Abstract
In this study, morphological and molecular characterizations of twenty-four Heterodera populations (cereal cyst nematodes, CCNs) collected from wheat production fields in Turkey were carried out. Light microscopy, species-specific markers, RFLP, and ITS sequencing were used to identify the nematode populations. The obtained CCN populations were identified as Heterodera avenae, H. filipjevi, and H. latipons according to the morphometric analysis, which was confirmed by the molecular techniques. The ITS region sequencing analysis confirmed the species identification, and phylogenetic analysis of this region grouped the populations with representative Heterodera populations from different origin countries deposited in GenBank. The simulation of four restriction enzymes, Alul, PstI, BsuRI (HaeIII), and Rsal, employed the ITS sequences of isolates to discriminate the Turkish Heterodera populations. ITS-RFLP patterns produced by endonuclease enzymes provided variations among Heterodera species. There was no intraspecific variation in populations of each Heterodera species in the ITS-RFLP analyses. The species-specific primers, AvenF-COI/AvenR-COI, HfF/HfR, and H-LatF/H-LatR, yielded 109 bp, 646 bp, and 204 bp products for H. avenae, H. filipjevi, and H. latipons populations, respectively. This is the first research to provide conclusive diagnostic tests for cyst nematode populations isolated from Turkey. These assays provide a sensitive, practical, and quick method for detecting Heterodera species and, therefore, have the potential to be utilized in the early identification of populations and monitoring of infestations without morphometric studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Small cereal grains such as rice, wheat, and corn have been an important part of human nutrition for thousands of years and have played a vital role in the formation of human civilization (Erenstein et al. 2022). More than 50% of the global daily calorie intake comes directly from grain consumption (Awika 2011). Wheat (Triticum aestivum L.) is an essential food crop in many countries and accounts for almost a third of the total food grain production in the world. In Turkey, wheat farming is practiced intensively and occupies an average of 8.5 million hectares (FAOSTAT 2021). Total wheat grain production reached 20 million tons in 2021, and this is 5% lower than that of 2016 production and 15% below the previous 5 years average (2012–2016). Unfortunately, this production is still inadequate and does not meet the needs of the growing population (Hoover et al. 2010). Cereal cultivation in Turkey is often subject to various restrictions, including insufficient fertilization and irrigation, soil-borne diseases, and several plant-parasitic nematodes (Shroyer et al. 1990; Dababat and Fourie 2018; Seid et al. 2021).
There are 12 cereal cyst nematode (CCN) species in the Heterodera avenae group, among the most important plant-parasitic nematodes hindering cereal yield worldwide (Dababat and Fourie 2018; Dababat et al. 2020; Mehalaine et al. 2020). Heterodera avenae Wollenweber, H. filipjevi (Madzhidov) Stelter, and H. latipons (Franklin) are the most damaging plant-parasitic nematodes in winter wheat, barley, oat, and rye (Subbotin et al. 2010; İmren et al. 2021). Heterodera avenae is widely distributed in temperate wheat-producing regions throughout the world (Imren et al. 2015; Dababat and Fourie 2018). Heterodera filipjevi is found in eastern and northern Europe, central and west Asia, the Middle East, the Indian subcontinent, and North America (Talatschian et al. 1976; Stoyanov 1982; Rivoal et al. 2003; Smiley et al. 2005; Toktay et al. 2015; Dababat and Fourie 2018; Özarslandan et al. 2020; Imren et al. 2021). Heterodera latipons occurs mainly in the Mediterranean region but also in Asia and Europe (Sewell 1973; Abidou et al. 2005; Sabova et al. 1988; Smiley and Nicol 2009; Imren et al. 2018).
Due to the similar morphology and small details that often distinguish species in the genus Heterodera (Turner and Subbotin 2013), the polymerase chain reaction-internal transcribed spacer-restriction fragment length polymorphism (PCR-ITS-RFLP) has been developed to differentiate between different species (Subbotin et al. 2003; Maafi et al. 2003). For example, the enzyme responsible for restriction PstI strongly distinguished H. filipjevi from the other species of the H. avenae group (Subbotin et al. 1999). By comparing the patterns obtained from DNA fingerprinting with random amplified polymorphic DNA, species-specific fragments may be discovered and used to generate species-specific primers. The sequence characterized amplified region PCR analysis has been successfully utilized to differentiate the species Globodera rostochiensis and G. pallida (Fullaondo et al. 1999) and H. glycines (Fullaondo et al. 1999; Ou et al. 2008). Heterodera avenae and other cyst-forming nematodes have also been distinguished by using species-specific markers (Qi et al. 2012).
