phytoplasma subgroup
Two subgroups of phytoplasmas (16SrI-B and 16SrVI-D) were identified associated with cherry cultivars in the present study.
Discussion
Sweet cherry is one of the major stone fruit crops mainly grown in temperate regions of the world. ‘Ca. P. asteris’ in cvs. Stella and Sunburst and ‘Ca. P. trifolii’ in cvs. BN, BNG and CITH-Cherry-9 were detected in Jammu and Kashmir, the major sweet cherry cultivating regions in India.
It was noticed that very faint amplifications of phytoplasma DNA were achieved from sweet cherry cvs Stella and Sunburst with primer pairs specific to 16Sr RNA gene with 3For/3Rev primer pairs (Manimekalai et al. 2010) while they amplified very efficiently employing secA-, tuf- and rp-specific primers. Similarly, very faint amplifications were produced in three cvs BN, BNG and CITH-Cherry-9 symptomatic cherry cultivars by employing another set of 16S rRNA gene with R16F2n/R16R nested primer pairs (Gundersen and Lee 1996). With all the other specific primers (secA, secY, tuf and rp genes), the intensity of phytoplasma-specific DNA amplifications was very clear and all the amplified products were cloned and sequenced. Results of the present study suggest that secA, secY, tuf and rp genes are better performing in detection of phytoplasmas in symptomatic cherry cultivars. Further, these results also confirmed the validity of these molecular markers for characterization of phytoplasma strains in cherry and may be used to detect phytoplasma presence in other stone fruit species. Both of the identified phytoplasmas have been reported as widespread in India infecting several agricultural crops of economic importance (Rao 2021).
Phytoplasmas are primarily identified based on the 16S rRNA genes, but the use of housekeeping genes is also very useful in the characterization of specific strains (Martini et al. 2019). Phytoplasma strains associated with several diseases of economically important crops in India were characterized using multilocus genes, such as brinjal little leaf (Azadvar and Baranwal 2012), coconut lethal wilt (Babu et al. 2021), chickpea stunt (Reddy et al. 2021), rose and marigold phyllody (Rihne et al. 2021; Panda et al. 2021).
Stone fruits (Prunus persica, P. domestica, P. armeniaca and P. avium) are being encouraged to be cultivated in larger areas in temperate locations of Himachal Pradesh, Uttarakhand and Jammu and Kashmir regions of India. Phytoplasma diseases are one of the major factors reducing the yield of these fruit species worldwide (Fiore et al. 2018; Hemmati et al. 2021). Very limited reports are available of phytoplasma presence in peach and apricot in India (Rao 2021); therefore, more extensive studies are required to investigate further the phytoplasma presence in stone fruits in India. In the recent years, sweet cheery plantations are getting increased importance and demand in the domestic market as well as in exports. In the present study, sweet cherry has been reported as host of two phytoplasma strains in the study. These phytoplasma diseases would affect adversely the yield parameters in addition to affecting the production leading to serious economic losses to the growers in India. Also, being perennial crop, sweet cherry plantations could best serve as a natural reservoir of the reported phytoplasma strains, which may increase the risk of transmission to other host plants through leafhoppers (Rao 2021). The timely and accurate detection of phytoplasmas in cherry utilizing multigene phytoplasma-specific primers would further enable the designing of appropriate diagnostic protocols which permit early reliable detection and better management of these diseases starting from screening cherry germplasm stocks.
The previous reports of ‘Ca. P. asteris’ in cherry were from China and Czech Republic (Fránová and ˇSpak 2013; Gao et al. 2011; Fránová et al. 2018). The presence of both ‘Ca. P. prunorum’ and ‘Ca. P. asteris’ in sour cherry showing small leaves, reduced vigor and dieback was reported in Czech Republic and Turkey (Navràtil et al. 2001; Çaglayan et al. 2013). Besides, the association of 16SrI-B and 16SrI-Q phytoplasmas subgroups association was also reported in sour cherry trees in Lithuania and Prunus Mahaleb in Hungary (Valiūnas et al. 2007; Valiunas et al. 2009; Varga et al. 2001). The presence of ‘Ca. P. asteris,’ ‘Ca. P. phoenicium’ and ‘Ca. P. australasia’-related strains in plum, sweet cherry, almond and apricot has also been described in Iran (Zirak et al. 2009a, b, 2010, 2021; Rasoulpour et al. 2019). Other reports of phytoplasmas associated with sweet cherry trees were a decline disease in Italy associated with the presence of ‘Ca. P. prunorum’ (Paltrinieri et al. 2001; Landi et al. 2007) that was also detected in Poland, in sweet and sour cherry trees with stunting, chlorotic leaf roll, short internodes, wilting and branch dieback in East Bohemia, Czech Republic (Cieślińska 2011; Ludvikova et al. 2011). The report of ‘Ca. P. asteris’ in sweet cherry is the first report from India.