Definitive molecular diagnosis often requires sequencing the ITS-rRNA locus with universal or specific developed primers. At present, the Heterodera molecular diagnostic outline has been generated for 40 species, but another 40 species have not been molecularly characterized yet (Turner and Subbotin 2013). Additionally, phylogenetic trees can be constructed using molecular data to represent the historical relationship between groups of organisms or taxa, thereby reconstructing the genealogical ties between organisms, and estimating the time of divergence between them, i.e., when they last shared a common ancestor. According to Jacob et al. (2008), these trees can be based primarily on morphological differences, but with the advances made by molecular studies, it is now possible to use DNA and protein sequences to reconstruct phylogenetic relationships.
Therefore, the objectives of this study were to (i) collect data on the occurrence and distribution of CCN species in wheat fields in Hatay, Adana, Osmaniye, Kahramanmaraş, Gaziantep, Kilis, Mardin, and Bolu provinces in Turkey, (ii) conduct a morphological and morphometrical analysis of cysts and second-stage juveniles (J2) of Turkish CCN populations, and (iii) get a better knowledge of the genetic variance among members of the H. avenae group; the molecular features of Turkish populations will be compared to those of populations originated from different countries.
Material and methods
Sampling and nematode extraction
A total of 24 samples were collected from different wheat-growing fields located in Hatay, Adana, Osmaniye, Kahramanmaraş, Gaziantep, Kilis, Mardin, and Bolu provinces of Turkey, in 2019. Soil samples were collected at the maturity stage/harvesting time at a depth of 0–30 cm. The samples were run to extract nematodes from 100 cm3 of each soil sample as described by Hooper (1986) with decanting and sieving. Extractions were poured onto filter paper disks to drain excess water, and then the dry extracted cysts were inspected under a Discovery V20 stereomicroscope at 20 × magnification (Carl Zeiss AG, Oberkochen, Germany) for the presence of cysts. At least 20 full cysts from each sample were collected and transferred to a refrigerator and stored at 4 °C until morphological and molecular analysis.
Morphological and morphometrical examinations
Morphological examination and diagnosis of cysts were performed to the species level with the aid of the Discovery V20 stereomicroscope at 20 × magnification. Vulval cone specimens were prepared through fixation of ten cysts per population in a formalin-glycerol fixative, then mounted on glycerol (Mulvey and Golden 1983). The prepared slides were examined under a Primo Star light microscope (Carl Zeiss AG, Oberkochen, Germany), according to Hooper (1986). Ten J2 derived from a single cyst were gently heated, fixed in triethanolamine/formalin, and embedded in glycerol to prepare permanent slides (Hooper, 1986).
The Heterodera populations were identified as per the previously reported morphological descriptions and diagnostic criteria of cysts and second stage juveniles (J2) (Franklin 1969; Mulvey and Golden 1983; Handoo and Ibrahim 2002). The length of the vulval slit, width of the vulval bridge, width of the fenestra, length of the fenestra, and the length of the underbridge were measured (Mulvey and Golden 1983). Additionally, the lack or presence of underbridge and bullae were performed on the cyst’s perineal pattern (Handoo and Ibrahim 2002).
The body length, stylet length, distance from the anterior region to the esophagus junction, body width, distance from the anterior region to the base of the esophageal bulb, tail length and width, and length of the hyaline portion of the tail were all measured as the most critical characters for J2 identification. The morphometrical features of J2 were also determined, including the a, b′, c, and c′ ratios. A total of ten cysts and ten J2 per population were included in the morphological measurements. Using an Axio Lab. A1 microscope (Carl Zeiss AG, Oberkochen, Germany), one cyst and 10 J2 were viewed and photographed for each population using an Axiocam ERc5s digital camera, and measurements were approximated using ZEN lite software (Carl Zeiss AG, Oberkochen, Germany).
To evaluate any meaningful difference among the populations (P ≤ 0.05), ANOVA was used to analyze data, software SPSS 10.0 for Windows (SPSS Inc., Chicago, IL, USA). The standard test of means was used to detect if there were statistically significant differences in variance across populations (P ≤ 0.05).
Molecular identification
DNA extraction and PCR
For each population, one cyst was crushed, and juveniles were split into minute pieces using a sterile scalpel blade. They were hand-selected and placed in a clean PCR tube containing a 10 µl drop of 1 × PCR reaction buffer (Thermo Fisher Scientific Inc., Waltham, MA, USA) containing 75 mM Tris–HCl (pH 8.8), 20 mM (NH4)2SO4, and 0.01%Tween 20. Proteinase K solution (> 600 mAU/ml, Qiagen GmbH, Hilden, Germany) was added to the mix. To inactivate the proteinase K, the tube was heated to 60 °C for 15 min and then to 95 °C for 5 min. The lysate was stored at − 20 °C for ITS sequencing and RFLP.
The universal primer sets, AB28 primer (5′-CGTAACAAGGTAGCTGTAG-3′) and TW81 primer (5′-TCCTCCGCTAAATGATATG-3′), were used to amplify the ITS sections, including the 5.8S ribosomal gene, as well as small parts of the 18S and 28S rDNA. A PCR reaction containing 0.5 µl of nematode lysate, 2 µl of 10 × ammonium buffer (Tris–HCl pH 8.5, (NH4)2S04, 15 mM MgCl2, 1% Tween 20), 0.5 mM primers, 0.2 mM dNTPs, and 1 unit of Ampliqon TEMPase Hot Start DNA polymerase (Berntsen, Rdovre, Denmark) was carried out in a total volume of 20 µl. Amplification reactions were carried out in a T100 thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA) with the following settings: one cycle of 12 min at 94 °C, 35 cycles of 20 s at 94 °C, 30 s at 55 °C, 45 s at 72 °C, and a final extension step of 10 min at 72 °C. Then, on a 1.6% agarose gel, the amplification products were separated. As a size reference, a 100 bp DNA Ladder (Solis BioDyne, Tartu, Estonia) was employed. A G:Box F3 gel documentation system was used to visualize and document the DNA banding profiles (Syngene, Cambridge, UK). The confirmed amplicons were sent to the Macrogen Inc. Sequencing Service (Seoul, Korea) for bidirectional sequencing.
Phylogenetic analysis
The sequencing and phylogenetic analysis of the ITS sequences were run for molecular diagnosis of the cyst populations from Hatay, Adana, Osmaniye, Kahramanmaraş, Gaziantep, Kilis, Mardin, and Bolu provinces. DNA sequences were produced with primers TW81 and AB28 to identity of the cyst populations. The obtained sequences were aligned using ClustalX 2.1, a multiple sequence alignment technique with default parameters, to produce the best formation for phylogenetic connections (Thompson et al. 2000). Phylogenies based on the alignment sequences of populations from the current study and those of reference isolates from GenBank were reconstructed using Maximum-Likelihood (ML) analysis with MEGA7 (Kumar et al. 2018). To create the ML phylogeny based on the General Time Reversible model (Nei and Kumar 2000), Nearest-Neigbour-Interchange (NNI) was used as the heuristic method for tree inference with bootstrap re-sampling analysis for 1,000 replicates to estimate the confidence of tree topologies (Felsenstein 1985).
PCR–RFLP analysis
To generate virtual PCR–RFLP gel electrophoresis images, SnapGene Gel Simulator is used (software from GSL Biotech; available at snapgene.com) with the sequences obtained from this study. This program processes DNA sequences with Alul, PstI, BsuRI (HaeIII), or Rsal restriction enzymes (Subbotin et al. 2003; Maafi et al. 2003) and stimulates gel electrophoresis and produces virtual gel images. For all populations, approximately 1050 bp fragments were amplified using the primer combination of TW81 and AB28 (Joyce et al., 1994).
Species-specific primer amplification
To identify H. avenae group populations, the species-specific primer pairs were used to amplify the species-specific products. Eight H. avenae individuals were used to amplify the species-specific 109 bp fragment. Each primer set contains 50 µl of PCR reaction mixes, which include 2 µl of DNA template, 21 µl ddH2O, 25 µl 2 DreamTaq PCR Master Mix (Thermo Fischer Scientific, Waltham, MA, USA), and 1 µM of each primer. PCR amplification using AVEN-COIF and AVEN-COIR primers was conducted with an initial denaturation step at 95 °C for 3 min, 30 cycles at 95 °C for 30 s, 58 °C for 30 s and 72 °C for 45 s, followed by 72 °C for 8 min (Toumi et al. 2013). The amplification with HfF and HfR was performed in the T100 thermal cycler with the following cycling profile: 3 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 56 °C, and 1 min at 72, followed by 72 °C for 8 min (Peng et al. 2013a, b). PCR amplification for H-LatF-COI and H-LatR-COI primers was done as follows: 95 °C for 5 min, then 30 cycles of 30 s at 94 °C, 45 s at 50 °C, and 45 s at 72 °C, with a final extension at 72 °C for 8 min (Toumi et al. 2013). The PCR products were separated on a 1.4% standard agarose gel. The consensus sequences were generated and finally used as BLAST queries against the NCBI nucleotide database. The cyst population species from Hatay, Adana, Osmaniye, Kahramanmaraş, Gaziantep, Kilis, Mardin, and Bolu provinces were compared with closely related cyst samples in GenBank. The phylogenetic tree of the cyst nematode populations from Hatay, Adana, Osmaniye, Kahramanmaraş, Gaziantep, Kilis, Mardin, and Bolu provinces is shown in Fig. 3. The phylogenetic similarity of Heterodera populations was also compared to international populations. Sequence alignments were used to build the phylogenetic tree, which was then reorganized globally using random replications (Table 1).