The ‘Ca. P. trifolii’ (16SrVI-D subgroup) in sweet cheery cvs was reported as a dominant strain infecting several horticultural crops and weeds in India and other Asian countries (Rao et al. 2018; Hemmati et al. 2021; Rao 2021). So far, the presence of ‘Ca. P. trifolii’ (16SrVI-A subgroup) has only been reported in sweet cherry from Israel (Weintraub et al. 2007). But no report is available of occurrence of 16SrVI-D subgroup in sweet cherry from India and abroad; hence, this is a new record in world.
The prevalence of phytoplasma strains in commercial cherry cultivars in Indian disease suggests that large-scale exchange of nursery plant materials may play an important role in the emergence of epidemic of phytoplasmas in cherry-growing areas. Another possibility is that the identified phytoplasma strains in cherry in Jammu and Kashmir province may be transmitted through insect vectors. Thus, the percentage of symptomatic cherry infected plants was not too high (5.8–25%) and very likely correlated with infection of similar phytoplasma strains reported earlier in other fruit trees, maize, sesame, ornamentals in the Jammu and Kashmir province (Rao et al. 2017; Singh et al. 2018). In India, different species of leafhoppers (Hishimonus phycitis, Exiniatus indicus, Empoasca species, Orosius olbicinctus, Recilia dorsalis, Amrasca bigutella) and planthoppers (Laodelphax striatellus, Logotella koloshon) have also been identified as efficient vectors of ‘Ca. P. asteris’ and ‘Ca. P. trifolii’ (Rao 2021), which may be responsible for natural spread of the identified phytoplasma strains in cherry and needs further investigation to manage the spread of phytoplasma disease to newer locations.
Proper attention for the establishment of phytoplasma-free cherry germplasm and issuance of import permit through strict quarantine regulation should be employed for avoiding the entry of new phytoplasma strains infected vegetative material from other states of India and abroad. Studies on resistance sources of cherry germplasm to phytoplasma disease are also not available; therefore, a careful monitoring of phytoplasma-free nurseries and uprooting of infected cherry plants should be suggested to avoid severe and wide epidemics of phytoplasma infection associated with cherry and other stone fruit crops in India.
References
Ahrens U, Seemüller E (1992) Detection of DNA of plant pathogenic mycoplasma like organisms by a polymerase chain reaction that amplifies a sequence of the 16S rRNA gene. Phytopathology 82:828–832
Avramov Z, Contaldo N, Bertaccini B, Sakalieva D (2011) First report of “stolbur” phytoplasmas in Prunus avium in Bulgaria. Bull Insectol 64(Supplement):S71–S72
Azadvar M, Baranwal VK (2012) Multilocus sequence analysis of phytoplasma associated with brinjal little leaf disease and its detection in Hishimonus phycitis in India. Phytopath Mollicut 2(1):15–21
Babu M, Thangeswari S, Josephrajkumar A, Krishnakumar V, Karthikeyan A, Selvamani V, Karun A (2021) First report on the association of ‘Candidatus Phytoplasma asteris’ with lethal wilt disease of coconut (Cocos nucifera L.) in India. J Gen Plant Pathol 87(1):16–23
Bertaccini A, Lee I-M (2018) Phytoplasmas: an update. In: Rao GP, Bertaccini A, Fiore N, Liefting LW (eds) Phytoplasmas: phytoplasmas: plant pathogenic bacteria-I: characterization and epidemiology of phytoplasma-associated diseases. Springer, Singapore, pp 1–29
Bertaccini A (2022) Plants and phytoplasmas: when bacteria modify plants. Plants 11:1425
Blando F, Dave Oomah B (2019) Sweet and sour cherries: Origin, distribution, nutritional composition and health benefits. Trends in Food Sci Technol 86:517–529
Çaglayan K, Gazel M, Küçükgöl C, Paltrineri S, Contaldo N, Bertaccini A (2013) First report of ‘Candidatus Phytoplasma asteris’(group 16SrI-B) infecting sweet cherries in Turkey. Jour Pl Pathol 95(4):4–77