Results
Morphological and morphometrical studies
The Turkish cyst populations and J2s obtained from a single cyst for each population were identified morphologically using international standard descriptions and measurements. Among the Turkish populations studied, variations in cyst vulval cone, J2 forms, and dimensions were observed (Table 2). CCN populations observed in wheat fields in the provinces of Hatay, Adana, Osmaniye, Kahramanmaraş, Gaziantep, Kilis, Mardin, and Bolu were classified as H. avenae, H. filipjevi, and H. latipons based on these morphological traits.
The cysts of H. avenae had heavy prominent bullae that surrounded the vulval cone and no underbridge. H. filipjevi had less prominent bullae and a clear but thin underbridge when compared to H. avenae. Heterodera latipons differed from the others two Heterodera species by having a strong underbridge and a lack of distinct bullae in the vulval cone. In the current study, the cyst of H. avenae had heavy prominent bullae without underbridge, while H. filipjevi also had less prominent bullae with a slight underbridge. However, H. latipons having a strong underbridge without bullae in the vulval cone differed from H. avenae and H. filipjevi populations.
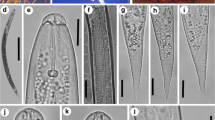
The H. avenae and H. filipjevi populations were bi-fenestrate, with relative semifenestral lengths ranging from 41.2 to 53.9 µl (Fig. 1), comparable to the H. latipons populations (Table 2). Heterodera avenae and H. filipjevi populations had slight changes in cyst vulval cone and J2 features and measures. There was a significant variation when H. latipons compared with H. avenae and H. filipjevi. In general, no underbridge was found in the vulval cone structures of any of the examined populations of H. avenae, while H. filipjevi cysts had a well-developed underbridge in the cyst vulval cones. However, the underbridge of H. latipons was strong with a few to absent bullae. Heterodera avenae populations had vulval bridge lengths ranging from 24.5 to 39.2 µm. In addition, the vulval cone had a small vulval slit, no underbridge, and thick bullae. With 480–576 µm and 20.5–23.5 µm, respectively, J2 body length and diameter were comparable to H. avenae populations (Fig. 2, Table 2). Tail length, tail diameter at the anus, and hyaline tail region length were all near to H. avenae population measures, ranging from 58.8 to 70.6 µm, 15.7 to 17.6 µm, and 39.2 to 46.1 µm, respectively. Heterodera filipjevi was also bi-fenestrate, but with slightly longer semifenestral length and breadth, ranging from 53.9 to 63.7 µm and 23.5 to 29.4 µm, respectively (Fig. 2, Table 2). The vulval slit was shorter, ranging in length from 6.9 to 10.8 µm, and an underbridge with medium development, ranging in length from 69.4 to 89.0 µm, as well as heavy bullae (Fig. 1).
The J2 body lengths of H. filipjevi populations were shorter, ranging from 490 to 552 µm with a diameter of 20.6 to 23.5 µm (Fig. 2, Table 2). The anal body diameter varied from 13.7 to 17.6 µm, while the tail length was substantially shorter, ranging from 49.0 to 63.7 µm with the rounded termination. The vulval bridge lengths of H. latipons populations ranged from 24.5 to 39.2 µm. The vulval cone has a robust underbridge as well as few bullae 480–576 µm and 20.5–23.5 µm, respectively, J2 body length and diameter were in the same range as H. latipons populations (Table 2). Tail length, tail diameter at the anus, and hyaline tail region length were all near to H. latipons population measures, ranging from 58.8 to 70.6 µm, 15.7 to 17.6 µm, and 39.2 to 46.1 µm, respectively.
Molecular identification
All samples produced an expected DNA band of approximately 1050 bp. No PCR product was generated in the no-template controls. The majority of cyst populations were 99–100% identical to that cyst samples in the GenBank database. Twenty-four cyst samples were identified based on their ITS sequences as H. avenae, H. filipjevi, and H. latipons. All accession numbers for ITS nucleotide sequences were deposited in GenBank (Table 1).
Phylogenetic analysis
Minor intraspecific variability was found in Heterodera species populations, which could be grouped into three major groups based on the Heterodera species level in the phylogenetic tree and representative GenBank populations, with a reasonable bootstrap value (Fig. 3). For CCN species-level phylogenetic study, one consensus sequence was obtained from bidirectional sequences and utilized for extensive phylogenetic analysis: H. avenae, H. filipjevi, and H. latipons. Based on genetic variations in the ITS sequences, a phylogenetic tree grouping the CCN species was created. The first cluster of H. avenae included samples from the four provinces of Hatay, Adana, Gaziantep, and Osmaniye. The isolates of CCN obtained from two provinces, Kahramanmaraş and Bolu were grouped as a second H. filipjevi cluster. The third cluster of H. latipons was found in the Kilis and Mardin populations. Each Heterodera species exhibited a slight intraspecific polymorphism, clustering the populations into three groups in the phylogenetic tree.