Cieślińska M (2011) European stone fruit yellows disease and its causal agent ‘Candidatus Phytoplasma prunorum’. J Pl Prot Res 51(4):441–447
Cieślińska M, Smolarek T (2015) Molecular diversity of phytoplasmas infecting cherry trees in Poland. Phytopath Mollicut 5(1-Supplement):S31–S32
Cieślińska M, Smolarek T (2019) Multilocus sequence analysis of phytoplasmas detected in cherry trees in Poland. Zemdirbyste-Agriculture 106(1):73–80
Deng S, Hiruki C (1991) Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J Microbiol Methods 14:53–61
FAOSTAT (2020) Agriculture data, agricultural statistics databases. Organization of the United Nations, Rome, Italy. http://faostat.fao.org
Fialová R, Navrátil M, Válová P, Lauterer P, Kocourek F, Poncarová-Vorácková Z (2004) Epidemioogy of European stone fruit yellows phytoplasma in the Czech Republic. Acta Hort 657:483–487
Fiore N, Bertaccini A, Bianco PA, Cieślińska M, Ferretti L, Hoat TX, Quaglino F (2018) Fruit crop phytoplasmas. In: Rao GP, Bertaccini A, Fiore N, Liefting LW (eds) Phytoplasmas: plant pathogenic bacteria-I. Springer, Singapore, pp 153–190
Fránová J, ˇSpak J (2013) First report of a 16SrI-C phytoplasma infecting celery (Apium graveolens) with stunting, bushy top and phyllody in the Czech Republic. J Phytopath 161:666–670
Fránová J, Lenz O, Pribylova J, Spak J, Koloniuk I, Such´a J, Paprstein F (2018) “Candidatus Phytoplasma asteris” and “ Candidatus Phytoplasma mali” strains infecting sweet and sour cherry in the Czech Republic. J Phytopath 166:59–66
Gao R, Wang J, Zhao W, Li XD, Zhu SF, Hao YJ (2011) Identification of a phytoplasma associated with cherry virescence in China. J Pl Phytopath 93:465–469
Gundersen DE, Lee I-M (1996) Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopath Mediterr 35:144–151
Hemmati C, Nikooei M, Al-Subhi AM, Al-Sadi AM (2021) History and current status of phytoplasma diseases in the Middle East. Biol 10(3):226
Hodgetts J, Boonham N, Mumford R, Harrison N, Dickinson M (2008) Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of Candidatus Phytoplasma. Int J Syst Evol Microbiol 58:1826–1837
Hu T, Subbiah V, Wu H, Amrit BK, Rauf A, Alhumaydhi FA, Suleria HAR (2021) Determination and characterization of phenolic compounds from Australia-grown sweet cherries (Prunus avium L.) and their potential antioxidant properties. ACS Omega 6(50):34687–34699
IRPCM (2004) ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int J Syst Evol Microbiol 54:1243–1255
Jomantiene R, Yhao Z, Lee I-M, Davis RE (2011) Phytoplasmas infecting sour cherry and lilac represent two distinct lineages having close evolutionary affinities with clover phyllody phytoplasma. Eur J Pl Pathol 130:97–107
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Landi F, Prandini A, Paltrinieri S, Mori N, Bertaccini A (2007) Detection of different types of phytoplasmas in stone fruit orchards in northern Italy. Bull Insectol 60(2):163
Lee I-M, Gundersen-Rindal DE, Bertaccini A (1998) Phytoplasma: ecology and genomic diversity. Phytopathology 88(12):1359–1366
Lee I-M, Martini M, Bottner KD, Dane RA, Black MC, Troxclair N (2003) Ecological implications from a molecular analysis of phytoplasmas involved in an aster yellows epidemic in various crops in Texas. Phytopathology 93(11):1368–1377
Lee I-M, Bottner-Parker KD, Zhao Y, Davis RE, Harrison NA (2010) Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. Int J Syst Evol Microbiol 60(12):2887–2897
Li HH, Qiu BS, Shi CL, Jin KX, Zhou Q, Huang XJ (1997) PCR amplification of 16S rDNA of phytoplasma associated with cherry fascinated disease and RFLP analysis. For Res 10:478–481
Lim PO, Sears BB (1992) Evolutionary relationships of a plant-pathogenic mycoplasma like organism and Acholeplasma laidlawii deduced from two ribosomal protein gene sequences. J Bacteriol 174(8):2606–2611
Ludvikova H, Franova J, Sucha J (2011) Phytoplasmas in apricot, peach and sour cherry orchards in East Bohemia, Czech Republic. Bull Insectol 64(suppl.):S67–S68