Phylogenetic trees of ITS regions of Heterodera species obtained in this study and the closest species. Sequences were analyzed using maximum likelihood method. Numbers at nodes indicate bootstrap values. The detailed information about isolates obtained in this study is shown in Table 1
PCR–RFLP analysis
Differentiation of the investigated populations was possible as supported by polymorphic PCR–RFLP patterns. All the investigated species produced RFLPs after digestion with six endonuclease restriction enzymes: Alul, PstI, BsuRI, and Rsal (Fig. 4). For each species population, the restriction enzymes produced identical RFLP patterns. Although no single enzyme was able to distinguish all examined species, a combination of the patterns generated by multiple separate enzymes was able to distinguish most of the species and populations studied.
Four enzymes, Alul, PstI, BsuRI, and Rsal, allowed differentiation among the CCN species: H. avenae, H. filipjevi, and H. latipons (Fig. 4). The endonuclease enzyme of AluI distinguished H. avenae (566 and 484 bp) from H. filipjevi (571 and 483 bp), and H. latipons (343, 170, 25, and 18 bp). The restriction enzyme PstI clearly differentiated H. filipjevi (713, 211, and 130 bp) from the other two Heterodera species. Moreover, the endonuclease enzyme RsaI separated H. latipons (1020 and 21 bp) from H. avenae (708, 320, and 21 bp) and H. filipjevi (707, 326, and 21 bp). Also, BsuRI distinguished H. latipons (532, 408, 77, and 24 bp) from H. avenae (420, 353, 176, 52, and 24 bp) and H. filipjevi (424, 378, 176, 52, and 24 bp).
Species-specific primer amplification
The species-specific primer pairs, AVEN-COIF and AVEN-COIR, yielded specific products for Hatay, Adana, Osmaniye, and Gaziantep populations (Fig. 5). Other cyst nematode species, H. filipjevi and H. latipons, yielded no amplification with these primers. A band of DNA was created for the Bolu and Kahramanmaraş populations using the species-specific primer pairs (HfF1 and HfR1). Eight H. filipjevi individuals were used to amplify the species-specific 646 bp fragment, while 204 bp fragment was amplified from eight H. latipons individuals.
The band profiles with cyst nematodes. 1–8, Heterodera avenae; 9–16, Heterodera filipjevi; 17–24, Heterodera latipons; Nt, nontemplate DNA. The order of the populations (from 1 to 24) in the agarose gel is as in Table 1. M, 100 bp DNA Ladder (Solis BioDyne, Tartu, Estonia), up to 1000 bp
Discussion
The cereal cyst nematodes, H. avenae, H. filipjevi, and H. latipons, are global pests attacking cereal crops leading to a significant loss in gain quality and quantity. These nematodes are closely related and exhibit few morphological and ITS sequence variations (Bekal et al. 1997; Tamura and Nei 1993; Subbotin et al. 1999, 2003; Umarao and Vangapandu 2008; Yan and Smiley 2008).
This result is convenient with the previous study based on morphological and morphometric characteristics. It is known that the underbridge and bullae in the vulval cone are the key morphological distinctions among H. avenae, H. filipjevi, and H. latipons, which are consistent with previous results from other studies (Subbotin et al. 1999, 2003; Yan and Smiley 2008). The second stage juveniles of H. avenae have longer tail, stylet and hyaline when compared to both H. filipjevi and H. latipons. Besides, H. filipjevi has a longer tail compared to H. latipons which is separated from those of two H. avenae and H. filipjevi populations which have small morphological characters (Madzhidov 1981; Valdeolivas and Romero 1990; Wouts et al. 1995).
This result is consistent with that of Subbotin et al. (2003), who reported that the ITS sequence alignment of H. filipjevi populations from Iran and Russia clustered together with a nucleotide similarity of 100% using the minimum evolution method. Imren et al. (2012) found that H. filipjevi populations from Iran and Turkey were phylogenetically aggregated into a single group. The Hatay, Adana, Gaziantep, and Osmaniye populations were identified as belonging to the first cluster of H. avenae. The Kahramanmaraş and Bolu populations were classified into the second cluster of H. filipjevi, whereas Kilis and Mardin populations consisted of the third cluster of H. latipons. Subbotin et al. (2001) reported that different cyst nematode species were thought to be phylogenetically examined based on ITS sequences, and this region was thought to be useful in identifying species (Subbotin et al. 2001).
According to Subbotin et al. (2003), restrictions by Alul and RsaI separated European (type A) from Asian (type B) populations. According to Maafi et al. (2003), Alul exhibited variability in the ITS region of multiple H. avenae populations. Turkish H. filipjevi was easily distinguished from other H. avenae and H. latipons using the restriction enzyme PstI. According to Subbotin et al. (1999), the restriction enzyme PstI clearly distinguished H. filipjevi from other Heterodera members. Also, BsuRI distinguished Turkish H. latipons from H. avenae and H. filipjevi. Maafi et al. (2003) reported that the restriction enzyme BsuRI distinctly separated H. latipons from other members of the Heterodera species. Our results demonstrated that the restriction profiles generated by PCR–RFLP might be used to differentiate between the Turkish populations of H. avenae, H. filipjevi, and H. latipons. A comparative investigation of numerous populations of Heterodera species should be conducted, and rDNA-RFLP can be used to distinguish species and populations.
Although PCR–RFLP can assist in species identification, however, the ones utilized to date still have some limitations. PCR–RFLP occasionally requires the use of costly restriction enzyme combinations. Due to polymorphisms and the technique’s limitations, it remains difficult to differentiate Heterodera species; also, these procedures are time-consuming and arduous. As a result, we tested a diagnostic test based on markers to complement current detection methods. In this study, the primer pairs designed for species-specific fragments were successfully used to detect H. avenae, H. filipjevi and H. latipons populations. The primer set AvenF-COI/AvenR-COI clearly differentiated H. avenae from H. filipjevi and H. latipons. Toumi et al. (2013) stated that the species-specific primers AvenF-COI and AvenR-COI clearly differentiated H. avenae from other members of the Heterodera. While in our study, the primers HfF and HfR, clearly differentiated H. filipjevi from other members of the Heterodera. Peng et al. (2013a, b) reported that HfF and HfR, clearly separated H. filipjevi population from the other Heterodera species. Also, H-LatF and H-LatR primers distinguished Turkish H. latipons from H. avenae and H. filipjevi. Toumi et al. (2013) reported that the H-LatF and H-LatR primers distinctly separated H. latipons from other members of the Heterodera species. The findings indicated that restriction profiles formed from species-specific markers derived from RAPD fragments might be used to differentiate between Turkish populations of H. avenae, H. filipjevi, and H. latipons. A comparative investigation of many populations of Heterodera species should be conducted, and species-specific markers can be used to distinguish Heterodera species and populations.
In this study, we reported comprehensively morphological, morphometric, and molecular data set for H. avenae, H. filipjevi, and H. latipons. These assays, including species-specific markers, ITS sequencing, and ITS-RFLP, provide a sensitive and practical method for detecting H. avenae, H. filipjevi, and H. latipons, and they may be used for early identification and monitoring of Heterodera infestations in the field.
Data availability
The data that support the findings of the current study are available from the corresponding author upon request.
References
Abidou H, El-Ahmed A, Nicol JM, Bolat N, Rivoal R, Yahyaoui A (2005) Occurrence and distribution of species of the Heterodera avenae group in Syria and Turkey. Nematologia Mediterranea 33:195–201
Awika JM (2011) Major cereal grains production and use around the world. In: M. Awika JM, Piironen V, Bean S (Eds) Advances in cereal science: implications to food processing and health promotion. American Chemical Society, Washington, DC, pp 1–13
Bekal S, Gauthier JP, Rivoal R (1997) Genetic diversity among a complex of cereal cyst nematodes inferred from RFLP analysis of the ribosomal internal transcribed spacer region. Genome 40:479–486
Dababat AA, Fourie H (2018) Nematode parasites of cereals. In: Sikora RA, Coyne D, Hallmann J, Timper P (eds) Plant parasitic nematodes in subtropical and tropical agriculture. CAB International, Wallingford, pp 163–221
Dababat AA, İmren M, Özer G, Mokrini F, Duman N, Paulıtz T (2020) Genetic and pathogenic variation in Heterodera latipons populations from Turkey. Nematology 23:47–56
Erenstein O, Poole N, Donovan J (2022) Role of staple cereals in human nutrition: separating the wheat from the chaff in the infodemics age. Trends in Food Science and Technology 119:508–513
Food and Agriculture Organization of the United Nations (FAOSTAT) (2021) Rome, Italy (http://faostat.fao.org) (Access date: December 27, 2021).