Manimekalai R, Soumya VP, Sathish Kumar R, Selvarajan R, Reddy K, Thomas GV, Baranwal VK (2010) Molecular detection of 16SrXI group phytoplasma associated with root (wilt) disease of coconut (Cocos nucifera) in India. Plant Dis 94(5):636
Martini M, Lee I-M, Bottner KD, Zhao Y, Botti S, Bertaccini A, Harrison NA, Carraro L, Marcone C, Khan J, Osler R (2007) Ribosomal protein gene-based phylogeny for finer differentiation and classification of phytoplasmas. Int J Syst Evol Microbiol 57:2037–2051
Martini M, Quaglino F, Bertaccini A (2019) Multilocus genetic characterization of phytoplasma. In: Bertaccini A, Oshima K, Kube M, Rao GP (eds) Phytoplasmas: plant pathogenic bacteria-III, genomics, host pathogen interactions and diagnosis. Springer, Singapore, pp 161–200
Mehle N, Brzin J, Boben J, Hren M, Frank J, Petrovič N, Gruden K, Dreo T, Žežlina I, Seljak G, Ravnikar M (2007) First report of ‘Candidatus Phytoplasma mali’ in Prunus avium, P. armeniaca and P. domestica. Pl Pathol 56:721
Navràtil M, Válová P, Fialová R, Petrová K, Fránová J, Nebesarova J, Karesová R (2001) Survey for stone fruit phytoplasmas in the Czech Republic. Acta Hort 550:377–382
Paltrinieri S, Martini M, Stefani E, Pondrelli M, Fideghelli C, Bertaccini A (2001) Phytoplasma infection in peach and cherry in Italy. Acta Hort 550:365–370
Paltrinieri S, Bertaccini A, Lugaresi C (2008) Phytoplasmas in declining cherry plants. Acta Hort 781:409–415
Panda P, Debnath P, Mall S, Nigam A, Rao GP (2021) Multilocus genes based characterization of phytoplasma strains associated with Mexican and French marigold species in India. Eur J Pl Pathol 161(2):313–330
Rao GP (2021) Our understanding about phytoplasma research scenario in India. Indian Phytopath 74:371–401
Rao GP, Thorat V, Manimekalai R, Tiwari AK, Yadav A (2017) A century progress of research on phytoplasma diseases in India. Phytopath Mollicut 7(1):1–38
Rao GP, Alvarez E, Yadav A (2018) Phytoplasma diseases of industrial crops. In: Rao GP, Bertaccini A, Fiore N, Liefting LW (eds) Phytoplasmas: plant pathogenic bacteria-I. Springer, Singapore, pp 91–121
Rasoulpour R, Salehi M, Bertaccini A (2019) Association of a ‘Candidatus Phytoplasma aurantifolia’-related strain with apricot showing European stone fruit yellows symptoms in Iran. 3 Biotech 9:65
Reddy MG, Baranwal VK, Sagar D, Rao GP (2021) Molecular characterization of Chickpea chlorotic dwarf virus and peanut witches’ broom phytoplasma associated with chickpea stunt disease and identification of new host crops and leafhopper vectors in India. 3 Biotech 11(3):1–23
Rihne T, Singh KP, Singh MK, Talukdar A, Rao GP (2021) Multilocus gene typing, mixed infection of phytoplasma strains associated with rose genotypes and confirmation of their natural reservoir sources. Trop Pl Pathol 46(6):596–607
Schneider B, Seemüller E, Smart CD, Kirkpatrick BC (1995) Phylogenetic classification of plant pathogenic mycoplasma like organisms or phytoplasmas. In: Razin S, Tully JG (eds) Molecular and diagnostic procedures in mycoplasmology, Vol-1. Academic Press, pp 369–380
Schneider B, Gibb KS, Seemüller E (1997) Sequence and RFLP analysis of the elongation factor Tu gene used in differentiation and classification of phytoplasmas. Microbiol 143:3381–3389
Sholberg AP, Kappel F (2008) Integrated management of stone fruit diseases. In: Ciancio A, Mukerji K (eds) Integrated management of diseases caused by fungi, phytoplasma and bacteria. Integrated management of plant pests and diseases, vol 3. Springer, Dordrecht, pp 3–25
Shreenath YS, Singh AK, Kumar PVD, Watpade S, Singh KP, Rao GP (2022) Characterization and distribution of phytoplasma strains associated with temperate stone fruits and their possible natural reservoirs in the north-western Himalayan states of India. Eur J Pl Pathol 164:93–108