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Franklin MT (1969) Heterodera latipons n. sp., a cereal cyst nematode from the Mediterranean region. Nematologica 15:535–542
Fullaondo A, Barrena E, Viribay M, Barrena I, Salazar A, Ritter E (1999) Identification of potato cyst nematode species Globodera rostochiensis and G. pallida by PCR using specific primer combinations. Nematology 1:157–163
Handoo ZA, Ibrahim IKA (2002) Description and SEM observations of a new species of cyst nematode Heterodera goldeni (Nematoda: Heteroderidae) attacking Panicum coloratum in Egypt. Journal of Nematology 34:312
Hooper DJ (1986) Extraction of free-living stages from soil. In: Southey JF (ed) Labarotory methods for work with plant and soil nematodes. Her Majesty’s Stationery Office, London, pp 100–148
Hoover K, Uzunovic A, Gething B, Dale A, Leung K, Ostiguy N, Janowiak JJ (2010) Lethal temperature for pinewood nematode, Bursaphelenchus xylophilus, in infested wood using microwave energy. Journal of Nematology 42:101
Imren M, Waeyenberge L, Viaene N, Toktay H, Dababat A, Elekçioğlu İH (2012) Molecular characterisation of cereal cyst nematodes from the South Anatolian Region in Turkey using ITS-rDNA sequences. Entomological Society of Turkey 36:491–499
Imren M, Waeyenberge L, Viaene N, Elekçioğlu İH, Dababat A (2015) Morphological and molecular identification of cereal cyst nematodes from the Eastern Mediterranean region of Turkey. Turkish Journal of Agriculture and Forestry 39:91–98
Imren M, Yildiz Ş, Toktay H, Duman N, Dababat AA (2018) Morphometric and genetic variability among Mediterranean cereal cyst nematode (Heterodera latipons) populations in Turkey. Turkish Journal of Zoology 42:625–636
İmren M, Özer G, Paulitz T, Morgounov A, Dababat AA (2021) Plant-parasitic nematode associated with wheat in central, eastern, and south-eastern Kazakhstan. Plant Disease 105:2299–2305
Jacob J, Mitreva M, Vanholme B, Gheysen G (2008) Exploring the transcriptome of the burrowing nematode Radopholus similis. Molecular Genetics and Genomics 280:1–17
Joyce SA, Reid A, Driver F, Curran J (1994) Application of polymerase chain reaction (PCR) methods to identification of entomopathogenic nematodes. In: Burnell AM, Ehlers RU, Masson JP (eds) Cost 812 Biotechnology: Genetics of Entomopathogenic Nematode-Bacterium Complexes, Kildare, Ireland. Office for Official Publications of the European Communities, Brussels, Belgium, pp 178–187
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35:1547–1549
Maafi ZT, Subbotin S, Moens M (2003) Molecular identification of cyst-forming nematodes (Heteroderidae) from Iran and a phylogeny based on ITS-rDNA sequences. Nematology 5:99–111
Madzhidov AR (1981) Bidera filipjevi n. sp. (Heteroderina: Tylenchida) in Tadzhikistan. Izvestiia Akademii Nauk Tadzhikskoi SSR: Otdelenie Biologicheskikh Nauk 2:40–44
Mehalaine K, Imren M, Özer G, Hammache M, Dababat AA (2020) Molecular identification and phylogenetic diversity of cereal cyst nematodes (Heterodera spp.) populations from Algeria. Nematropica 50:134–143
Mulvey RH, Golden AM (1983) An illustrated key to the cyst-forming genera and species of Heteroderidae in the western hemisphere with species morphometrics and distribution. Journal of Nematology 15:1
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford USA
Ou S, Peng D, Liu X, Li Y, Moens M (2008) Identification of Heterodera glycines using PCR with sequence characterised amplified region (SCAR) primers. Nematology 10:397–403
Özarslandan A, Imren M, Özer G, Karaca C, Dababat A (2020) Identification and genetic diversity of the Mediterranean cereal cyst nematode, Heterodera latipons Franklin, 1969 (Tylenchida: Heteroderidae) in cereal production areas of Northern Cyprus. Turkish Journal of Entomology 44:273–281
Peng H, Gao BL, Kong LA, Yu Q, Huang WK, He XF, Long H, Peng DL (2013a) Correction: Exploring the host parasitism of the migratory plant-parasitic nematode Ditylenchus destuctor by expressed sequence Tags Analysis. PLoS ONE 9:e69579
Peng H, Qi X, Peng D, Long H, He X, Huang W, He W (2013b) Sensitive and direct detection of Heterodera filipjevi in soil and wheat roots by species-specific SCAR-PCR assays. Plant Disease 97:1288–1294
Qi XL, Peng DL, Peng H, Long HB, Huang WK, He WT (2012) Rapid molecular diagnosis based on SCAR marker system for cereal cyst nematode. Scientia Agricultura Sinica 45:4388–4395
Rivoal R, Valette S, Bekal S, Gauthier JP, Yahyaoui A (2003) Genetic and phenotypic diversity in the graminaceous cyst nematode complex, inferred from PCR-RFLP of ribosomal DNA and morphometric analysis. European Journal of Plant Pathology 109:227–241
Sabova M, Valocka B, Liskova M, Vargova V (1988) The first finding of Heterodera latipons Franklin, 1969 on grass stands in Czechoslovakia. Helminthologia 25:201–206
Seid A, İmren M, Ali MA, Toumi F, Paulitz T, Dababat AA (2021) Genetic resistance of wheat towards plant-parasitic nematodes: current status and future prospects. Biotech Studies 30:43–62
Sewell R (1973) Plant-parasitic nematodes from Canada and abroad, 1971. Canadian Plant Disease Survey 53:34–35
Shroyer JP, Ryan J, Monem MA, Mourid ME (1990) Production of fall planted cereals in Morocco and technology for its improvement. Journal of Agronomic Education 19:32–40
Smiley RW, Whittaker RG, Gourlie JA, Easley SA, Ingham RE (2005) Plant-parasitic nematodes associated with reduced wheat yield in Oregon: Heterodera avenae. Journal of Nematology 37:297
Smiley RW, Nicol JM (2009) Nematodes which challenge global wheat production. In: Carver BF (ed) Wheat science and trade, Wiley-Blackwell, Hoboken, New Jersey, United States, pp 171–187
Stoyanov D (1982) Cyst-forming nematodes on cereals in Bulgaria. EPPO Bulletin 12:341–344
Subbotin SA, Rumpenhorst HJ, Sturhan D (1996) Morphological and electrophoretic studies on populations of the Heterodera avenae complex from the former USSR. Russian Journal of Nematology 4:29–38
Subbotin SA, Waeyenberge L, Molokanova I, Moens M (1999) Identification of Heterodera avenae group species by morphometrics and rDNA-RFLPs. Nematology 1:195–207
Subbotin SA, Vierstraete A, De Ley P, Rowe J, Waeyenberge L, Moens M, Vanfleteren JR (2001) Phylogenetic relationships within the cyst-forming nematodes (Nematoda, Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Molecular Phylogenetics and Evolution 21:1–16
Subbotin S, Sturhan D, Rumpenhorst HJ, Moens M (2003) Molecular and morphological characterisation of the Heterodera avenae species complex (Tylenchida: Heteroderidae). Nematology 5:515–538
Subbotin SA, Mundo-Ocampo M, Baldwin JG (2010) Description and Diagnosis of Heterodera species. In: Hunt DJ, Perry RN (eds) Systematics of cyst nematodes (Nematoda: Heteroderinae). Brill, Leiden, pp 7–349
Talatschian P, Akhiani A, Grayeli Z, Shahmohammadi M, Teymouri F (1976) Survey on cyst forming nematodes in Iran in 1975 and their importance. Iranian Journal of Plant Pathology 12:42–43
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512–526
Thompson GJ, Miller LR, Lenz M, Crozier RH (2000) Phylogenetic analysis and trait evolution in Australian lineages of drywood termites (Isoptera, Kalotermitidae). Molecular Phylogenetics and Evolution 17:419–429
Toktay T, Imren M, Ocal A, Waeyenberge L, Viaene N (2015) Incidence of cereal cyst nematodes in the East Anatolia Region in Turkey. Russian Journal of Nematology 23:29–40
Toumi F, Waeyenberge L, Viaene N, Dababat A, Nicol JM, Ogbonnaya F, Moens M (2013) Development of two species-specific primer sets to detect the cereal cyst nematodes Heterodera avenae and Heterodera filipjevi. European Journal of Plant Pathology 136:613–624
Turner SJ, Subbotin SA (2013) Cyst nematodes. In: Perry RN, Moens M (eds) Plant nematology. CAB International, Wallingford, pp 109–143
Umarao GT, Vangapandu S (2008) Molecular characterisation of Indian populations of Heterodera filipjevi in tomato using PCR-RFLP of rDNA. International Journal Nematology 18:118–122
Valdeolivas A, Romero MD (1990) Morphometric relationships of some members of the Heterodera avenae complex (Nematoda: Heteroderidae). Nematologica 36:292–303
Wouts WM, Schoemaker A, Sturhan D, Burrows PR (1995) Heterodera spinicauda sp. N. (Nematoda: Heteroderidae) from mud flats in the Netherlands, with a key to the species of the H. avenae group. Nematologica 41:575–583
Yan GP, Smiley RW (2008) First detection of the cereal cyst nematode Heterodera filipjevi in North America. Phytopathology 98:176
Author information
Authors and Affiliations
Contributions
The experiments were designed by A. Dababat, Ş. Yıldız, and M. İmren, and were carried out and written by D. Dağlı, N. Duman, E. Yüksel, G. Özer, and M. İmren; and M. İmren is responsible for supervision and review.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dağlı, D., Duman, N., Yüksel, E. et al. Characterization of cereal cyst nematodes in wheat using morphometrics, SCAR markers, RFLP, and rDNA-ITS sequence analyses. Trop. plant pathol. 48, 207–216 (2023). https://doi.org/10.1007/s40858-022-00528-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-022-00528-7