Singh AK, Rao A, Goel S, Rao GP (2018) Identification of ‘Candidatus Phytoplasma asteris’ causing sesame phyllody disease and its natural weed host in Jammu, India. Indian Phytopathol 71(1):143–146
Thompson JD et al (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Uyemoto JK, Kirkpatrick BC (2011) X-disease phytoplasma. In: Hadidi A, Barba M, Candresse T, Jelkmann W (eds) Virus and virus like diseases of pome and stone fruits. APS, St. Paul, pp 243–245
Valiūnas D, Jomantienė R, Davis RE (2007) Phytoplasmas detected in cultivated fruit plants in Lithuania. Bull Insectol 60(2):139–140
Valiunas D, Jomantiene R, Ivanauskas A, Abraitis R, Staniene G, Zhao Y, Davis RE (2009) First report of a new phytoplasma subgroup, 16SrIII-T, associated with decline disease affecting sweet and sour cherry trees in Lithuania. Pl Dis 93(5):550–550
Varga K, Kőlber M, Ember I, Erdős Z, Biró E, Paltrinieri S, Martini M, Bertaccini A (2001) Identification of phytoplasmas infecting sour cherry in Hungary. Acta Hort 550:383–388
Wang J, Liu Q, Wei W, Davis RE, Tan Y, Lee M, Zhao Y (2018) Multilocus genotyping identifies a highly homogeneous phytoplasma lineage associated with sweet cherry virescence disease in China and its carriage by an erythroneurine leafhopper. Crop Prot 106:13–22
Weintraub PG, Zeidan M, Spiegel S, Gera A (2007) Diversity of the known phytoplasmas in Israel. Bull Insectol 60:143
Zhao Y, Wei W, Lee IM, Shao J, Suo X, Davis RE (2009) Construction of an interactive online phytoplasma classification tool, iPhyClassifier and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int J Syst Evol Microbiol 59:2582–2593
Zirak L, Bahar M, Ahoonmanesh A (2009a) Characterization of phytoplasmas associated with almond diseases in Iran. J Phytopath 157(11–12):736–741
Zirak L, Bahar M, Ahoonmanesh A (2009b) Molecular characterization of phytoplasmas related to peanut witches-broom and stolbur groups infecting plum in Iran. J Plant Pathol 91:713–716
Zirak L, Bahar M, Ahoonmanesh A (2010) Molecular characterization of phytoplasmas associated with peach diseases in Iran. J Phytopath 158:105–110
Zirak L, Khakvar R, Zarrini G, Hasanpour K (2021) Detection and molecular characterization of phytoplasmas associated with stone fruit trees in northwest of Iran. Crop Prot 142:105526
Acknowledgements
The authors are grateful for the financial help provided by the Director, ICAR-Indian Agricultural Research Institute, New Delhi, India. The authors are also thankful to the Director, ICAR-Central Institute of Temperate Horticulture, Srinagar, India, for providing help during survey and the samples collection at the institute experimental fields and also their help in recording disease incidence on cherry orchards. We would also like to thank the Head, Division of Plant Pathology, Indian Agricultural Research Institute, for providing laboratory facilities.
Author information
Authors and Affiliations
Contributions
The authors conceived the idea, analyzed data and drafted the manuscript for publication. The first author YSS did the survey, sample analysis, sequence analysis and sequence submission and also contributed significantly to the preparation of draft of the manuscript. SUN, GSM and KLK contributed to the survey and sample collection, and helped to record disease incidence and finalization of the manuscript, and GPR contributed significantly to the preparation of draft of the manuscript, finalization of the manuscript and formatting.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human studies and participants
The current study did not include any human or animal volunteers or animals.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shreenath, Y.S., Nabi, S.U., Madhu, G.S. et al. Identification and multilocus gene characterization of phytoplasmas associated with sweet cherry in India. 3 Biotech 12, 291 (2022). https://doi.org/10.1007/s13205-022-03357-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03357-2